Comprehensive Assembly and Comparative Analysis of Chloroplast Genome and Mitogenome of Prunus salicina var. cordata
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation and Sequencing
2.3. Mitogenome and Chloroplast Assembly and Annotation
2.4. Analysis of RSCU and Repeat Sequences
2.5. Phylogenetic Analyses
3. Results
3.1. Characteristics of the Mitogenomes of P. salicina var. cordata
3.2. The Amino Acids of P. salicina var. cordata
3.3. Phylogenetic Analysis
3.4. Characteristics of the Chloroplast of P. salicina var. cordata
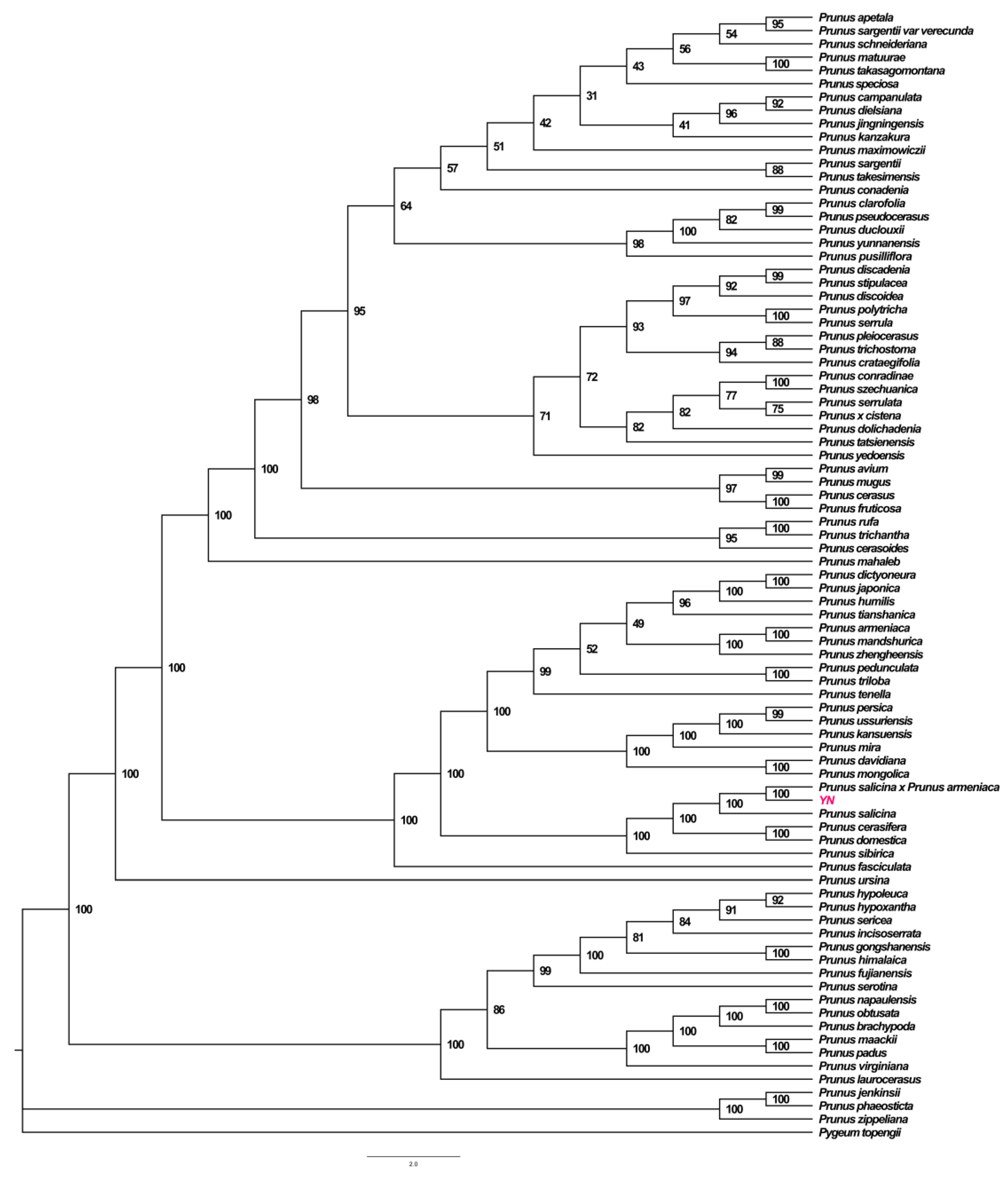
3.5. Phylogenetic Tree Analysis of P. salicina var. cordata Chloroplast Genome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Munekata, P.E.; Yilmaz, B.; Pateiro, M.; Kumar, M.; Domínguez, R.; Shariati, M.A.; Hano, C.; Lorenzo, J.M. Valorization of By-Products from Prunus Genus Fruit Processing: Oppor and Appli. Criti Revie Food Sci. Nutr. 2023, 63, 7795–7810. [Google Scholar] [CrossRef]
- Chukwuma, C. Antioxidative. Metabolic and Vascular Medicinal Potentials of Natural Products in the Non-Edible Wastes of Fruits Belonging to the Citrus and Prunus Genera: A Review. Plants 2024, 13, 191. [Google Scholar] [CrossRef]
