Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions
Abstract
1. Introduction
2. Methods
2.1. Plant Materials
2.2. RNA-Seq Sequencing and Transcriptome Assembly
2.3. Analysis of Differential Expressed Genes
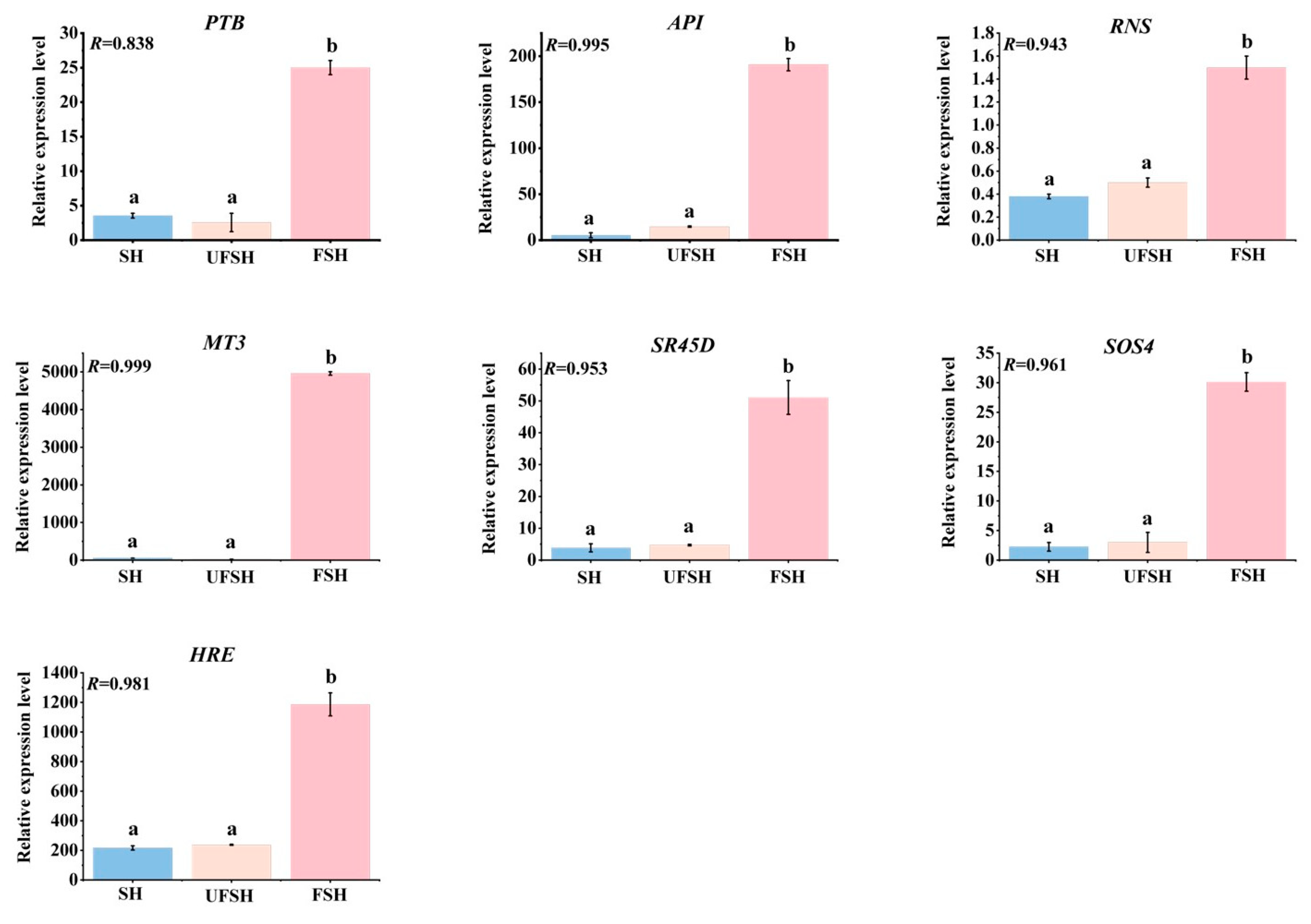
2.4. qRT-PCR Analysis
2.5. WGCNA Constructs the Co-Expression Module and Regulatory Network
2.6. Statistical Analysis
3. Results
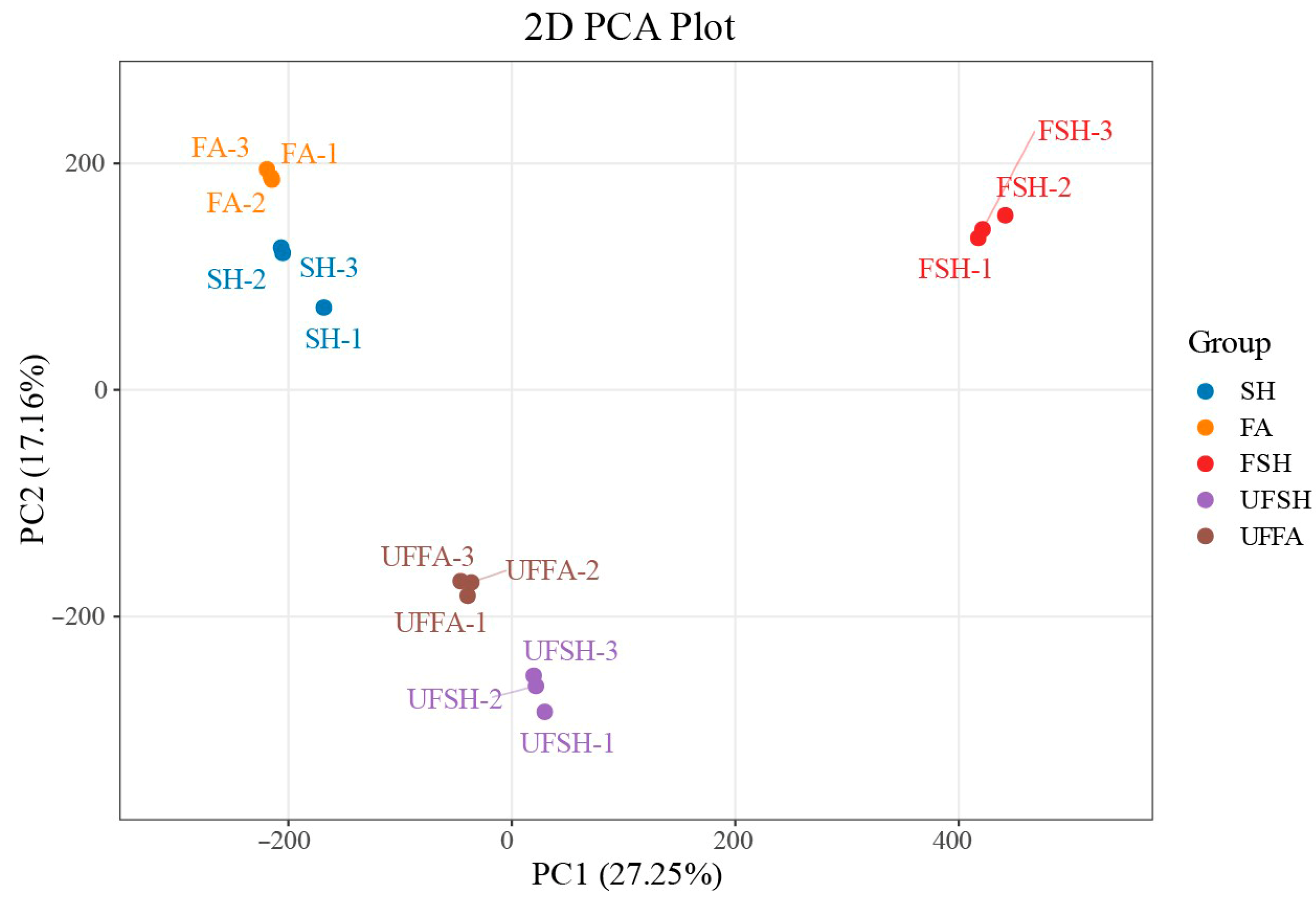
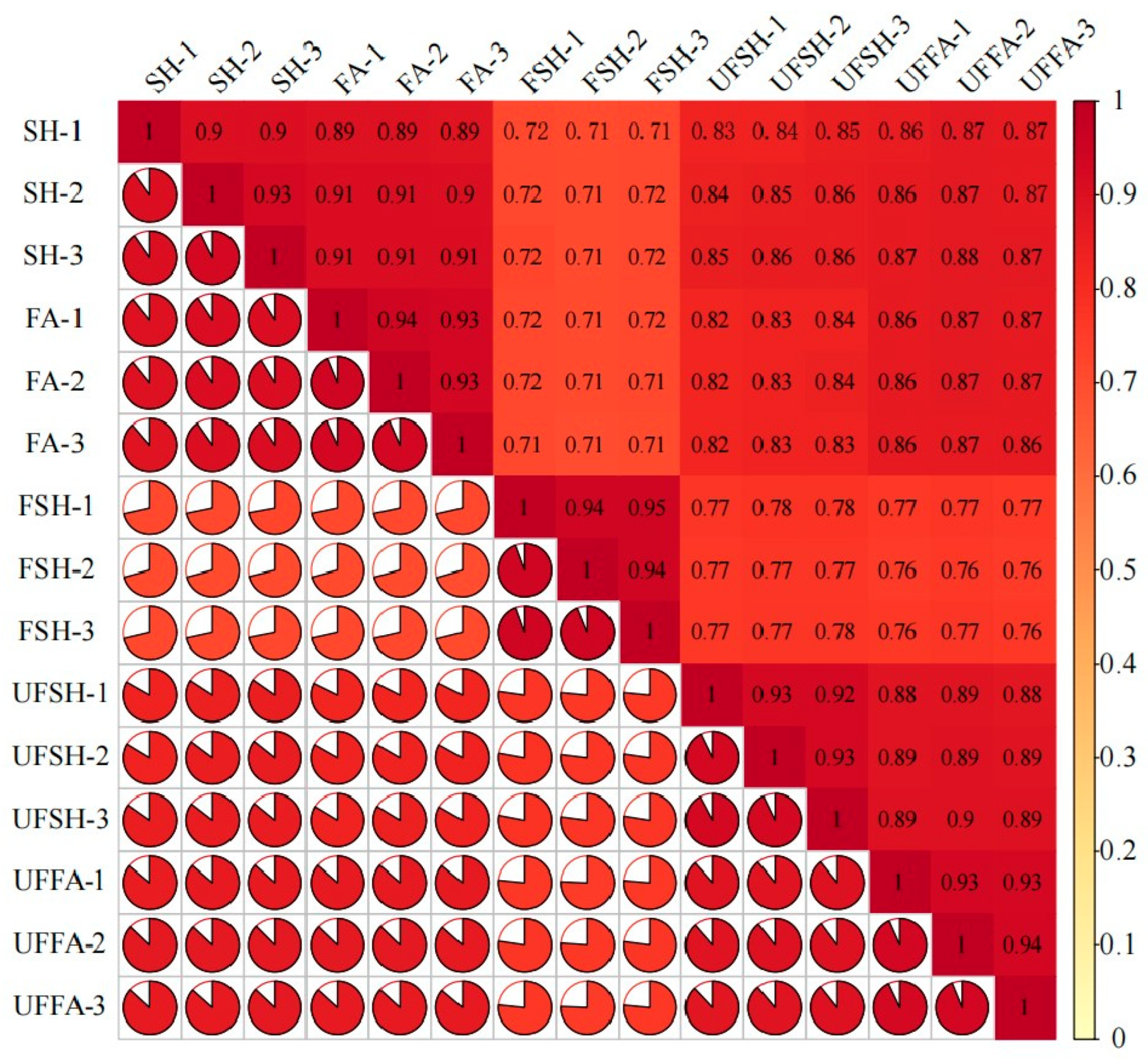
3.1. Identification of Total Unigene During the Development of Bambusaoldhamii
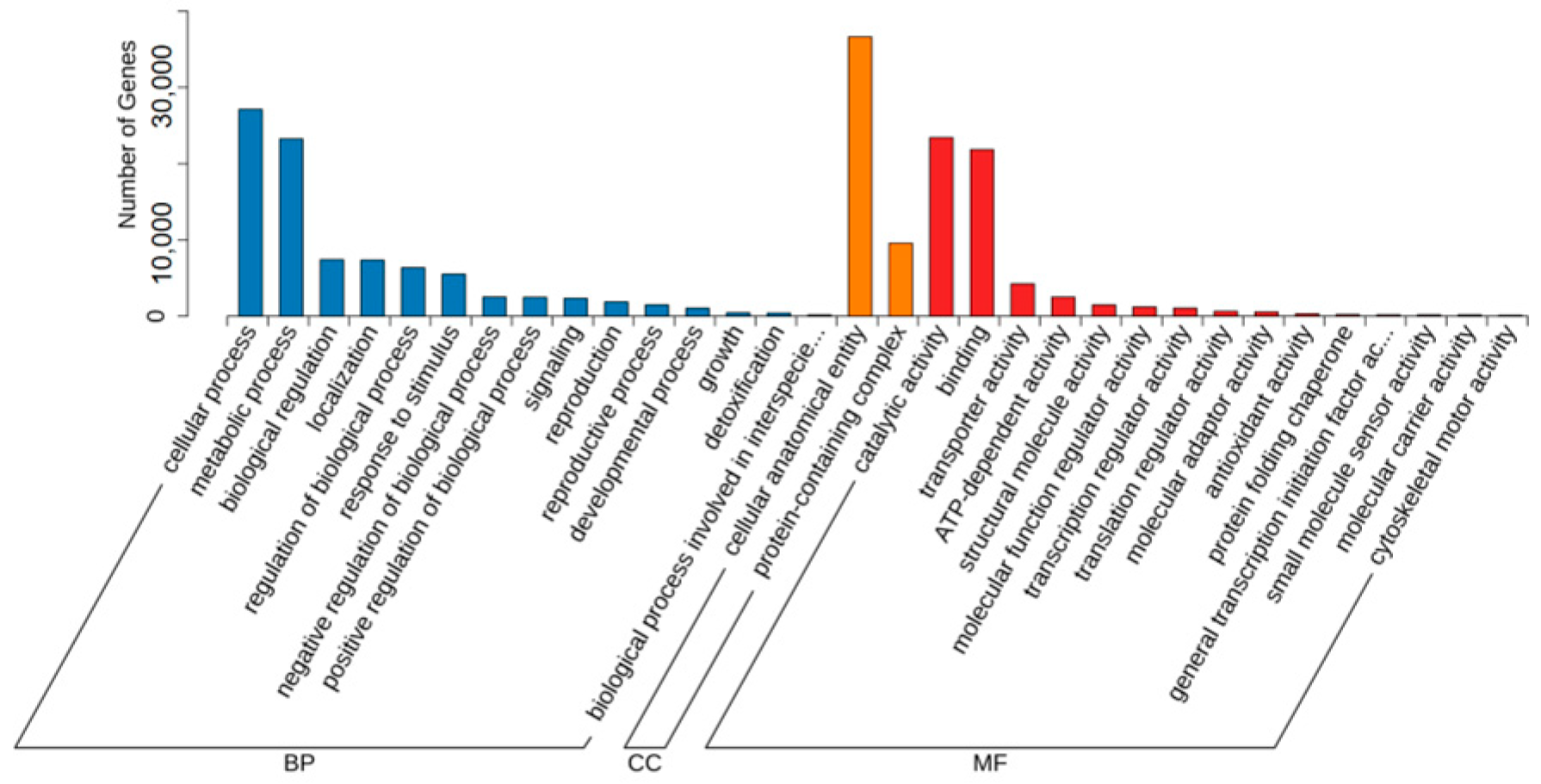
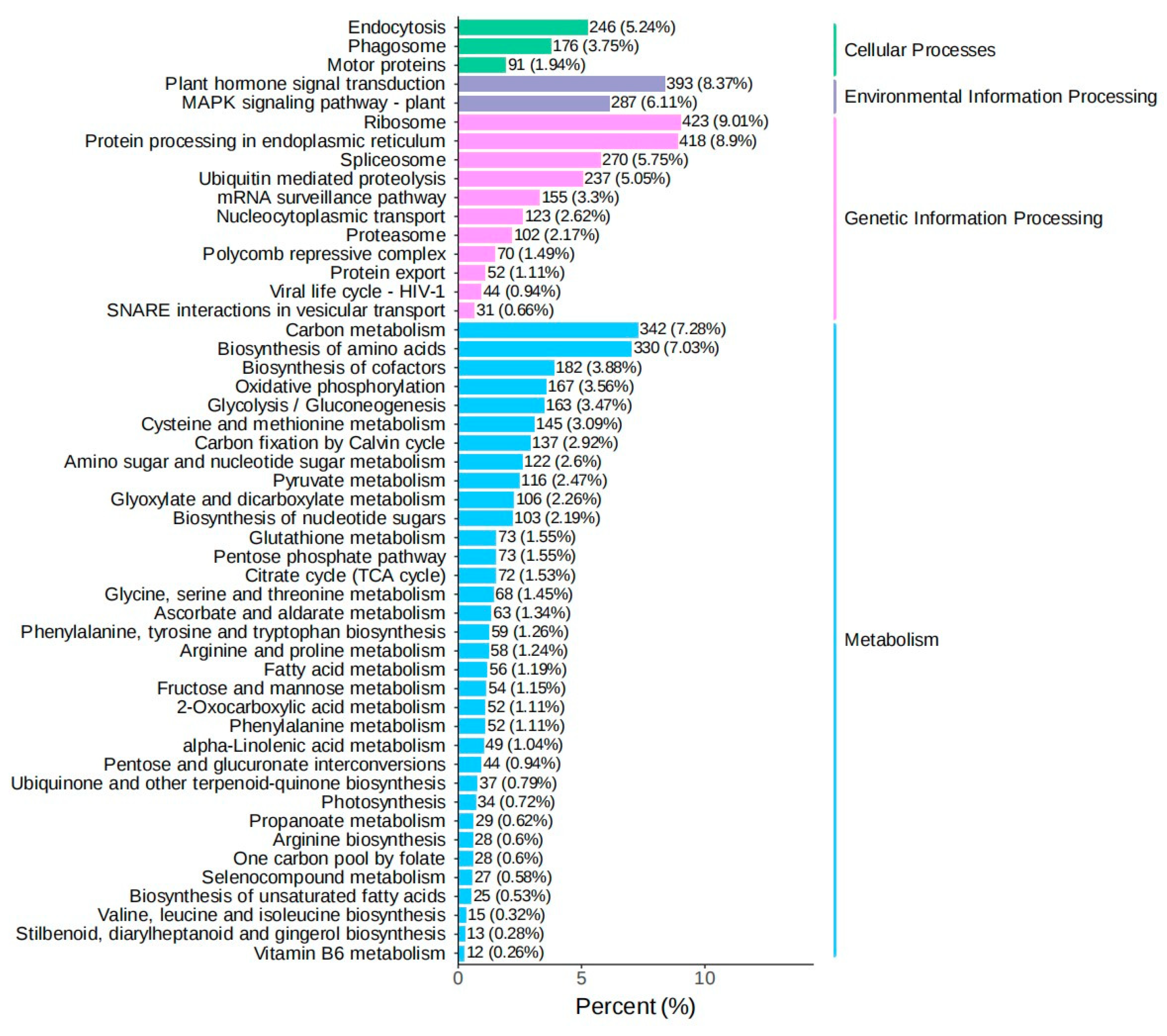
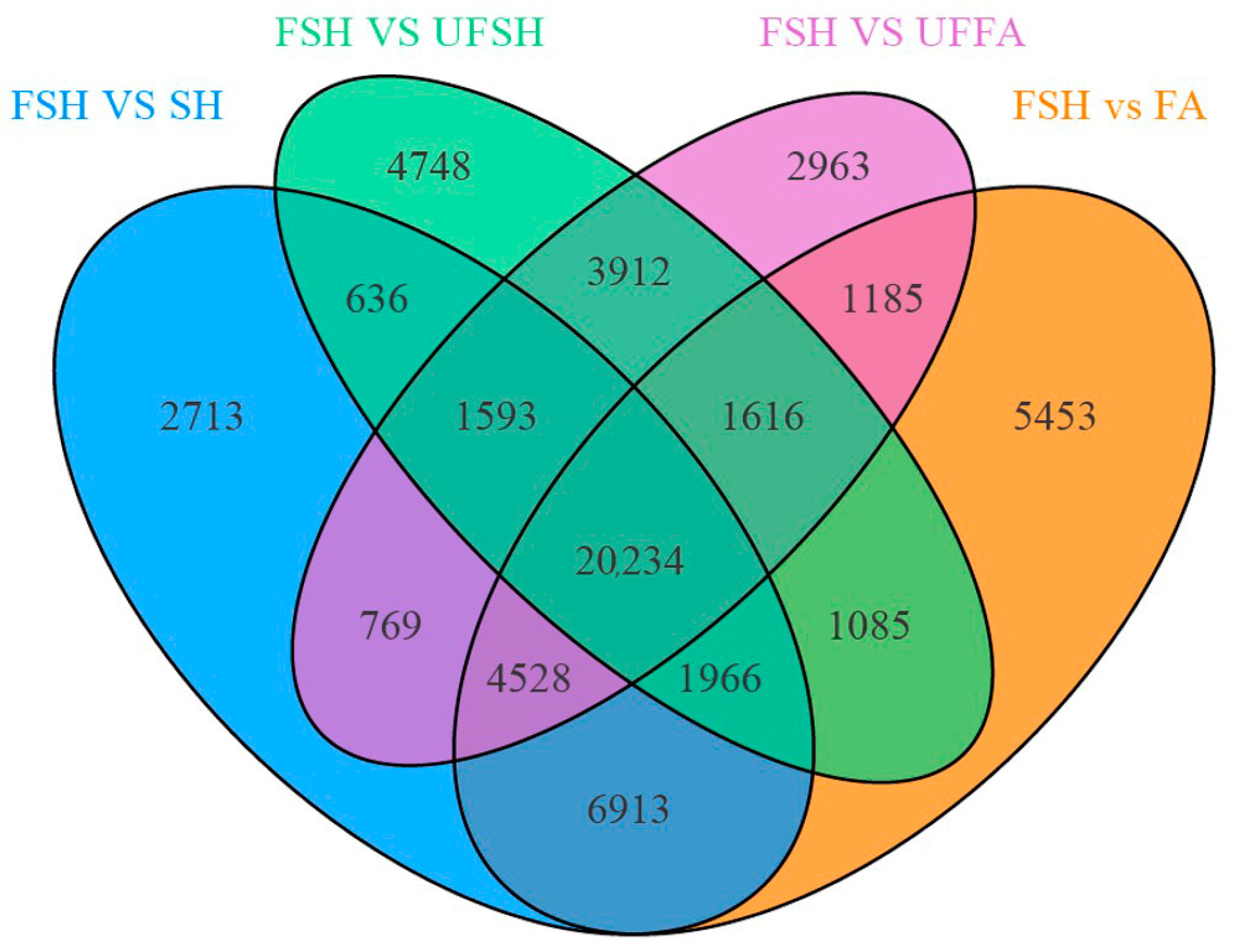
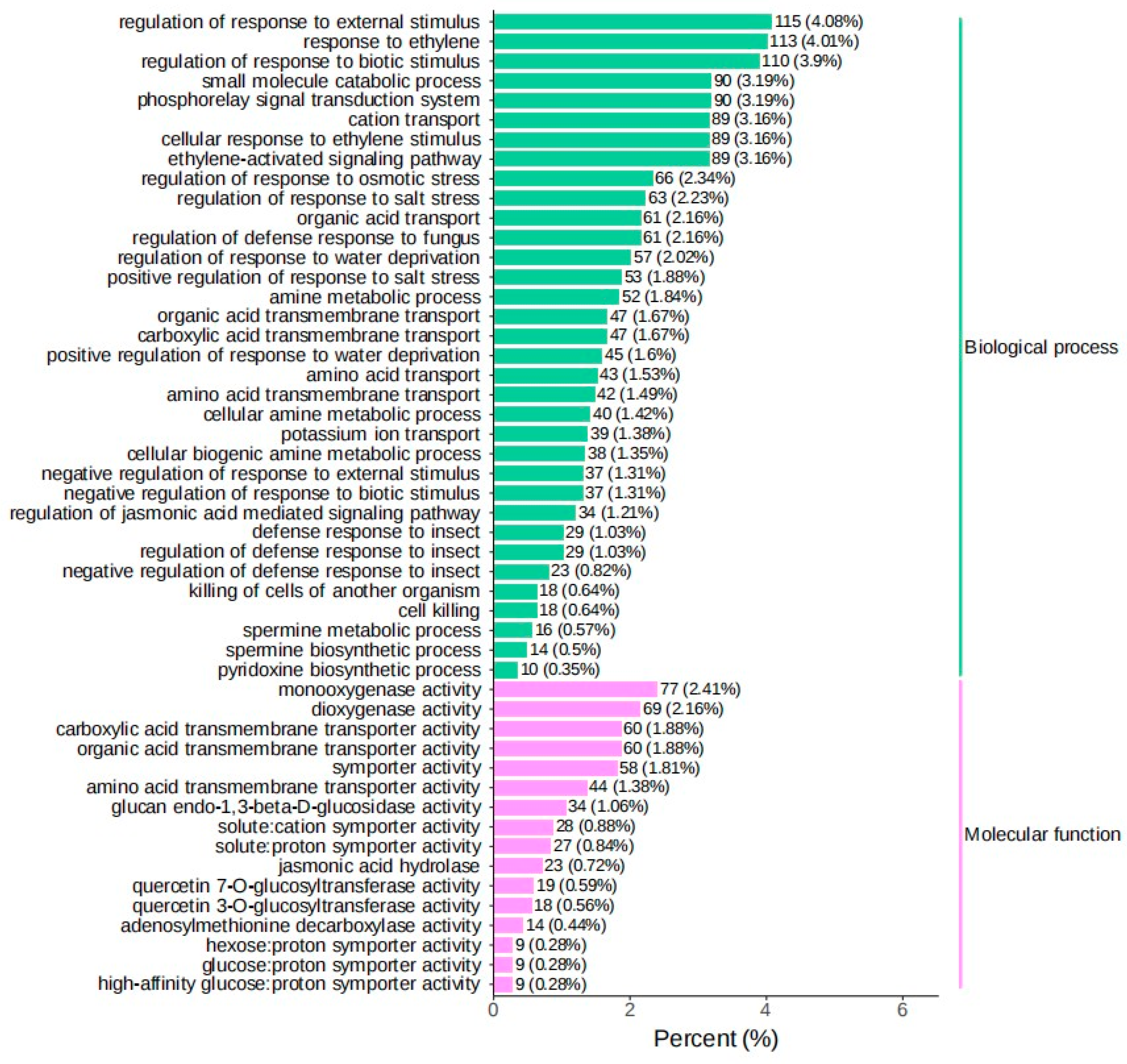
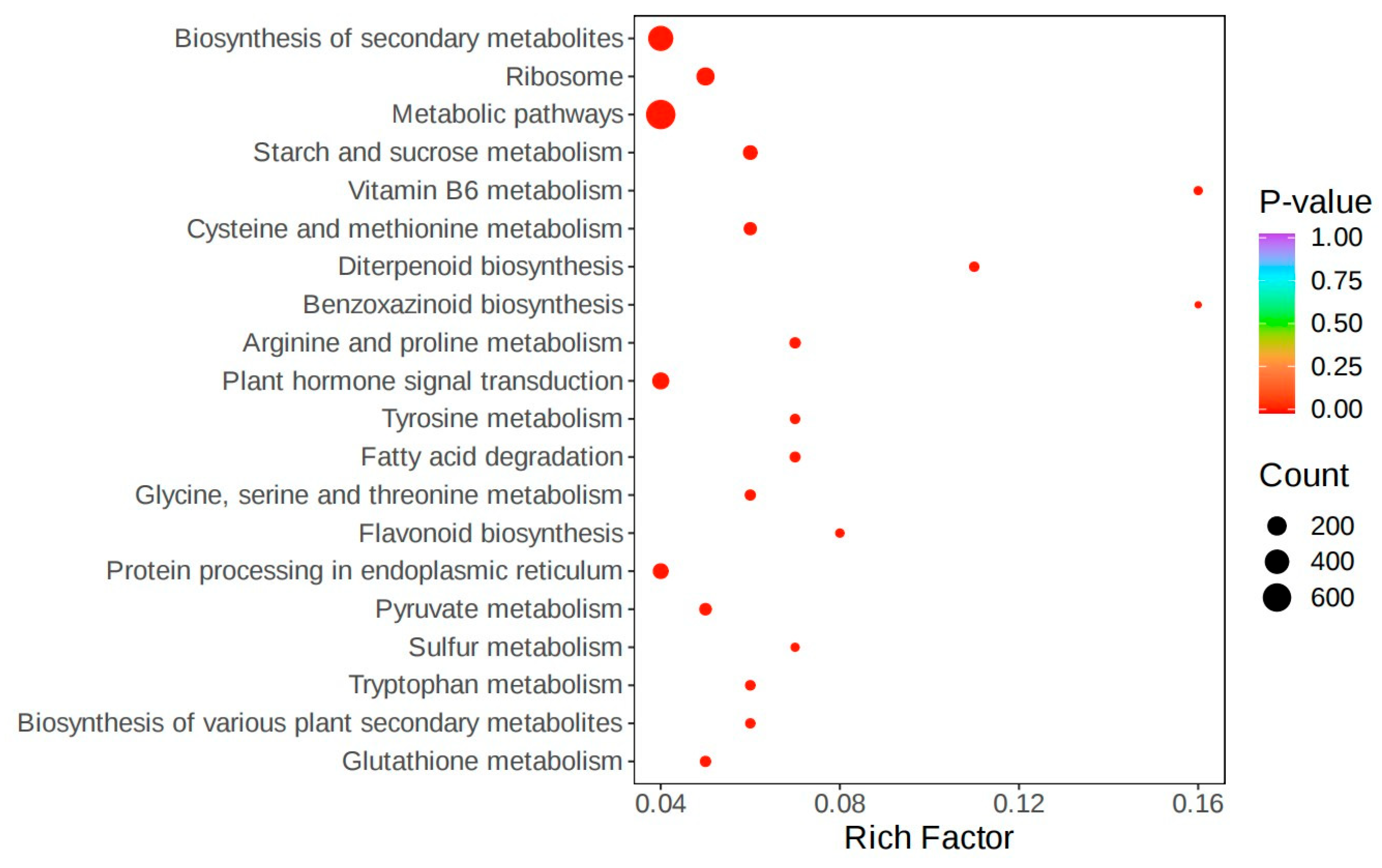
3.2. Screening of Differentially Expressed Genes in Introduced Cultivation and Enrichment Analysis of Their Biological Functions
3.3. External Stimuli Regulate DEGs, Causing Bambusaoldhamii to Flower
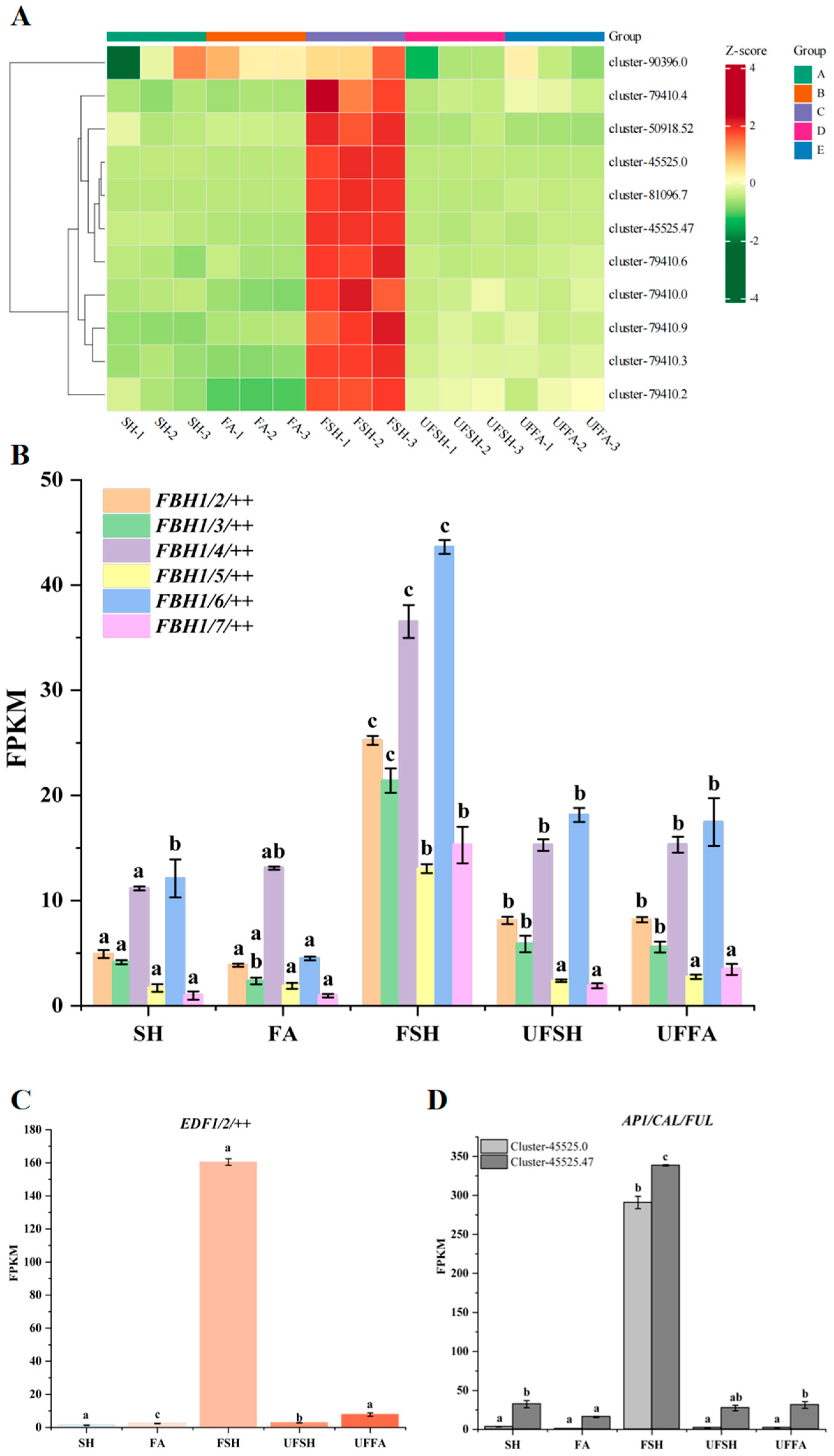
3.4. DEGs Related to Flower Organ Development Are Involved in the Flower Development of Bambusaoldhamii
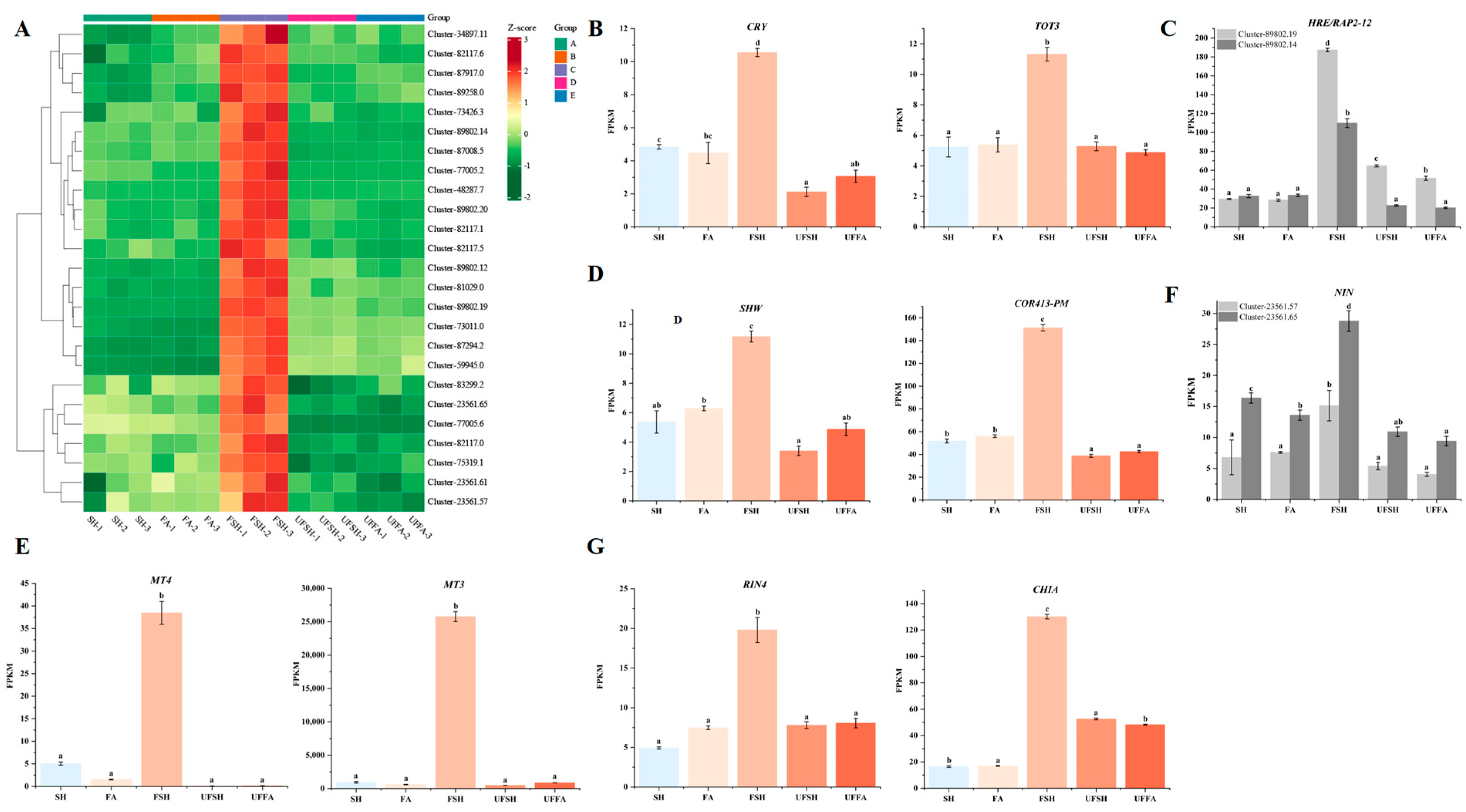
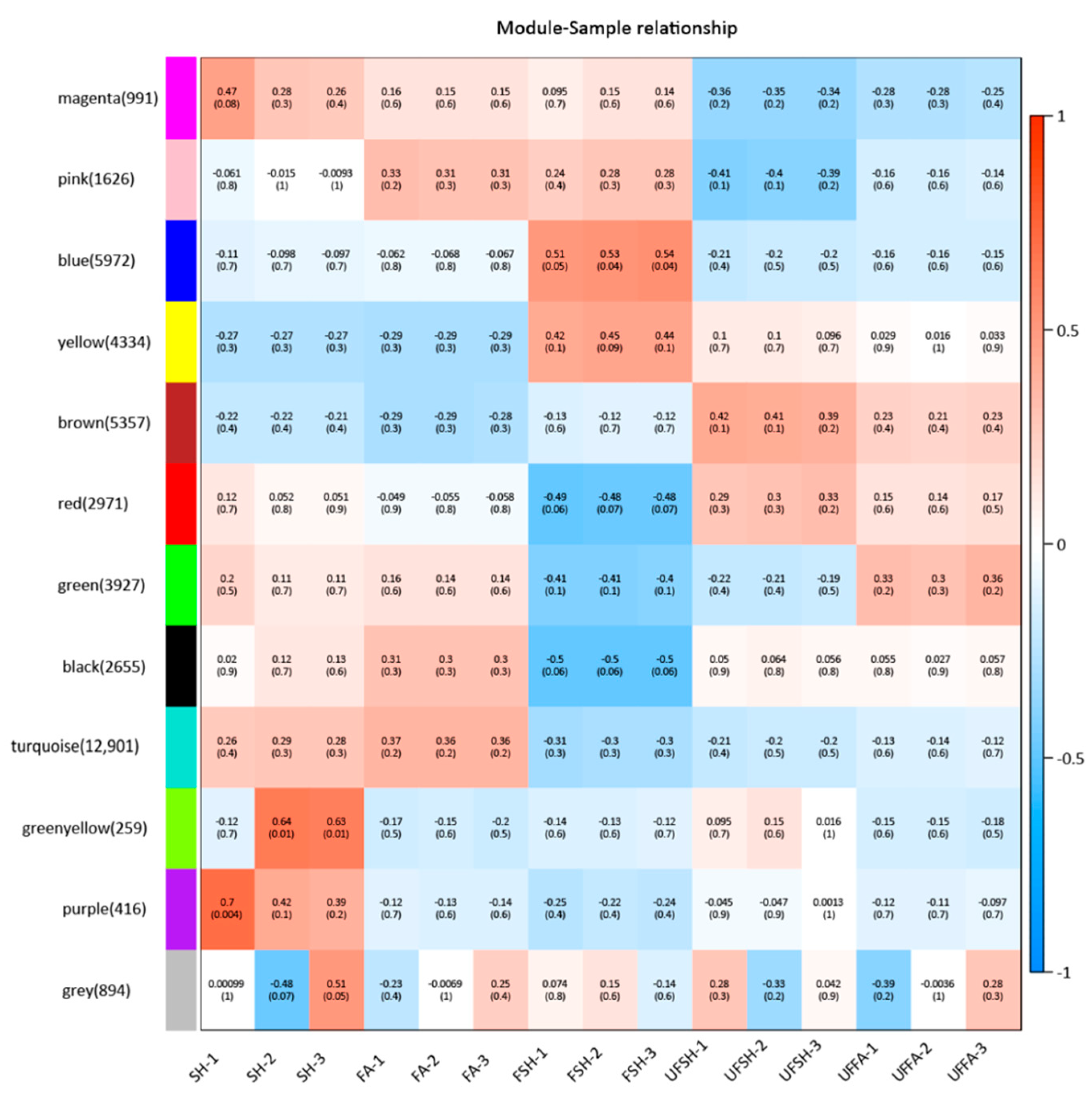
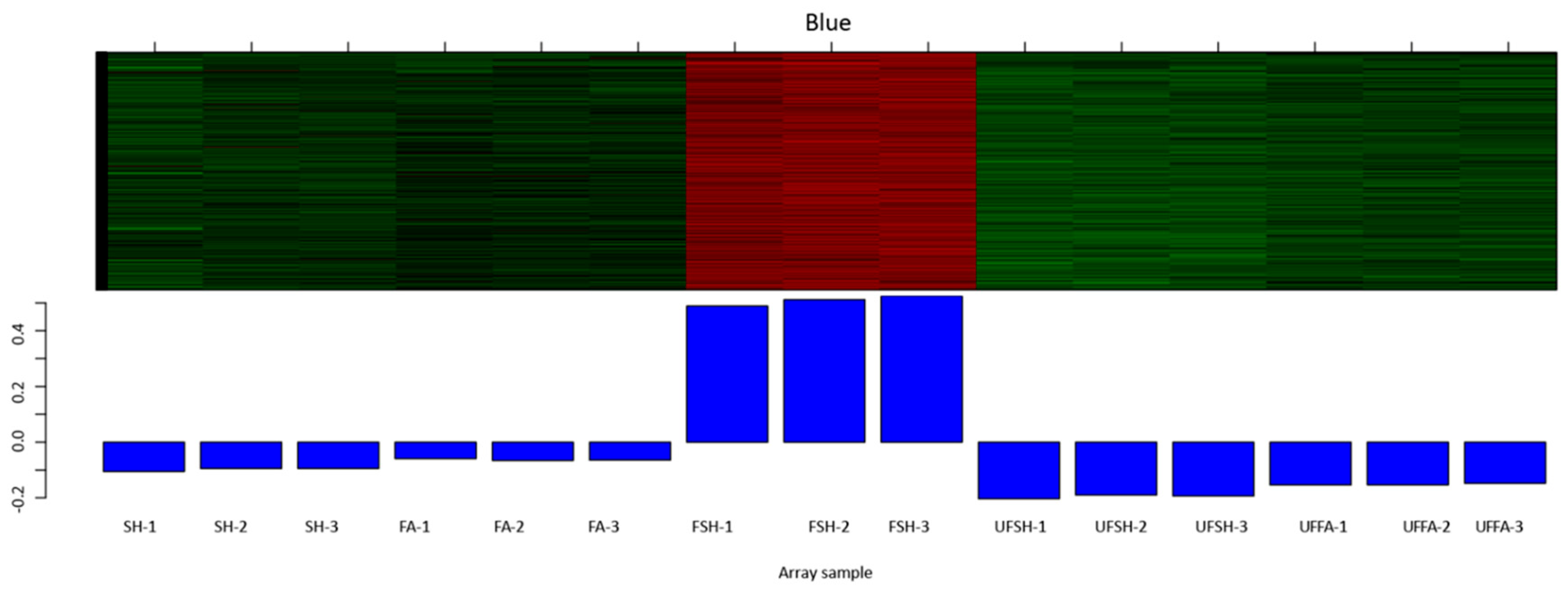
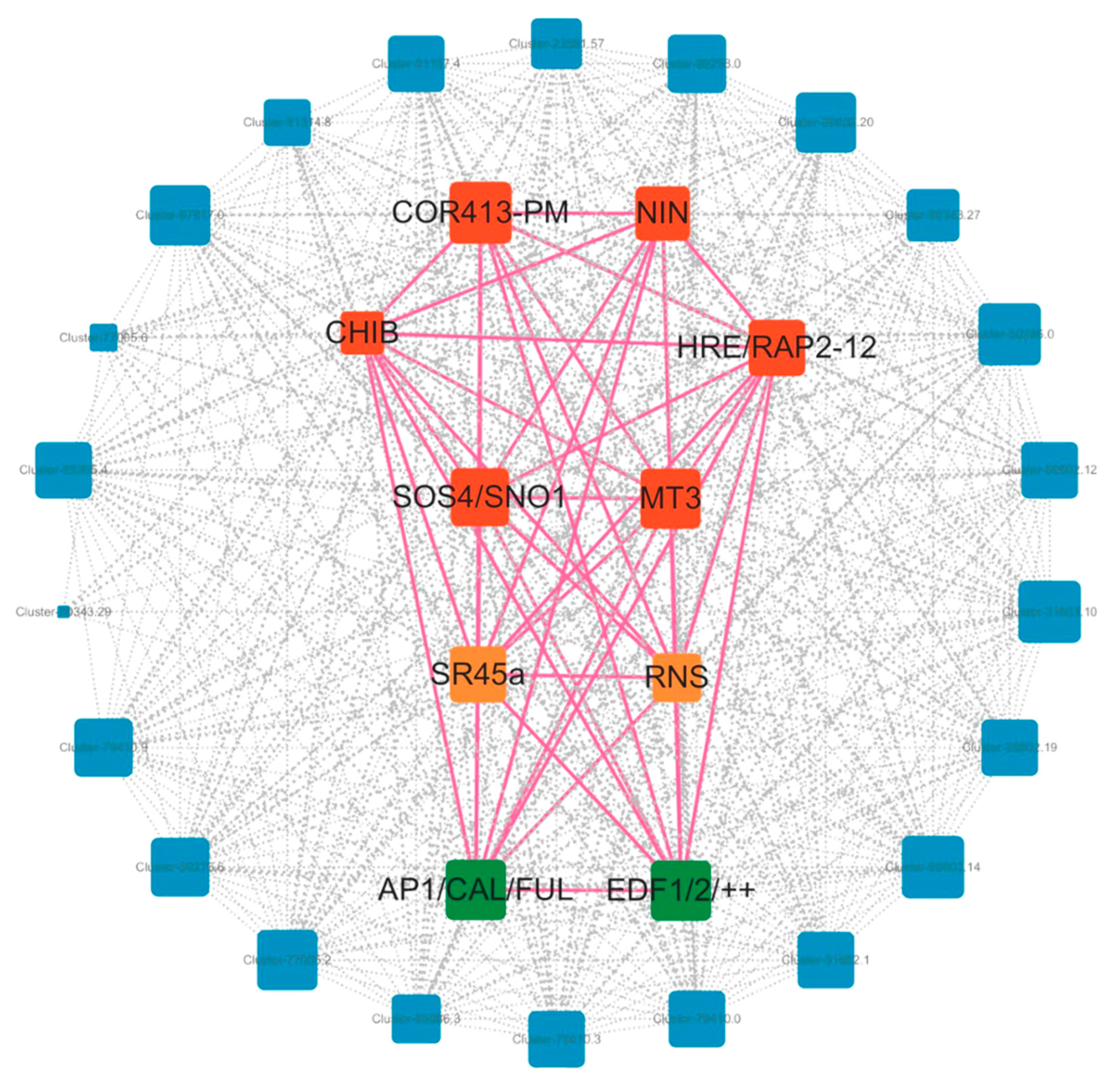
3.5. Construction of the Co-Expression Module and Regulatory Network of Flowering Genes in Bambusaoldhamii
3.6. The Gene Expression Was Verified by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Jesus, D.A.; Batista, D.M.; Monteiro, E.F.; Salzman, S.; Carvalho, L.M.; Santana, K.; André, T. Structural changes and adaptative evolutionary constraints in FLOWERING LOCUS T and TERMINAL FLOWER1-like genes of flowering plants. Front. Genet. 2022, 13, 954015. [Google Scholar] [CrossRef]
- Zheng, X.; Lin, S.; Fu, H.; Wan, Y.; Ding, Y. The Bamboo Flowering Cycle Sheds Light on Flowering Diversity. Front. Plant Sci. 2020, 11, 381. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, S.; Hu, J.; Zou, X.; Tie, L.; Li, Y.; Cui, X.; Huang, C.; Sardans, J.; Peñuelas, J. Variations and trade-offs in leaf and culm functional traits among 77 woody bamboo species. BMC Plant Biol. 2024, 24, 387. