Cotton STARD Gene Family: Characterization, Evolution, and Expression Profiles During Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of STARD Genes in Cotton
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Chromosomal Locations of STARDs from Four Types of Cotton
2.4. Gene Duplication and Synteny Analysis in Different Cotton Types
2.5. Calculation of Selection Pressure
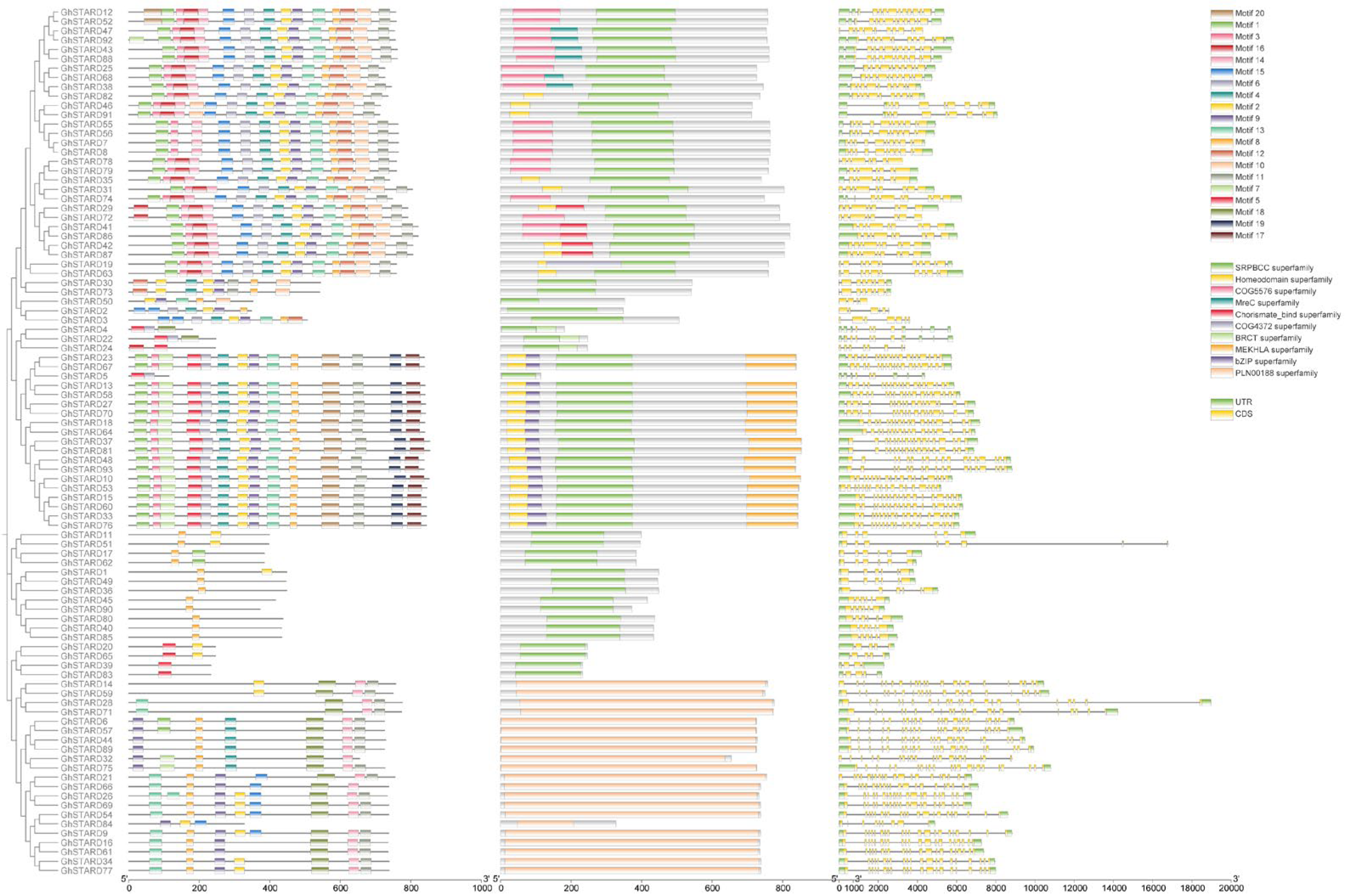
2.6. Analysis of the Conserved Protein Motifs and Gene Structure
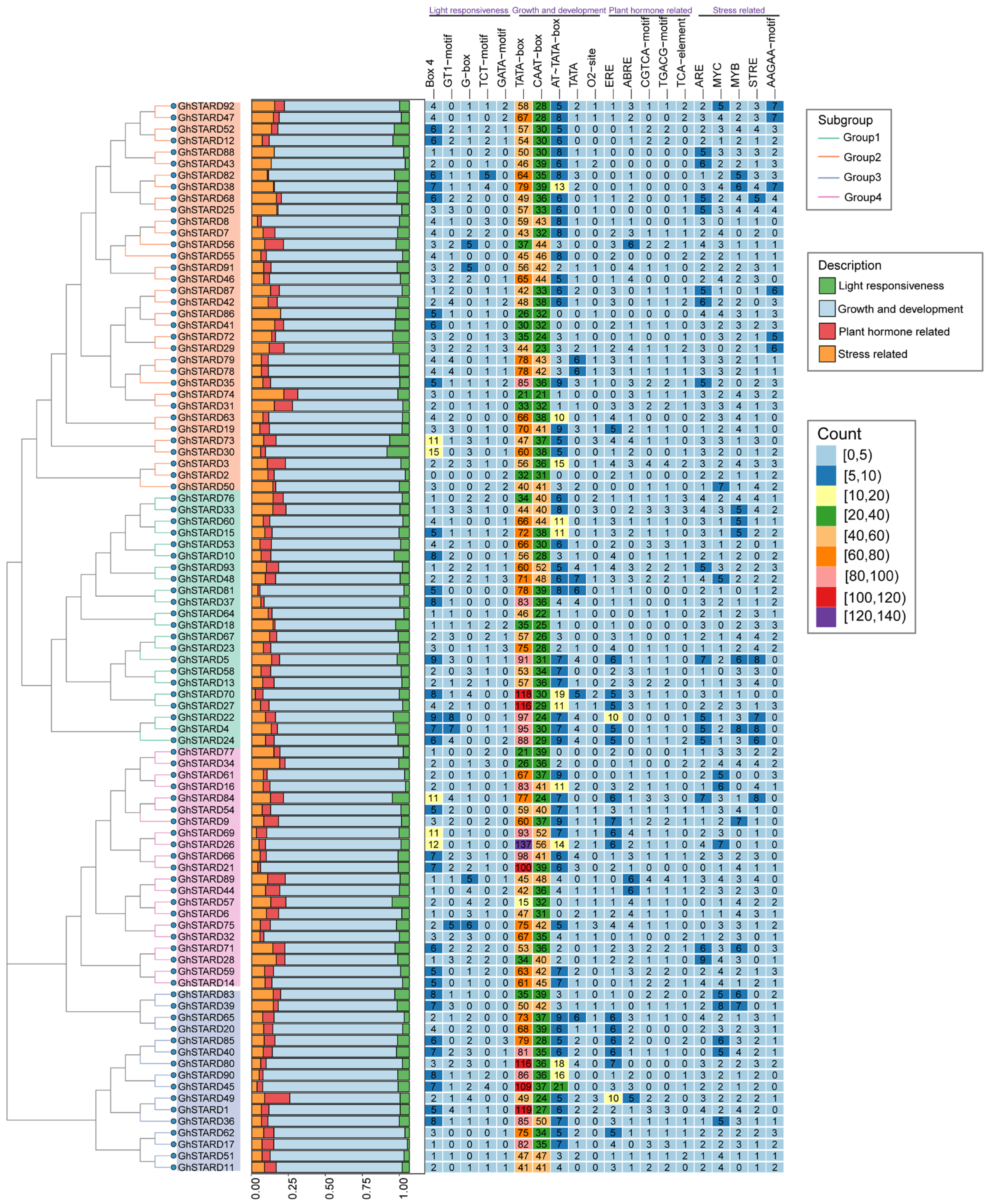
2.7. Analysis of STARDs Promoter Regions
