Detection of Selection Signatures and Genome-Wide Association Analysis of Body Weight Traits in Xianan Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Phenotypic Measurements
2.3. Resequencing Data and Variant Discovery
2.4. GWAS
2.5. Selective Sweep Analysis
2.6. Gene Function Annotation
2.7. Cell siRNA Interference Experiment
2.8. Statistical Analysis
3. Results
3.1. Descriptive Statistics
3.2. Quality Control and Population Structure
3.3. GWAS Result for Body Weight Trait
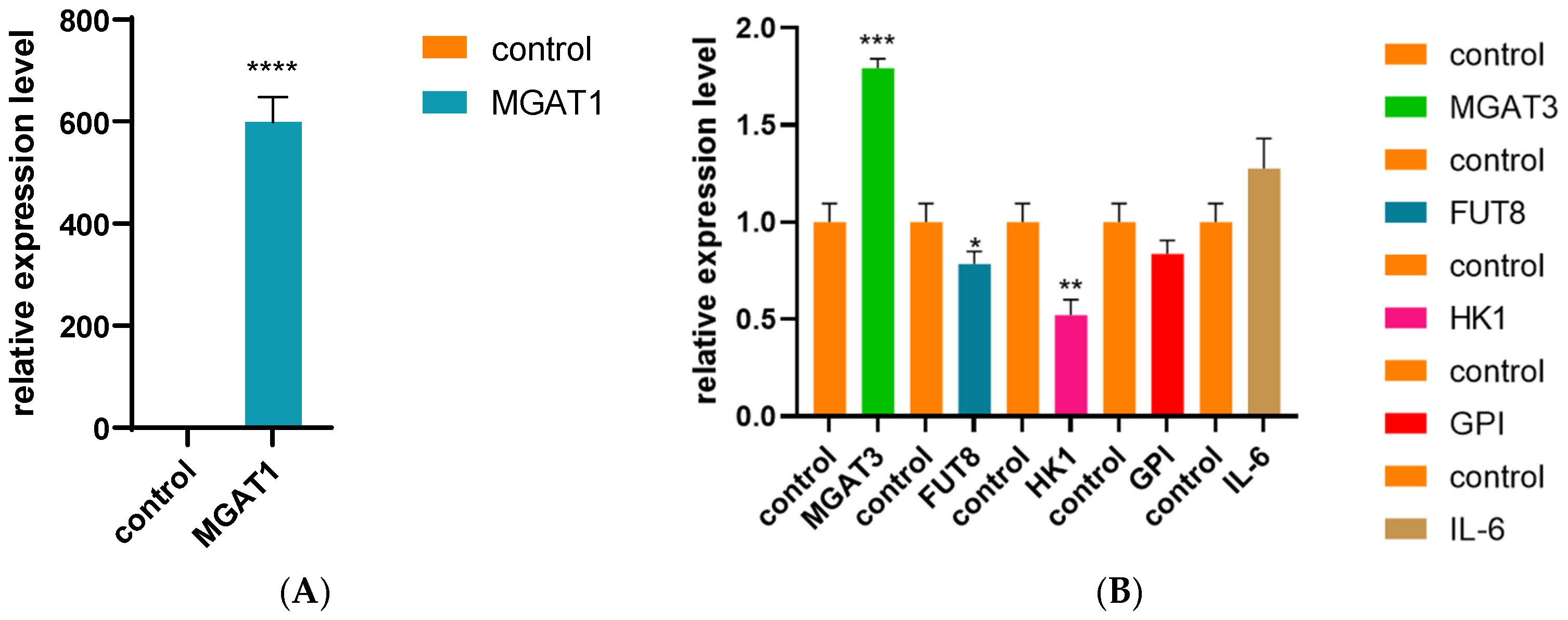
3.4. Cell siRNA Interference Experiment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primers | Sequence (5′ to 3′) | Base Number |
|---|---|---|
| MANEA-F | GATTCCCGGACCCTGCTAAA | 20 |
| MANEA-R | TGGTCTCAGCATTTTTAAACCCA | 23 |
| MGAT1-F | AAATGGAGTACTGGATGGGGG | 21 |
| MGAT1-R | GCAGCCATGCACCTTTCTTC | 20 |
| MGAT3-F | GCCGGAACCTCGTTGATGG | 19 |
| MGAT3-R | GCGTTTCATCTTCATCCCTGGC | 22 |
| FUT8-F | ATGGTGATCCTGCAGTGTGG | 20 |
| FUT8-R | CGTCTGACGTGGACTCCAAT | 20 |
| HK1-F | GAACGAATTTCCGCGTCCTG | 20 |
| HK1-R | TGTGGTCAAACAGCTCCTCC | 20 |
| GPI -F | GCTGGTGGACGTGGCTAAG | 19 |
| GPI -R | GCGTTTGATCGGTTCCGAAG | 20 |
| IL-6 -F | ACGAAAGAGAGCTCCATCTGC | 21 |
| IL-6 -R | AATGGAGTGAAGGCGCTTGT | 20 |
| Chromosome | Position | Genes | Ref | Alt | Region | Log10 (p-Value) |
|---|---|---|---|---|---|---|
| 9 | 54256092 | LOC783932 | C | T | intergenic region | 0.411667 |
| 3 | 27516835 | G | A | intergenic region | 0.444595 | |
| 3 | 27504770 | C | G | intergenic region | 0.479156 | |
| 3 | 27516216 | G | A | intergenic region | 0.481131 | |
| 9 | 54379055 | MANEA | T | C | 5_prime_UTR_variant | 0.484033 |
| 15 | 26953056 | A | G | intergenic region | 0.484294 | |
| 3 | 27509476 | G | C | intergenic region | 0.490252 | |
| 19 | 49112923 | TRNAE-UUC | A | G | upstream_gene_variant | 0.494642 |
| 19 | 49110492 | A | C | intron_variant | 0.494642 | |
| 19 | 49110226 | A | G | intron_variant | 0.494642 | |