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.; Zhou, L.; Zhang, A.; Wang, S.; Liu, Y.; Yang, D.; Wang, S. Influence of flowering on the anatomical structure, chemical components and carbohydrate metabolism of Bambusa tuldoides culms at different ages. Front. Plant Sci. 2023, 14, 1260302. [Google Scholar] [CrossRef]
- Chakraborty, S.; Biswas, P.; Dutta, S.; Basak, M.; Guha, S.; Chatterjee, U.; Das, M. Studies on Reproductive Development and Breeding Habit of the Commercially Important Bamboo Bambusa tulda Roxb. Plants 2021, 10, 2375. [Google Scholar] [CrossRef]
- Xie, N.; Chen, L.-N.; Wong, K.-M.; Cui, Y.-Z.; Yang, H.-Q.; Scali, M. Seed Set and Natural Regeneration of Dendrocalamus membranaceus Munro after Mass and Sporadic Flowering in Yunnan, China. PLoS ONE 2016, 11, e0153845. [Google Scholar] [CrossRef]
- Naithani, H.B.; Kharsati, E.F. Gregarious flowering of bamboos, Cephalostachyum capitatum and Neomicrocalamus prainii from Meghalaya, India. J. Non Timber For. Prod. 2023, 30, 29–31. [Google Scholar] [CrossRef]
- Isagi, Y.; Shimada, K.; Kushima, H.; Tanaka, N.; Nagao, A.; Ishikawa, T.; OnoDera, H.; Watanabe, S. Clonal structure and flowering traits of a bamboo Phyllostachys pubescens (Mazel) Ohwi stand grown from a simultaneous flowering as revealed by AFLP analysis. Mol. Ecol. 2004, 13, 2017–2021. [Google Scholar] [CrossRef]
- Wu, C.; Cheng, Z.; Gao, J. Mysterious Bamboo flowering phenomenon: A literature review and new perspectives. Sci. Total Environ. 2024, 911, 168695. [Google Scholar] [CrossRef]
- Mizuki, I.; Sato, A.; Matsuo, A.; Suyama, Y.; Suzuki, J.; Makita, A. Clonal structure, seed set, and self-pollination rate in mass-flowering bamboo species during off-year flowering events. PLoS ONE 2014, 9, e105051. [Google Scholar] [CrossRef]
- Ma, P.-F.; Liu, Y.-L.; Guo, C.; Jin, G.; Guo, Z.-H.; Mao, L.; Yang, Y.-Z.; Niu, L.-Z.; Wang, Y.-J.; Clark, L.G.; et al. Genome assemblies of 11 bamboo species highlight diversification induced by dynamic subgenome dominance. Nat. Genet. 2024, 56, 710–720. [Google Scholar] [CrossRef]