2.8. Gene Expression Pattern Analysis
2.9. RNA Extraction and RT qPCR Analysis
2.10. Protein–Protein Interaction (PPI) Analysis
3. Results
3.1. Identification of the STARD Proteins in Cotton
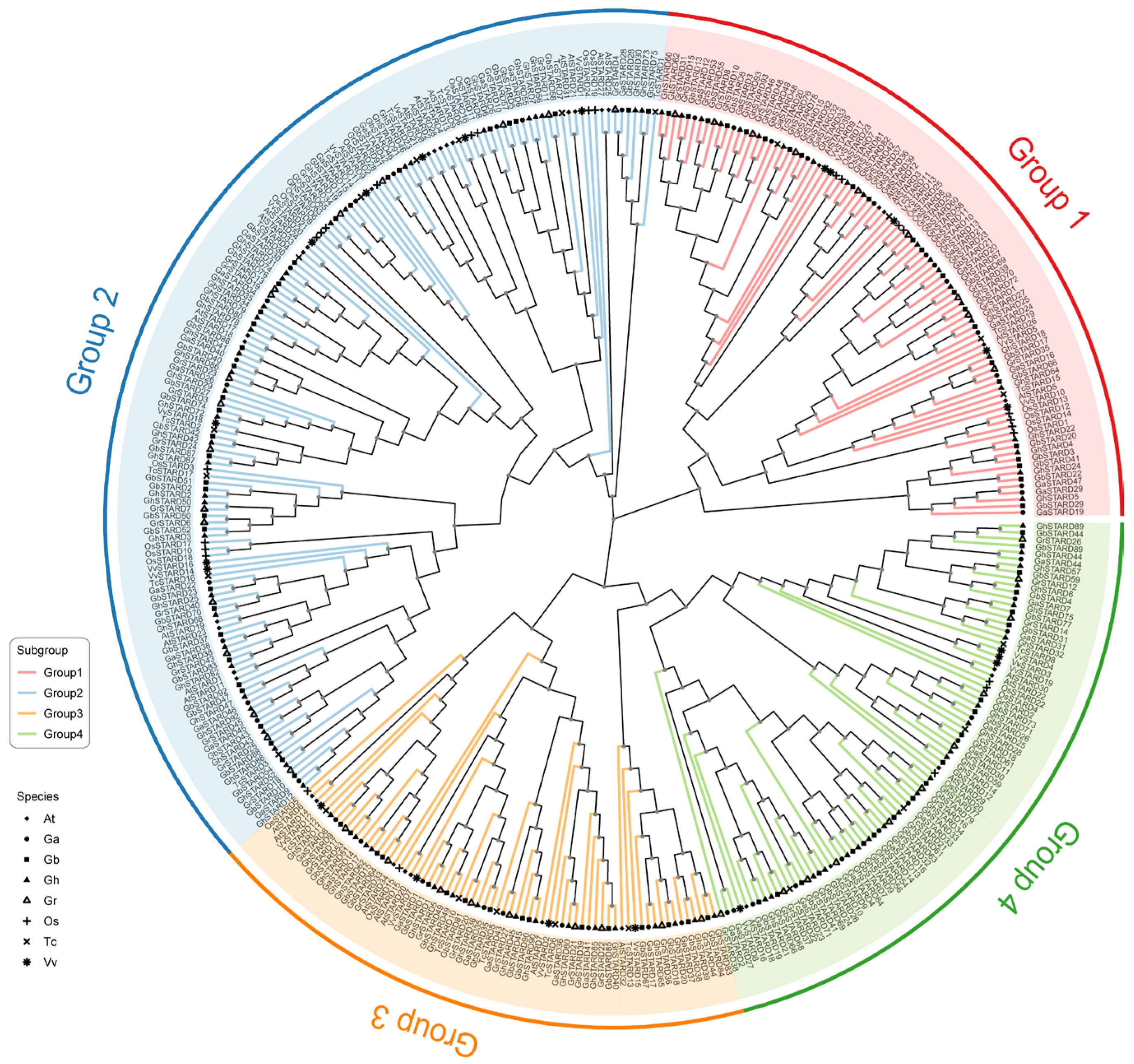
3.2. Phylogenetic Analysis of STARD Gene Family in Cotton
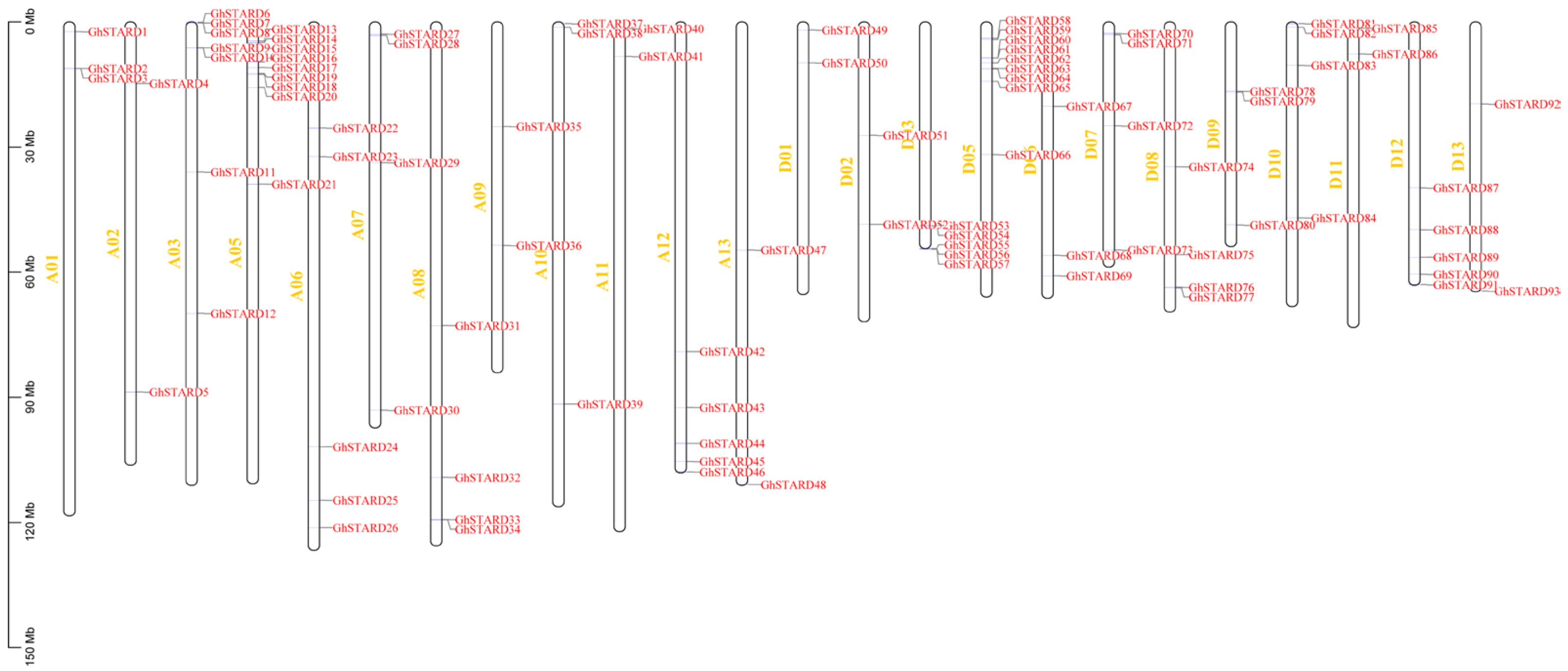
3.3. Chromosomal Localization of Cotton STARD Genes