- Milošević, T.; Milošević, N. Analysis of similarities and differences of accessions belonging to Prunus domestica L. and P. insititia L. using endocarp dimensions and shape variations. Vavilovskii Zhurnal Genet. Sel. 2025, 1, 44–54. [Google Scholar] [CrossRef]
- Ling, C.; Luo, A.; Gao, J. MrBayes 3.2.6 on Tianhe-1A: A High Performance and Distributed Implementation of Phylogenetic Analysis. In Proceedings of the 2016 IEEE 22nd International Conference on Parallel and Distributed Systems (ICPADS), Wuhan, China, 13–16 December 2016. [Google Scholar]
- Abrao, A.; Yu, M.; Gouvinhas, I.; Ferreira, L.; Silva, A.M.; Domínguez-Perles, R.; Barros, A. Prunus lusitanica L. Fruits: A Promising Underexploited Source of Nutrients with Potential Economic Value. Foods 2023, 12, 973. [Google Scholar] [CrossRef]
- Igwe, E.O.; Charlton, K.E. A Systematic Review on the Health Effects of Plums (Prunus domestica and Prunus salicina). Phyther. Res. 2016, 30, 701–731. [Google Scholar] [CrossRef]
- Weiwei, C.; Tao, L.; Huali, Y.; Yingyan, X.U.; Yijin, X.U.; Dongming, P.; Guixin, C. Isolation and Expression of WRKY Homologous Genes from Prunus salicina var. cordata. Acta Bot. Boreali-Occident. Sin. 2017, 37, 1493–1499. [Google Scholar]
- Suprun, I.I.; Stepanov, I.V.; Tokmakov, S.V.; Eremin, G.V. Study of Prunus domestica Genetic Diversity by Analysis of Microsatellite Loci. Russ. J. Genet. 2019, 55, 172–179. [Google Scholar] [CrossRef]