- Veller, C.; Nowak, M.A.; Davis, C.C. Extended flowering intervals of bamboos evolved by discrete multiplication. Ecol. Lett. 2015, 18, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Zhang, Y.; Cheng, Z.; Hou, D.; Li, X.; Gao, J. Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol. J. 2016, 15, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Pei, J.; Zhuo, J.; Tang, Q.; Hou, D.; Lin, X. The CONSTANS-LIKE gene PeCOL13 regulates flowering through intron-retained alternative splicing in Phyllostachys edulis. Int. J. Biol. Macromol. 2024, 274, 133393. [Google Scholar] [CrossRef]
- Yuan, J.L.; Yue, J.J.; Gu, X.P.; Lin, C.S. Flowering of Woody Bamboo in Tissue Culture Systems. Front. Plant Sci. 2017, 8, 1589. [Google Scholar] [CrossRef]
- Li, B.; Xiao, G.; Luo, K.; Wang, Z.; Mao, B.; Lin, X.; Guo, X. Overexpression of PvGF14c from Phyllostachys violascens Delays Flowering Time in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 105. [Google Scholar] [CrossRef]
- Lin, X.; Chow, T.; Chen, H.; Liu, C.; Chou, S.; Huang, B.; Kuo, C.; Wen, C.; Huang, L.; Fang, W. Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet. Mol. Res. 2010, 9, 1085–1093. [Google Scholar] [CrossRef]
- Wang, W.; Franklin, S.B.; Lu, Z.; Rude, B.J. Delayed Flowering in Bamboo: Evidence from Fargesia qinlingensis in the Qinling Mountains of China. Front. Plant Sci. 2016, 7, 151. [Google Scholar] [CrossRef]
- Jiao, Y.; Hu, Q.; Zhu, Y.; Zhu, L.; Ma, T.; Zeng, H.; Zang, Q.; Li, X.; Lin, X. Comparative transcriptomic analysis of the flower induction and development of the Lei bamboo (Phyllostachys violascens). BMC Bioinform. 2019, 20, 687. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.; Wang, Z.; Teng, Y. Nitrate Signaling and Its Role in Regulating Flowering Time in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 5310. [Google Scholar] [CrossRef]
- Kinoshita, A.; Richter, R. Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 2490–2504. [Google Scholar] [CrossRef] [PubMed]