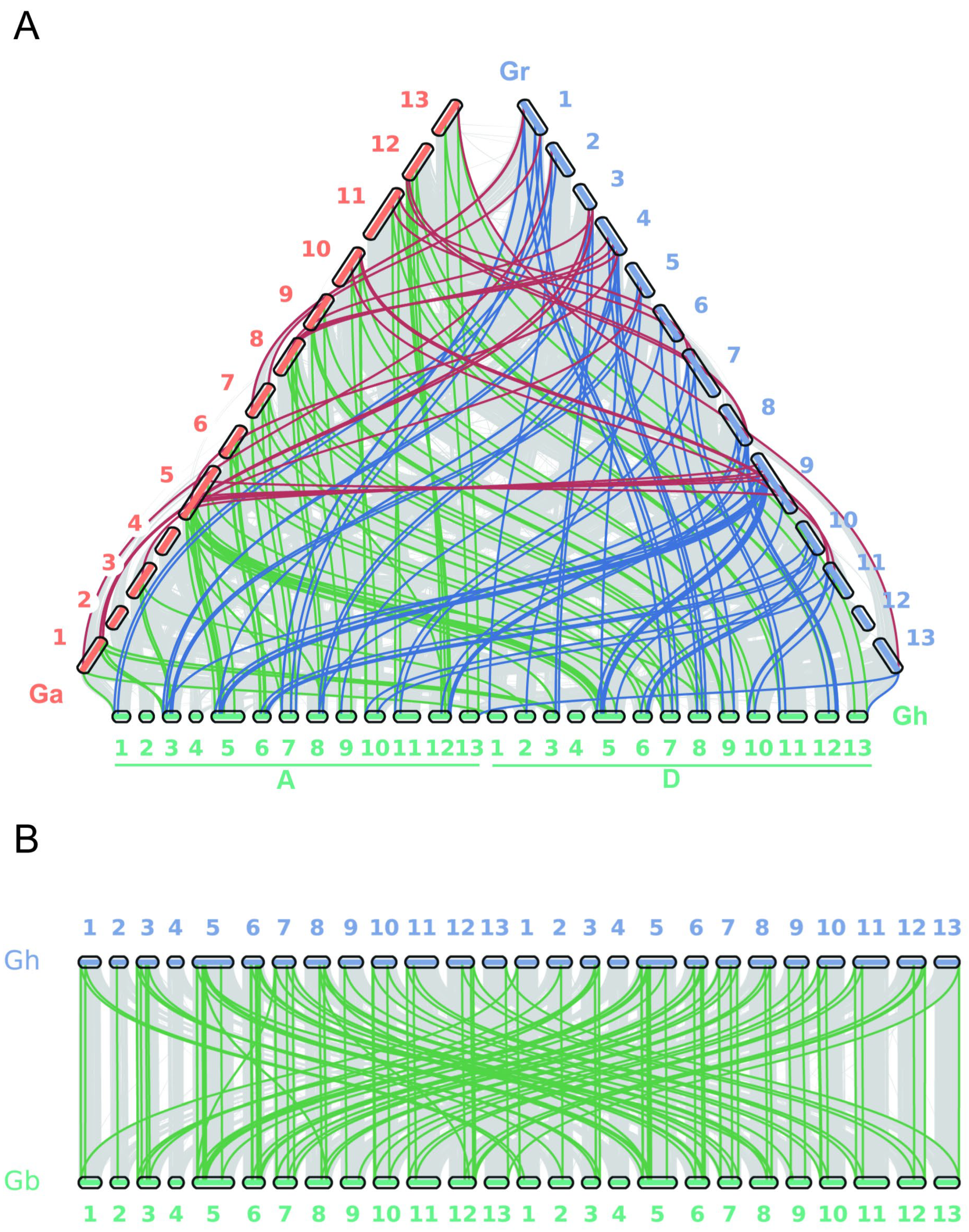
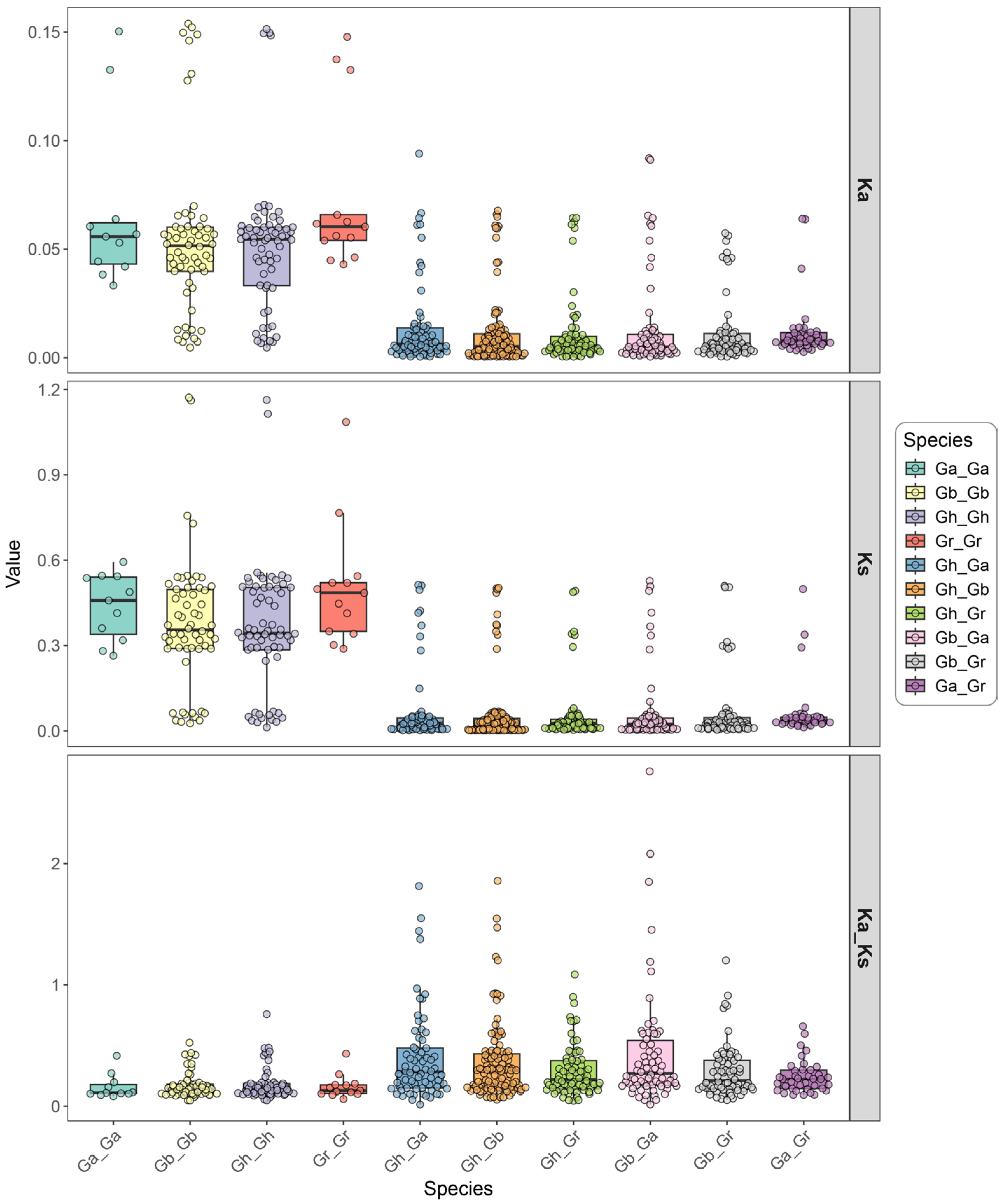
3.4. Collinearity Analysis and Selective Pressure Analysis