| 19 | 49108642 | G | A | intron_variant | 0.494642 | |
| 28 | 4287845 | T | G | intergenic_region | 0.503047 | |
| 15 | 26940189 | T | C | intergenic_region | 0.512241 | |
| 15 | 26938941 | T | C | intergenic_region | 0.512241 | |
| 6 | 111114247 | CD38 | C | T | intergenic_region | 0.51408 |
| 3 | 89849741 | TRNAW-CAA | G | A | intergenic_region | 0.514114 |
| Gene | Sample CT | 18S rRNA | Ref Gene CT | 2−ΔΔCt |
|---|---|---|---|---|
| NC | 28.098 | NC | 18.536 | 1.000 |
| NC | 28.284 | NC | 18.541 | —— |
| NC | 28.433 | NC | 18.604 | —— |
| MGAT1 | 18.393 | MGAT1 | 17.960 | 599.219 |
| MGAT1 | 18.739 | MGAT1 | 18.118 | —— |
| MGAT1 | 18.782 | MGAT1 | 18.387 | —— |
| NC | 33.232 | NC | 17.408 | 1.000 |
| NC | 32.515 | NC | 17.425 | —— |
| NC | 32.323 | NC | 17.489 | —— |
| MGAT3 | 31.995 | MGAT3 | 17.605 | 1.501 |
| MGAT3 | 31.767 | MGAT3 | 17.456 | —— |
| MGAT3 | 32.784 | MGAT3 | 17.472 | —— |
| NC | 22.822 | NC | 17.339 | 1.000 |
| NC | 22.968 | NC | 17.620 | —— |
| NC | 22.959 | NC | 17.352 | —— |
| FUT8 | 23.991 | FUT8 | 18.130 | 0.784 |
| FUT8 | 24.026 | FUT8 | 18.093 | —— |
| FUT8 | 24.039 | FUT8 | 18.341 | —— |
| NC | 23.635 | NC | 18.247 | 1.000 |
| NC | 23.421 | NC | 18.060 | —— |
| NC | 23.515 | NC | 18.122 | —— |
| HK1 | 26.633 | HK1 | 20.305 | 0.521 |
| HK1 | 26.744 | HK1 | 20.182 | —— |
| HK1 | 26.638 | HK1 | 20.525 | —— |
| NC | 21.382 | NC | 17.195 | 1.000 |
| NC | 21.547 | NC | 17.057 | —— |
| NC | 21.505 | NC | 17.517 | —— |
| GPI | 22.554 | GPI | 18.195 | 0.836 |
| GPI | 22.661 | GPI | 18.063 | —— |
| GPI | 22.690 | GPI | 18.241 | —— |
| NC | 25.159 | NC | 17.876 | 1.000 |
| NC | 25.569 | NC | 17.501 | —— |
| NC | 22.632 | NC | 17.525 | —— |
| IL-6 | 25.789 | IL-6 | 18.575 | 1.275 |
| IL-6 | 25.779 | IL-6 | 18.640 | —— |
| IL-6 | 25.916 | IL-6 | 18.430 | —— |