- Visentin, I.; Ferigolo, L.F.; Russo, G.; Korwin Krukowski, P.; Capezzali, C.; Tarkowská, D.; Gresta, F.; Deva, E.; Nogueira, F.T.S.; Schubert, A.; et al. Strigolactones promote flowering by inducing the miR319-LA-SFT module in tomato. Proc. Natl. Acad. Sci. USA 2024, 121, e2316371121. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-H.; Li, P.-F.; Chen, M.-K.; Lee, Y.-I.; Yang, C.-H. Forever Young Flower Negatively Regulates Ethylene Response DNA-Binding Factors by Activating an Ethylene-Responsive Factor to Control Arabidopsis Floral Organ Senescence and Abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef]
- Calonje, M.; Cubas, P.; MartínEz-Zapater, J.M.; Carmona, M.J. Floral meristem identity genes are expressed during tendril development in grapevine. Plant Physiol. 2004, 135, 1491–1501. [Google Scholar] [CrossRef]
- Adrian, J.; Torti, S.; Turck, F. From decision to commitment: The molecular memory of flowering. Mol. Plant 2009, 2, 628–642. [Google Scholar] [CrossRef]
- Qiao, Z.; Deng, F.; Zeng, H.; Li, X.; Lu, L.; Lei, Y.; Li, L.; Chen, Y.; Chen, J. MADS-Box Family Genes in Lagerstroemia indica and Their Involvement in Flower Development. Plants 2024, 13, 709. [Google Scholar] [CrossRef]
- Long, L.; Yu, M.; Wenjing, Y.; Yulong, D.; Shiqi, L. Research advance in growth and development of bamboo organs. Ind. Crops Prod. 2023, 205, 117428. [Google Scholar] [CrossRef]
- Louis, B.; Waikhom, S.D.; Goyari, S.; Jose, R.C.; Roy, P.; Talukdar, N.C. First proteome study of sporadic flowering in bamboo species (Bambusa vulgaris and Dendrocalamus manipureanus) reveal the boom is associated with stress and mobile genetic elements. Gene 2015, 574, 255–264. [Google Scholar] [CrossRef]
- Cheng, Z.; Hou, D.; Liu, J.; Li, X.; Xie, L.; Ma, Y.; Gao, J. Characterization of moso bamboo (Phyllostachys edulis) Dof transcription factors in floral development and abiotic stress responses. Genome 2018, 61, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Dror, D.; Weitzman, G.; Rog, I.; Kafri-Amit, T.; Klein, T. Physiological effects of mature tree transplanting characterize the roles of the soil-root interface in the field. Agric. For. Meteorol. 2020. [Google Scholar] [CrossRef]