- Bella, Y.A.; Bouda, S.; Khachtib, Y.; Haddioui, A. Genetic Variability of Cultivated Plum (Prunus domestica L. & Prunus salicina Lindl.) in Morocco Assessed by ISSR Markers. Aust. J. Crop Sci. 2021, 15, 948–954. [Google Scholar]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, Z.; Tian, C.; Yang, Y.; Liu, L.; Feng, Y.; Li, Z. Complete Mitochondrial Genome of the Endangered Prunus pedunculata (Prunoideae, Rosaceae) in China: Characterization and Phylogenetic Analysis. Front. Plant Sci. 2023, 14, 1266797. [Google Scholar] [CrossRef]
- Wang, M.; Yu, W.; Yang, J.; Hou, Z.; Li, C.; Niu, Z.; Niu, Z.; Zhang, B.; Xue, Q.; Liu, W.; et al. Mitochondrial genome comparison and phylogenetic analysis of Dendrobium (Orchidaceae) based on whole mitogenomes. BMC Plant Biol. 2023, 23, 586. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, D.; Yu, Z.; Zeng, B. Assembly and Analysis of the Complete Mitochondrial Genome of the Chinese Wild Dwarf Almond (Prunus tenella). Front. Genet. 2023, 14, 1329060. [Google Scholar] [CrossRef]
- Cui, H.; Ding, Z.; Zhu, Q.; Wu, Y.; Qiu, B.; Gao, P. Comparative analysis of nuclear, chloroplast, and mitochondrial genomes of watermelon and melon provides evidence of gene transfer. Sci. Rep. 2021, 11, 1595. [Google Scholar] [CrossRef]