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; Grace, D.; Njuki, J.; Johnson, N.; Enahoro, D.; Silvestri, S.; Rufino, M.C. The Roles of Livestock in Developing Countries. Animal 2013, 7, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lu, Y.; Chong, Y.; Li, M.; Hong, J.; Wu, J.; Wu, D.; Xi, D.; Deng, W. Beef Cattle Genome Project: Advances in Genome Sequencing, Assembly, and Functional Genes Discovery. Int. J. Mol. Sci. 2024, 25, 7147. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yue, B.; Yang, Y.; Tang, J.; Yang, S.; Qi, A.; Qu, K.; Lan, X.; Lei, C.; Wei, Z.; et al. Distribution of Copy Number Variation in SYT11 Gene and Its Association with Growth Conformation Traits in Chinese Cattle. Biology 2022, 11, 223. [Google Scholar] [CrossRef]
- Yang, D.; Chen, H.; Wang, X.; Tian, Z.; Tang, L.; Zhang, Z.; Lei, C.; Zhang, L.; Wang, Y. Association of Polymorphisms of Leptin Gene with Body Weight and Body Sizes Indexes in Chinese Indigenous Cattle. J. Genet. Genom. 2007, 34, 400–405. [Google Scholar] [CrossRef]
- Liu, S.; Ma, X.; Yue, T.; Wang, Z.; Qi, K.; Li, J.; Lin, F.; Rushdi, H.E.; Gao, Y.; Fu, T.; et al. Transcriptome-Wide m6A Analysis Provides Novel Insights Into Testicular Development and Spermatogenesis in Xia-Nan Cattle. Front. Cell Dev. Biol. 2021, 9, 791221. [Google Scholar] [CrossRef]
- Rot, C.; Creutzinger, K.; Goetz, H.; Winder, C.; Morrison, J.; Conboy, M.; Bajus, A.; Renaud, D.L. Factors Associated with Body Weight of Young Surplus Dairy Calves on Arrival to a Calf Rearing Facility. Prev. Vet. Med. 2022, 203, 105630. [Google Scholar] [CrossRef]
- Zuin, R.G.; Buzanskas, M.E.; Caetano, S.L.; Venturini, G.C.; Guidolin, D.G.F.; Grossi, D.A.; Chud, T.C.S.; Paz, C.C.P.; Lôbo, R.B.; Munari, D.P. Genetic Analysis on Growth and Carcass Traits in Nelore Cattle. Meat Sci. 2012, 91, 352–357. [Google Scholar] [CrossRef]
- Sieber, M.; Freeman, A.E.; Kelley, D.H. Effects of Body Measurements and Weight on Calf Size and Calving Difficulty of Holsteins. J. Dairy Sci. 1989, 72, 2402–2410. [Google Scholar] [CrossRef]