- Suchocka, M.; Swoczyna, T.; Kosno-Jończy, J.; Kalaji, H.M.; Subramanyam, R. Impact of heavy pruning on development and photosynthesis of Tilia cordata Mill. trees. PLoS ONE 2021, 16, e0256465. [Google Scholar] [CrossRef]
- Dutta, S.; Biswas, P.; Chakraborty, S.; Mitra, D.; Pal, A.; Das, M. Identification, characterization and gene expression analyses of important flowering genes related to photoperiodic pathway in bamboo. BMC Genom. 2018, 19, 190. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, Z.; Li, X.; Xie, L.; Bai, Y.; Peng, L.; Li, J.; Gao, J. Expression Analysis and Regulation Network Identification of the CONSTANS-Like Gene Family in Moso Bamboo (Phyllostachys edulis) Under Photoperiod Treatments. DNA Cell Biol. 2019, 38, 607–626. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Q.; Nguyen, P.; Lin, C. Cryptochrome-mediated light responses in plants. Enzymes 2014, 35, 167–189. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q.; Dai, S. Drought-Responsive Mechanisms in Plant Leaves Revealed by Proteomics. Int. J. Mol. Sci. 2016, 17, 1706. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, W.; Wu, Q.; Xie, Z.; Qi, K.; Zhang, S.; Wu, J.; Wang, P. Dual roles of pear early flowering 4-like genes in regulating flowering and leaf senescence. BMC Plant Biol. 2024, 24, 1117. [Google Scholar] [CrossRef]
- Lu, X.; Yu, S.; Lü, P.; Chen, H.; Zhong, S.; Zhou, B. Genome-Wide Transcriptomic Analysis Reveals a Regulatory Network of Oxidative Stress-Induced Flowering Signals Produced in Litchi Leaves. Genes 2020, 11, 324. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Ju, Y.; Kessler, S.A. Reactive Oxygen Species as Mediators of Gametophyte Development and Double Fertilization in Flowering Plants. Front. Plant Sci. 2020, 11, 1199. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Wen, X.; Deng, C.; Yang, X.; Dai, S. Genome-wide identification and characterization analysis of RWP-RK family genes reveal their role in flowering time of Chrysanthemum lavandulifolium. BMC Plant Biol. 2023, 23, 197. [Google Scholar] [CrossRef]