- Drogoudi, P.; Pantelidis, G.; Phenotypic, V. Peel Contribution to Fruit Antioxidant Contents in European and Japanese Plums. Plants 2022, 10, 1338. [Google Scholar] [CrossRef]
- Guerrero, B.I.; Guerra, M.E.; Rodrigo, J. Simple Sequence Repeat (SSR)-Based Genetic Diversity in Interspecific Plumcot-Type (Prunus salicina × Prunus armeniaca) Hybrids. Plants 2022, 11, 1241. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Lang, B.F.; Beck, N.; Prince, S.; Sarrasin, M.; Rioux, P.; Burger, G. Mitochondrial Genome Annotation with MFannot: A Critical Analysis of Gene Identification and Gene Model Prediction. Front. Plant Sci. 2023, 14, 1222186. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, H.; Ni, Y.; Wu, B.; Li, J.; Burzyński, A.; Liu, C. Plant Mitochondrial Genome Map (PMGmap): A Software Tool for the Comprehensive Visualization of Coding, Noncoding and Genome Features of Plant Mitochondrial Genomes. Mol. Ecol. Resour. 2024, 24, e13952. [Google Scholar] [CrossRef]
- Burgstaller-Muehlbacher, S.; Crotty, S.; Schmidt, H.; Reden, F.; Drucks, T.; Haeseler, A. ModelRevelator: Fast phylogenetic model estimation via deep learning. Mol. Phylogenet Evol. 2023, 188, 107905. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Zhai, T.; Zhao, Z.; Fu, C.; Huang, L.; Jiang, C.; Li, M.; Wang, Z.; Yang, X. De novo assembly and comparative analysis of cherry (Prunus subgenus Cerasus) mitogenomes. Front Plant Sci. 2025, 16, 1568698. [Google Scholar] [CrossRef]
- Fang, B.; Li, J.; Zhao, Q.; Yu, J. Assembly of the Complete Mitochondrial Genome of Chinese Plum (Prunus salicina): Characterization of Genome Recombination and RNA Editing Sites. Genes 2021, 12, 1970. [Google Scholar] [CrossRef]
- Richter, U.; Lahtinen, T.; Marttinen, P.; Suomi, F.; Battersby, B.J. Quality Control of Mitochondrial Protein Synthesis Is Required for Membrane Integrity and Cell Fitness. J. Cell Biol. 2015, 211, 373–389. [Google Scholar] [CrossRef]
- Sottile, F.; Caltagirone, C.; Giacalone, G.; Peano, C.; Barone, E. Unlocking Plum Genetic Potential: Where Are We At? Horticulturae 2022, 8, 128. [Google Scholar] [CrossRef]
- Rahemi, A.; Fatahi, R.; Ebadi, A.; Taghavi, T.; Hassani, D.; Gradziel, T.; Folta, K.; Chaparro, J. Genetic Diversity of Some Wild Almonds and Related Prunus Species Revealed by SSR and EST-SSR Molecular Markers. Plant Syst. Evol. 2012, 298, 173–192. [Google Scholar] [CrossRef]