- Igoshin, A.V.; Yudin, N.S.; Belonogova, N.M.; Larkin, D.M. Genome-wide Association Study for Body Weight in Cattle Populations from Siberia. Anim. Genet. 2019, 50, 250–253. [Google Scholar] [CrossRef]
- Yeo, G.S.H. The Role of the FTO (Fat Mass and Obesity Related) Locus in Regulating Body Size and Composition. Mol. Cell. Endocrinol. 2014, 397, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, F.; Gao, G.; Yan, X.; Liu, H.; Liu, Z.; Wang, Z.; He, L.; Lv, Q.; Wang, Z.; et al. Genome-Wide Association Study of Body Weight Traits in Inner Mongolia Cashmere Goats. Front. Vet. Sci. 2021, 8, 752746. [Google Scholar] [CrossRef] [PubMed]
- Bangar, Y.C.; Magotra, A. Meta-Analysis of SNP in Growth Hormone Gene Associated with Milk Traits in Dairy Cows. Trop. Anim. Health Prod. 2021, 53, 222. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Pham, M.; Tu, Y.; Lv, X. Accelerating BWA-MEM Read Mapping on GPUs. In Proceedings of the 37th International Conference on Supercomputing (ICS ‘23), Orlando, FL, USA, 21 June 2023; ACM: New York, NY, USA, 2023; pp. 155–166. [Google Scholar]
- Babadi, M.; Fu, J.M.; Lee, S.K.; Smirnov, A.N.; Gauthier, L.D.; Walker, M.; Benjamin, D.I.; Zhao, X.; Karczewski, K.J.; Wong, I.; et al. GATK-gCNV Enables the Discovery of Rare Copy Number Variants from Exome Sequencing Data. Nat. Genet. 2023, 55, 1589–1597. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, s13742-015-0047–0048. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore Technology and Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain w1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Farmer, L.J.; Farrell, D.T. Review: Beef-Eating Quality: A European Journey. Animal 2018, 12, 2424–2433. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.P.; Stein, M.B.; Kranzler, H.R.; Yang, B.Z.; Farrer, L.A.; Gelernter, J. The α-Endomannosidase Gene (MANEA) Is Associated with Panic Disorder and Social Anxiety Disorder. Transl. Psychiatry 2014, 4, e353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feng, Y.; Deng, Y.; Tang, Z.; Cai, S.; Li, J.; Liu, R.; Wan, J.; He, H.; Zeng, G.; Ye, J.; et al. Prognostic Implication of Heterogeneity and Trajectory Progression Induced by Enzalutamide in Prostate Cancer. Front. Endocrinol. 