- Lin, C.; Guo, X.; Yu, X.; Li, S.; Li, W.; Yu, X.; An, F.; Zhao, P.; Ruan, M. Genome-Wide Survey of the RWP-RK Gene Family in Cassava (Manihot esculenta Crantz) and Functional Analysis. Int. J. Mol. Sci. 2023, 24, 12925. [Google Scholar] [CrossRef]
- Liu, C.; Yuan, D.; Liu, T.; Xing, M.; Xu, W.; Zhang, H.; Jin, H.; Cai, C.; Li, S. Characterization and Comparative Analysis of RWP-RK Proteins from Arachis duranensis, Arachis ipaensis, and Arachis hypogaea. Int. J. Genom. 2020, 2020, 2568640. [Google Scholar] [CrossRef]
- Yan, D.; Nambara, E. Conserved and unique functions of NIN-like proteins in nitrate sensing and signaling. Plant Sci. Int. J. Exp. Plant Biol. 2023, 336, 111842. [Google Scholar] [CrossRef]
- Sámano, M.L.; Nanjareddy, K.; Arthikala, M.-K. NIN-like proteins (NLPs) as crucial nitrate sensors: An overview of their roles in nitrogen signaling, symbiosis, abiotic stress, and beyond. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2024, 30, 1209–1223. [Google Scholar] [CrossRef]
- Rai, P.K. Comparative assessment of soil properties after bamboo flowering and death in a tropical forest of Indo-Burma hot spot. Ambio 2009, 38, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.S.; Liu, F.; Liu, Z.Y.; Huang, Q.L.; Zhang, G.F.; Pan, H. Structural variability and differentiation of niches in the rhizosphere and endosphere bacterial microbiome of moso bamboo (Phyllostachys edulis). Sci. Rep. 2021, 11, 1574. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, K.; Yang, W.; Wu, F. Effects of Fargesia denudata density on soil nutrient pool. Chin. J. Appl. Ecol. 2005, 16, 996–1001. [Google Scholar]
- Fan, H.; Zhuo, R.; Wang, H.; Xu, J.; Jin, K.; Huang, B.; Qiao, G.; Brunner, A. A comprehensive analysis of the floral transition in ma bamboo (Dendrocalamus latiflorus) reveals the roles of DlFTs involved in flowering. Tree Physiol. 2022, 42, 1899–1911. [Google Scholar] [CrossRef]