- Decker, S.T.; Funai, K. Mitochondrial membrane lipids in the regulation of bioenergetic flux. Cell Metab. 2024, 36, 1963–1978. [Google Scholar] [CrossRef]
- Wang, X.; Chai, H.; Li, S.; Xu, Y.; Wu, Y.; Wang, J.; Yang, Z. Physiological characteristics and transcriptomic analyses of alfalfa root crown in wintering. Front. Plant Sci. 2024, 15, 1486564. [Google Scholar] [CrossRef]
- Takashi, A.; Toshio, H.; Hideaki, Y.; Gradziel, T.M.; Ryutaro, T. Genome-Wide View of Genetic Diversity Reveals Paths of Selection and Cultivar Differentiation in Peach Domestication. DNA Res. 2016, 23, 271–282. [Google Scholar]
- Naeem, H.; Awan, F.S.; Dracatos, P.M.; Sajid, M.W.; Zulfiqar, B. Population Structure and Phylogenetic Relationship of Peach [Prunus persica (L.) Batsch] and Nectarine [Prunus persica var. nucipersica (L.) C.K. Schneid.] Based on Retrotransposon Markers. Genet. Resour. Crop Evol. 2021, 68, 3011–3023. [Google Scholar] [CrossRef]
- Mustapha, S.B.; Ben Tamarzizt, H.; Baraket, G.; Abdallah, D.; Salhi-Hannachi, A. Cytoplasmic Polymorphism and Evolutionary History of Plum Cultivars: Insights from Chloroplast DNA Sequence Variation of TrnL-TrnF Spacer and Aggregated TrnL Intron & TrnL-TrnF Spacer. Genet. Mol. Res. 2015, 14, 3964–3979. [Google Scholar] [CrossRef]