2023, 14, 1148898. [Google Scholar] [CrossRef]
- Yengo, L.; Vedantam, S.; Marouli, E.; Sidorenko, J.; Bartell, E.; Sakaue, S.; Graff, M.; Eliasen, A.U.; Jiang, Y.; Raghavan, S.; et al. A Saturated Map of Common Genetic Variants Associated with Human Height. Nature 2022, 610, 704–712. [Google Scholar] [CrossRef]
- Qanbari, S.; Gianola, D.; Hayes, B.; Schenkel, F.; Miller, S.; Moore, S.; Thaller, G.; Simianer, H. Application of Site and Haplotype-Frequency Based Approaches for Detecting Selection Signatures in Cattle. BMC Genom. 2011, 12, 318. [Google Scholar] [CrossRef]
- Tapia-Rivera, J.C.; Baltazar-Rodríguez, L.M.; Cárdenas-Rojas, M.I.; Álvarez, A.; Bustos-Saldaña, R.; Delgado-Enciso, I.; Valdez-Velázquez, L.L.; Guzmán-Esquivel, J.; Ramírez-Flores, M. Polimorfismo rs4285184 del gen MGAT1 como factor de riesgo de obesidad en la población mexicana. Med. Clin. 2017, 148, 149–152. [Google Scholar] [CrossRef]

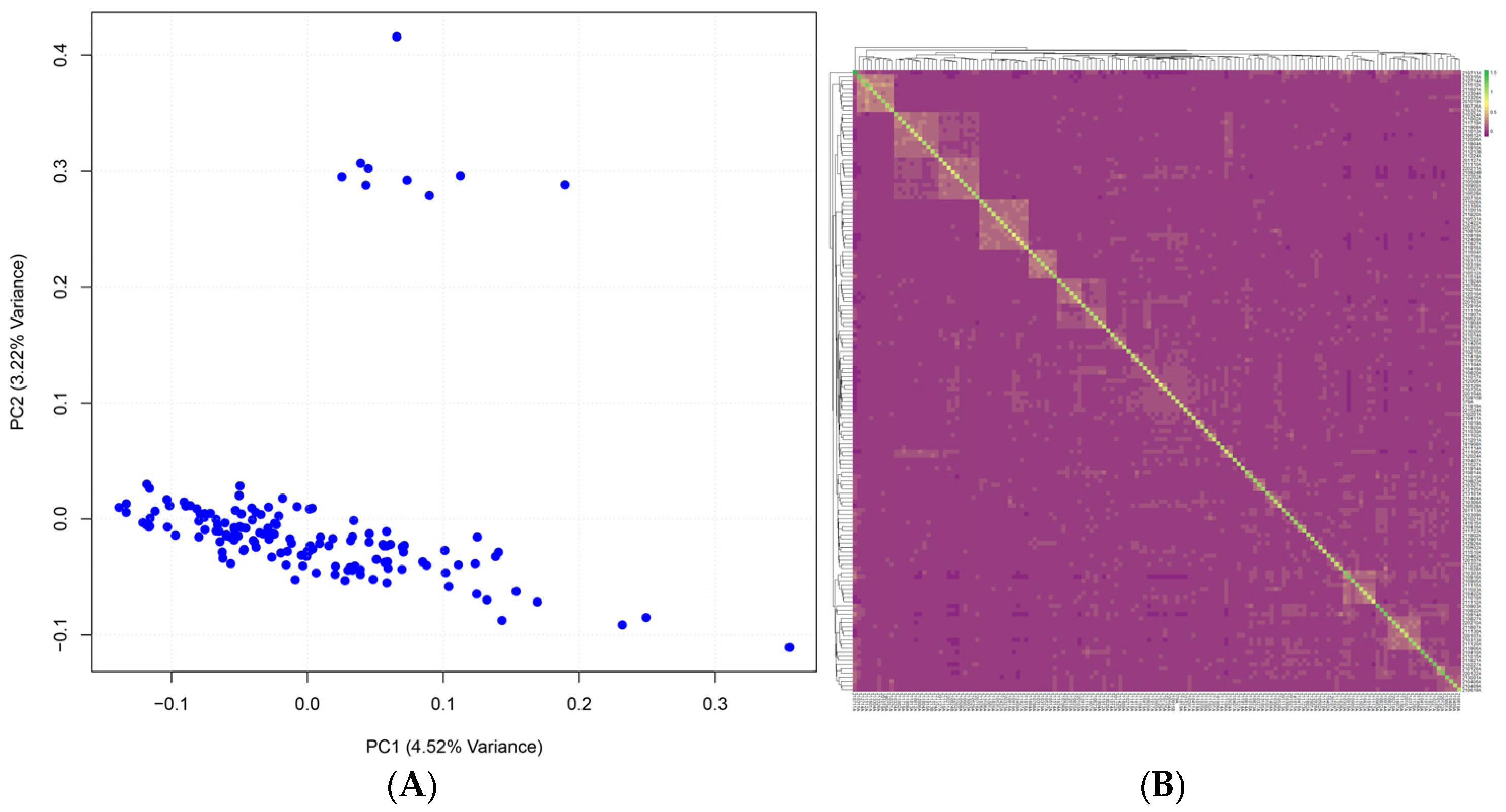


| Trait | N | Mean | SD | Min | Max | CV (%) |
|---|---|---|---|---|---|---|
| BW (kg) | 149 | 400.31 | 71.77 | 239 | 629.31 | 17.78 |
| Chromosome | Position | Genes | Ref | Alt | Region | p_Value |
|---|---|---|---|---|---|---|
| 9 | 54241895 | C | T | intergenic_region | 7.43 × 10−9 | |
| 9 | 54239625 | G | T | intergenic_region | 1.88 × 10−8 | |
| 9 | 54379055 | MANEA | T | C | 5_prime_UTR_variant | 3.45 × 10−8 |
| 9 | 54326869 | MANEA | T | C | intron_variant | 3.63 × 10−8 |
| 9 | 54246066 | T | C | intergenic_region | 3.63 × 10−8 | |
| 9 | 54246316 | A | G | intergenic_region | 6.22 × 10−8 | |
| 9 | 54242338 | C | T | intergenic_region | 6.73 × 10−8 | |
| 9 | 54393780 | A | G | intergenic_region | 7.05 × 10−8 | |