- Lou, Y.; Sun, H.; Zhu, C.; Yang, K.; Li, X.; Gao, Z. PeVDE, a violaxanthin de-epoxidase gene from moso bamboo, confers photoprotection ability in transgenic Arabidopsis under high light. Front. Plant Sci. 2022, 13, 927949. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Peng, J.; Li, F.; Ali, F.; Wang, Z. Regulation of seed germination: ROS, epigenetic, and hormonal aspects. J. Adv. Res. 2024, 71, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, K.; Zhang, T.; Chen, J.; Zhao, S.; Cui, Y.; Zhou, W.; Yu, Y.; Chen, S.; Wang, C.; et al. The RhWRKY33a-RhPLATZ9 regulatory module delays petal senescence by suppressing rapid reactive oxygen species accumulation in rose flowers. Plant J. 2023, 114, 1425–1442. [Google Scholar] [CrossRef]
- Jin, S.; Ahn, J.H. Regulation of flowering time by ambient temperature: Repressing the repressors and activating the activators. New Phytol. 2021, 230, 938–942. [Google Scholar] [CrossRef]
- Park, K.; Kim, S.; Jung, J. Analysis of temperature effects on the protein accumulation of the FT–FD module using newly generated Arabidopsis transgenic plants. Plant Direct 2023, 7, e552. [Google Scholar] [CrossRef]
- Nagalla, A.D.; Nishide, N.; Hibara, K.-I.; Izawa, T. High Ambient Temperatures Inhibit Ghd7-Mediated Flowering Repression in Rice. Plant Cell Physiol. 2021, 62, 1745–1759. [Google Scholar] [CrossRef]
- Liu, S.; Hou, D.; Vasupalli, N.; Lin, X. Overexpression of PvSVP1, an SVP-like gene of bamboo, causes early flowering and abnormal floral organs in Arabidopsis and rice. Acta Biochim. Biophys. Sin. 2023, 55, 237–249. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; García-Ponce, B.; Garay-Arroyo, A. Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated Arabidopsis thaliana floral MADS-box genes. J. Exp. Bot. 2006, 57, 3099–3107. [Google Scholar] [CrossRef] [PubMed]
- de Silva, K.K.; Dunwell, J.M.; Wickramasuriya, A.M.; Gerdol, M. Weighted Gene Correlation Network Analysis (WGCNA) of Arabidopsis Somatic Embryogenesis (SE) and Identification of Key Gene Modules to Uncover SE-Associated Hub Genes. Int. J. Genom. 2022, 2022, 7471063. [Google Scholar] [CrossRef]

| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| FA-1 | 62,102,718 | 57,080,886 | 8.56 | 0.01 | 97.86 | 93.62 | 53.5 |