- Zeinalabedini, M.; Khayam-Nekoui, M.; Grigorian, V.; Gradziel, T.M.; Martínez-Gómez, P. The Origin and Dissemination of the Cultivated Almond as Determined by Nuclear and Chloroplast SSR Marker Analysis. Sci. Hortic. 2010, 125, 593–601. [Google Scholar] [CrossRef]
- Kong, S.-G.; Yamazaki, Y.; Shimada, A.; Kijima, S.T.; Hirose, K.; Katoh, K.; Ahn, J.; Song, H.-G.; Han, J.-W.; Higa, T. CHLOROPLAST UNUSUAL POSITIONING 1 Is a Plant-Specific Actin Polymerization Factor Regulating Chloroplast Movement. Plant Cell 2024, 36, 1159–1181. [Google Scholar] [CrossRef]

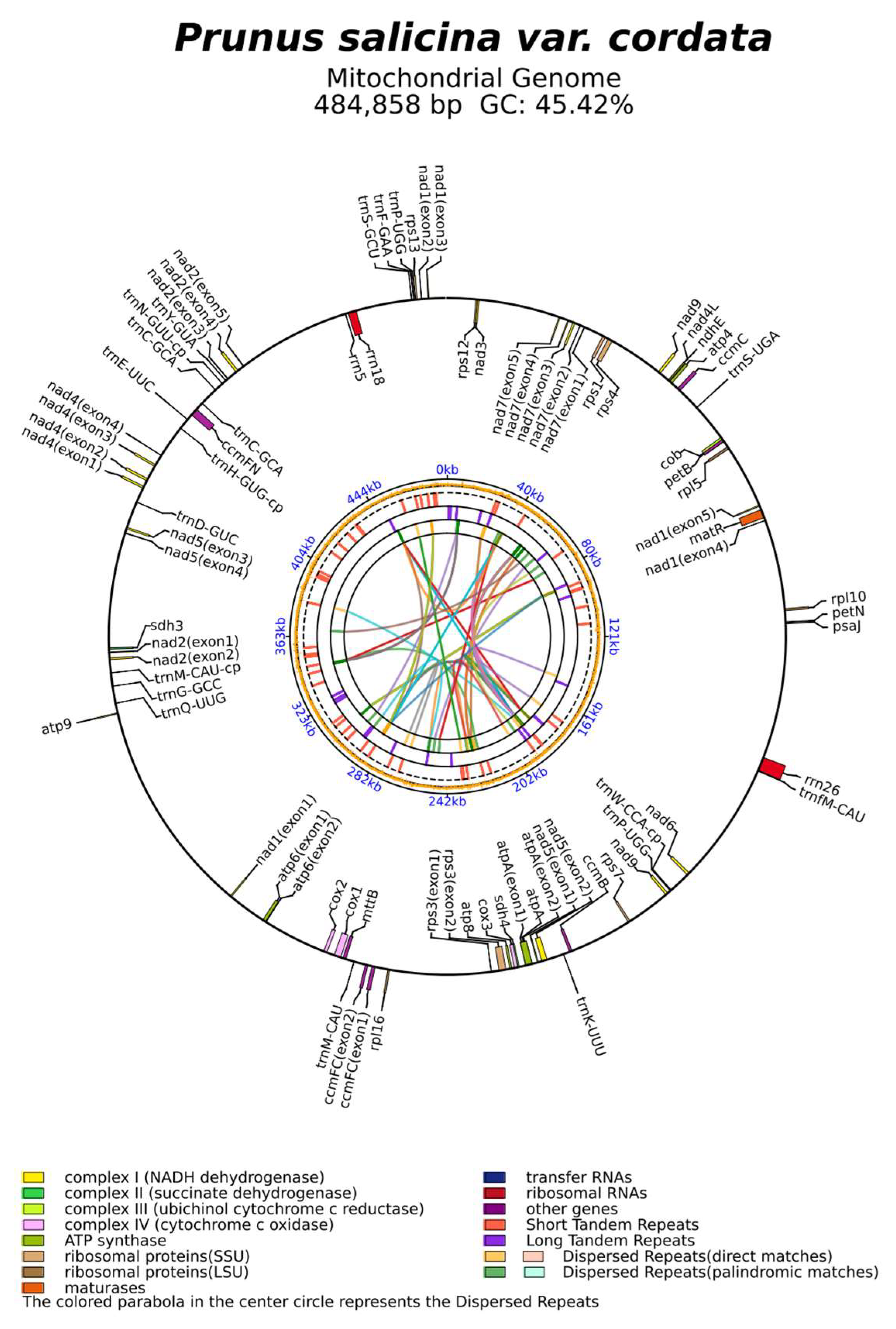
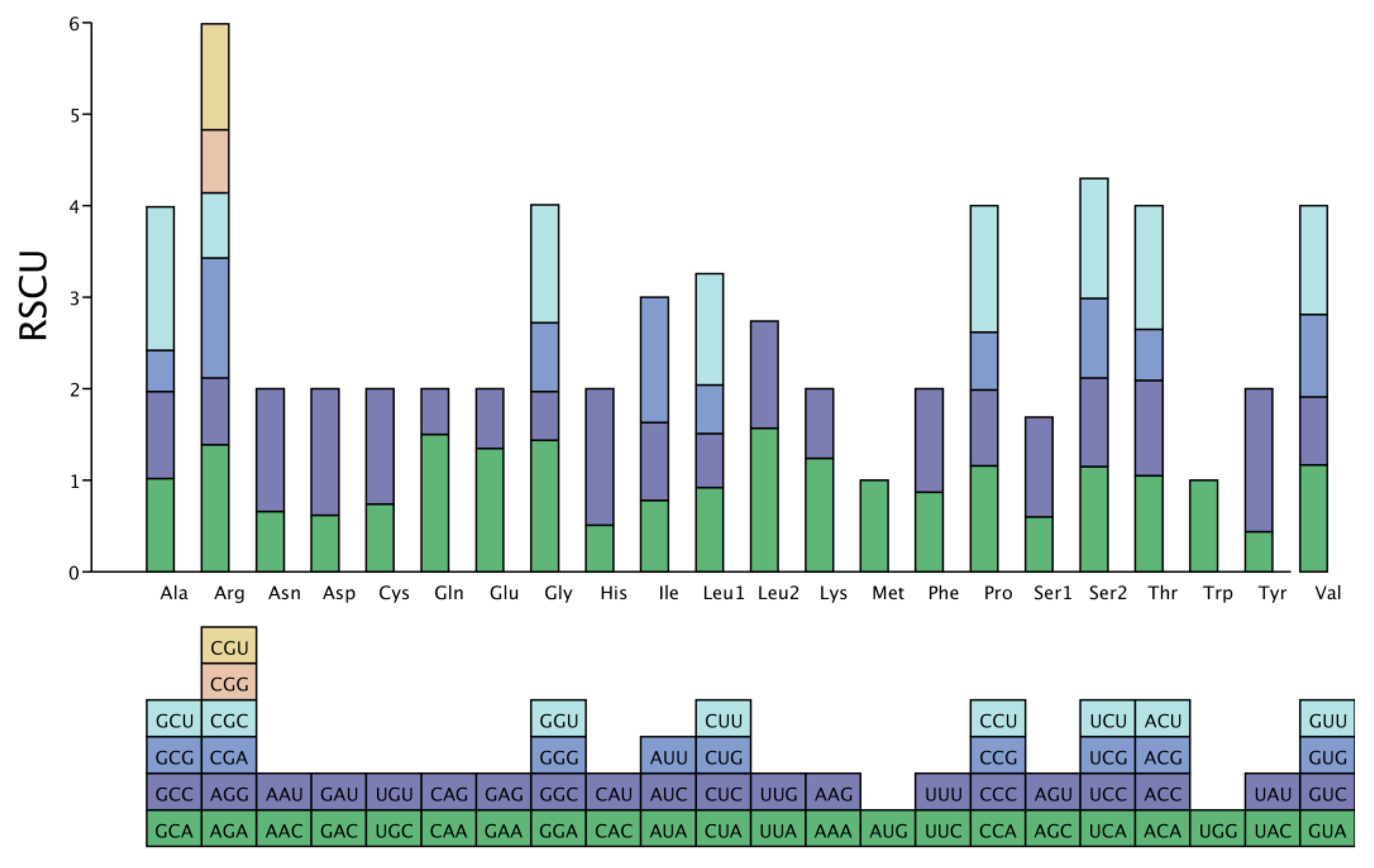
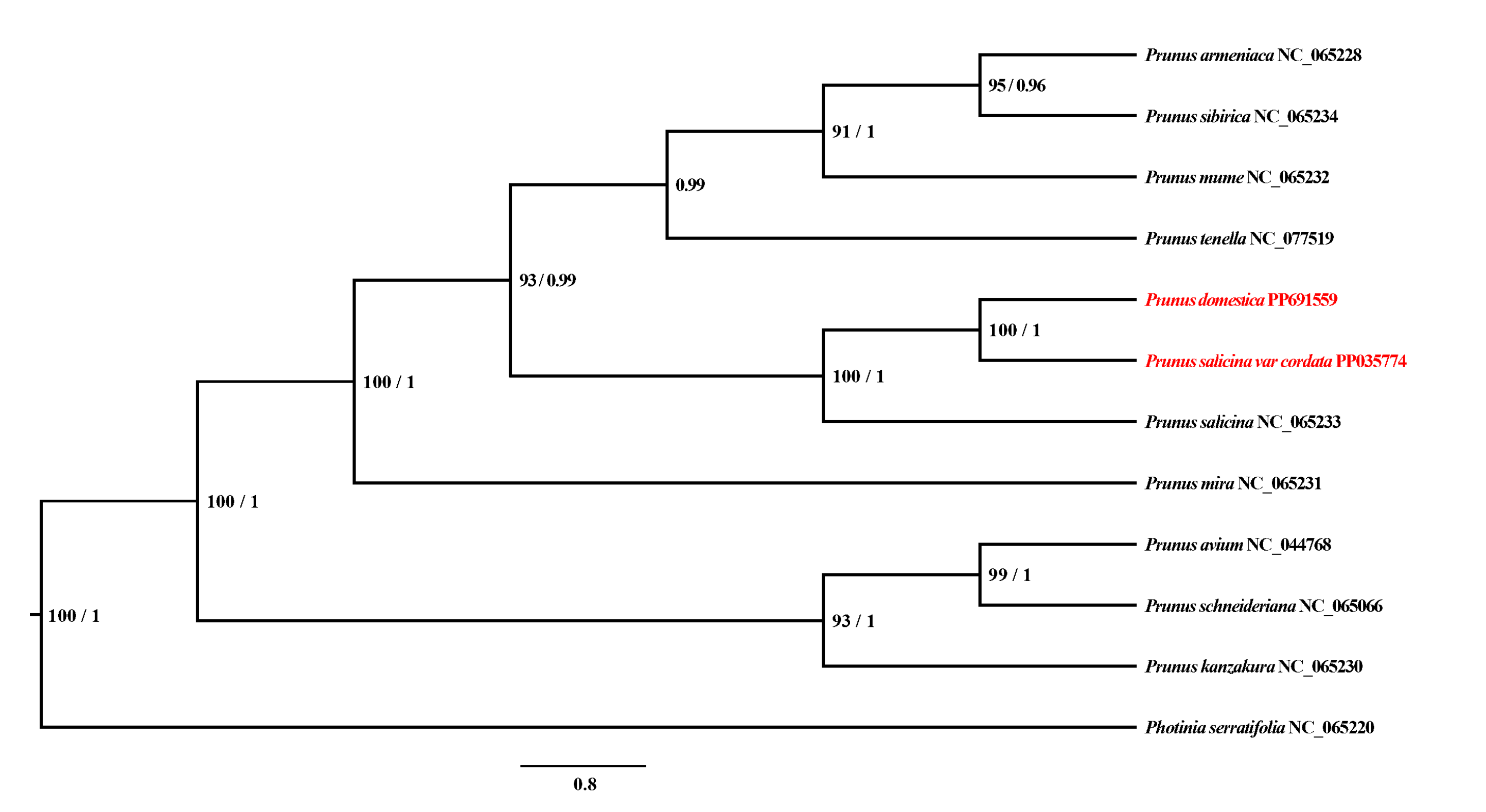
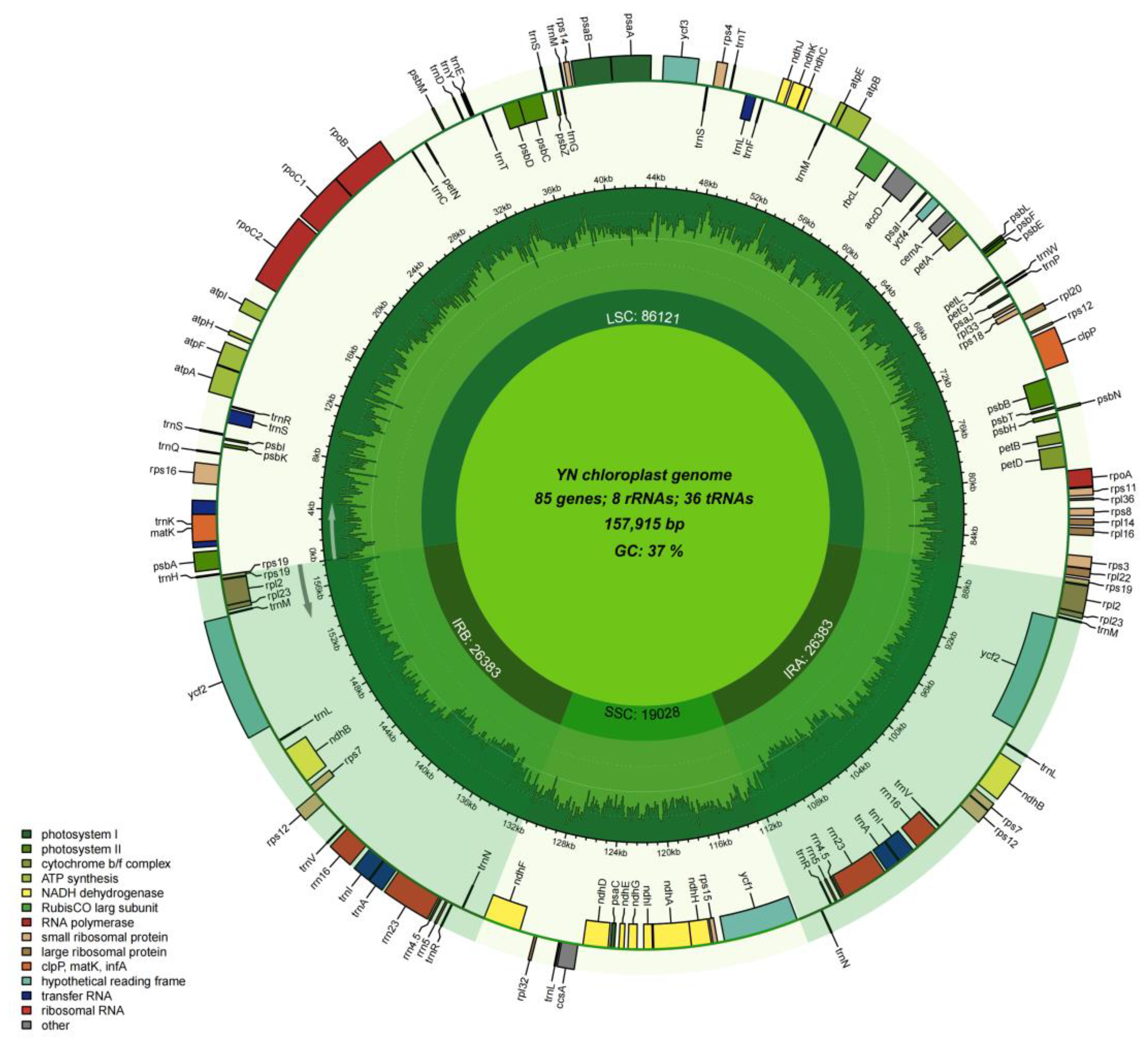
| Family | Genus | Organism | Length | AT% | GC% | ID |
|---|---|---|---|---|---|---|
| Gunneridae | Prunus | Prunus avium | 389,709 | 54.3 | 45.7 | NC_044768.1 |
| Prunus kanzakura | 422,215 | 54.4 | 45.6 | NC_065230.1 | ||
| Prunus mira | 429,732 | 54.4 | 45.6 | NC_065231.1 | ||
| Prunus schneideriana | 434,334 | 54.3 | 45.7 | NC_065066.1 | ||
| Prunus tenella | 452,158 | 54.4 | 45.6 | NC_077519.1 | ||
| P. domestica | 484,858 | 54.6 | 45.4 | PP691559 | ||
| P. salicina var. cordata | 484,858 | 54.6 | 45.4 | PP035774.1 | ||
| P. salicina | 508,005 | 54.5 | 45.5 | NC_065233.1 | ||