| 5 | 32474126 | HDAC7 | A | G | 5_prime_UTR_variant | 1.41 × 10−7 |
| 9 | 54256092 | C | T | intergenic_region | 1.58 × 10−7 | |
| 9 | 54261359 | A | G | intergenic_region | 2.55 × 10−7 | |
| 5 | 32517886 | RAPGEF3 | G | A | upstream_gene_variant | 2.54 × 10−7 |
| 1 | 121950885 | PLSCR2 | C | T | upstream_gene_variant | 2.54 × 10−7 |
| 9 | 54339270 | MANEA | T | A | intron_variant | 2.50 × 10−7 |
| 9 | 54390222 | G | A | intergenic_region | 6.45 × 10−7 | |
| 9 | 54266069 | G | A | intergenic_region | 6.41 × 10−7 | |
| 9 | 54354464 | MANEA | G | A | intron_variant | 6.40 × 10−7 |
| 5 | 32476523 | HDAC7 | C | T | intron_variant | 4.39 × 10−7 |
| 9 | 54315282 | MANEA | T | C | intron_variant | 6.29 × 10−7 |
| 9 | 54258450 | A | C | intergenic_region | 6.22 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Li, X.; Zhang, M.; Liu, S.; Zhang, Y.; Zheng, Y.; Wei, Z.; Han, M.; Huang, H.; Fu, T.; et al. Detection of Selection Signatures and Genome-Wide Association Analysis of Body Weight Traits in Xianan Cattle. Genes 2025, 16, 682. https://doi.org/10.3390/genes16060682
Zhu H, Li X, Zhang M, Liu S, Zhang Y, Zheng Y, Wei Z, Han M, Huang H, Fu T, et al. Detection of Selection Signatures and Genome-Wide Association Analysis of Body Weight Traits in Xianan Cattle. Genes. 2025; 16(6):682. https://doi.org/10.3390/genes16060682
Chicago/Turabian StyleZhu, Huaini, Xiaofeng Li, Man Zhang, Siyu Liu, Yan Zhang, Ying Zheng, Zhitong Wei, Mingpeng Han, Hetian Huang, Tong Fu, and et al. 2025. "Detection of Selection Signatures and Genome-Wide Association Analysis of Body Weight Traits in Xianan Cattle" Genes 16, no. 6: 682. https://doi.org/10.3390/genes16060682
APA StyleZhu, H., Li, X., Zhang, M., Liu, S., Zhang, Y., Zheng, Y., Wei, Z., Han, M., Huang, H., Fu, T., & Liang, D. (2025). Detection of Selection Signatures and Genome-Wide Association Analysis of Body Weight Traits in Xianan Cattle. Genes, 16(6), 682. https://doi.org/10.3390/genes16060682