| FA-2 | 58,318,738 | 53,194,242 | 7.98 | 0.01 | 98.01 | 94.07 | 53.42 |
| FA-3 | 53,872,404 | 48,781,900 | 7.32 | 0.01 | 97.96 | 93.92 | 53.24 |
| UFFA-1 | 54,141,882 | 50,014,552 | 7.5 | 0.01 | 98.04 | 94.11 | 53.56 |
| UFFA-2 | 57,137,426 | 51,287,204 | 7.69 | 0.01 | 98.15 | 94.44 | 53.95 |
| UFFA-3 | 56,744,142 | 52,810,016 | 7.92 | 0.01 | 97.91 | 93.75 | 53.91 |
| SH-1 | 58,328,184 | 53,256,984 | 7.99 | 0.01 | 97.85 | 93.65 | 54.51 |
| SH-2 | 48,324,260 | 43,677,018 | 6.55 | 0.01 | 97.9 | 93.78 | 53.48 |
| SH-3 | 55,391,694 | 50,735,326 | 7.61 | 0.01 | 97.88 | 93.69 | 53.38 |
| FSH-1 | 66,474,268 | 60,477,054 | 9.07 | 0.01 | 98.2 | 94.56 | 52.7 |
| FSH-2 | 64,482,610 | 57,003,080 | 8.55 | 0.01 | 98.1 | 94.28 | 52.58 |
| FSH-3 | 58,368,886 | 52,412,646 | 7.86 | 0.01 | 98.04 | 94.11 | 52.97 |
| UFSH-1 | 53,820,998 | 48,669,652 | 7.3 | 0.01 | 98.01 | 94.03 | 52.81 |
| UFSH-2 | 56,564,710 | 50,818,384 | 7.62 | 0.01 | 97.94 | 93.8 | 53.08 |
| UFSH-3 | 50,249,878 | 45,604,840 | 6.84 | 0.01 | 97.89 | 93.64 | 53.65 |
| Typle | Number | Mean Length | N 50 | N 90 |
|---|---|---|---|---|
| Transcript | 377,416 | 945 | 1492 | 390 |
| Unigene | 213,273 | 1174 | 1685 | 537 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, S.; Wei, X.; Chen, J.; Luo, S.; Jiang, T.; Yang, J.; Zheng, R.; Chen, S. Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions. Genes 2025, 16, 811. https://doi.org/10.3390/genes16070811
Ye S, Wei X, Chen J, Luo S, Jiang T, Yang J, Zheng R, Chen S. Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions. Genes. 2025; 16(7):811. https://doi.org/10.3390/genes16070811
Chicago/Turabian StyleYe, Shanwen, Xuhui Wei, Jiamei Chen, Suzhen Luo, Tingguo Jiang, Jie Yang, Rong Zheng, and Shuanglin Chen. 2025. "Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions" Genes 16, no. 7: 811. https://doi.org/10.3390/genes16070811
APA StyleYe, S., Wei, X., Chen, J., Luo, S., Jiang, T., Yang, J., Zheng, R., & Chen, S. (2025). Research on Key Genes for Flowering of Bambusaoldhamii Under Introduced Cultivation Conditions. Genes, 16(7), 811. https://doi.org/10.3390/genes16070811