| Prunus sibirica | 510,187 | 54.6 | 45.4 | NC_065234.1 | ||
| Prunus armeniaca | 510,346 | 54.5 | 45.5 | NC_065228.1 | ||
| Prunus mume | 535,727 | 54.5 | 45.5 | NC_065232.1 | ||
| Photinia | Photinia serratifolia | 473,561 | 54.8 | 45.2 | NC_065220.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, R.; Zhao, M.; Lan, Q.; Peng, S.; Lin, F.; Li, Z. Comprehensive Assembly and Comparative Analysis of Chloroplast Genome and Mitogenome of Prunus salicina var. cordata. Genes 2025, 16, 660. https://doi.org/10.3390/genes16060660
Liao R, Zhao M, Lan Q, Peng S, Lin F, Li Z. Comprehensive Assembly and Comparative Analysis of Chloroplast Genome and Mitogenome of Prunus salicina var. cordata. Genes. 2025; 16(6):660. https://doi.org/10.3390/genes16060660
Chicago/Turabian StyleLiao, Ruyu, Mengshi Zhao, Qin Lan, Song Peng, Fengqiang Lin, and Zhaolong Li. 2025. "Comprehensive Assembly and Comparative Analysis of Chloroplast Genome and Mitogenome of Prunus salicina var. cordata" Genes 16, no. 6: 660. https://doi.org/10.3390/genes16060660
APA StyleLiao, R., Zhao, M., Lan, Q., Peng, S., Lin, F., & Li, Z. (2025). Comprehensive Assembly and Comparative Analysis of Chloroplast Genome and Mitogenome of Prunus salicina var. cordata. Genes, 16(6), 660. https://doi.org/10.3390/genes16060660