3.5. Structure Analysis of Cotton STARD Genes
3.6. Cis-Element Analysis of GhSTARD Gene Promotors
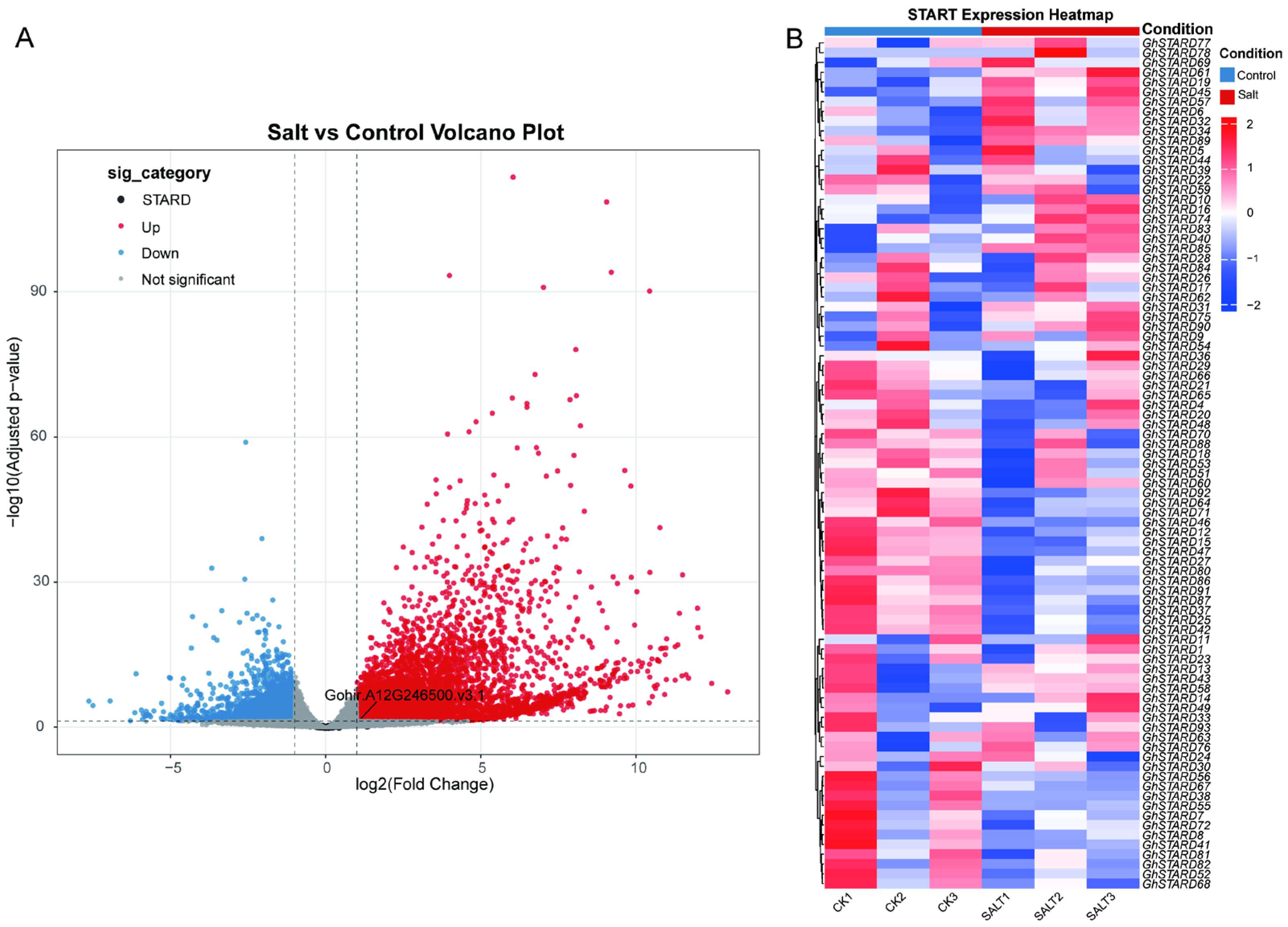
3.7. The Induced Expression of Cotton STARD Genes Under Salt Stress
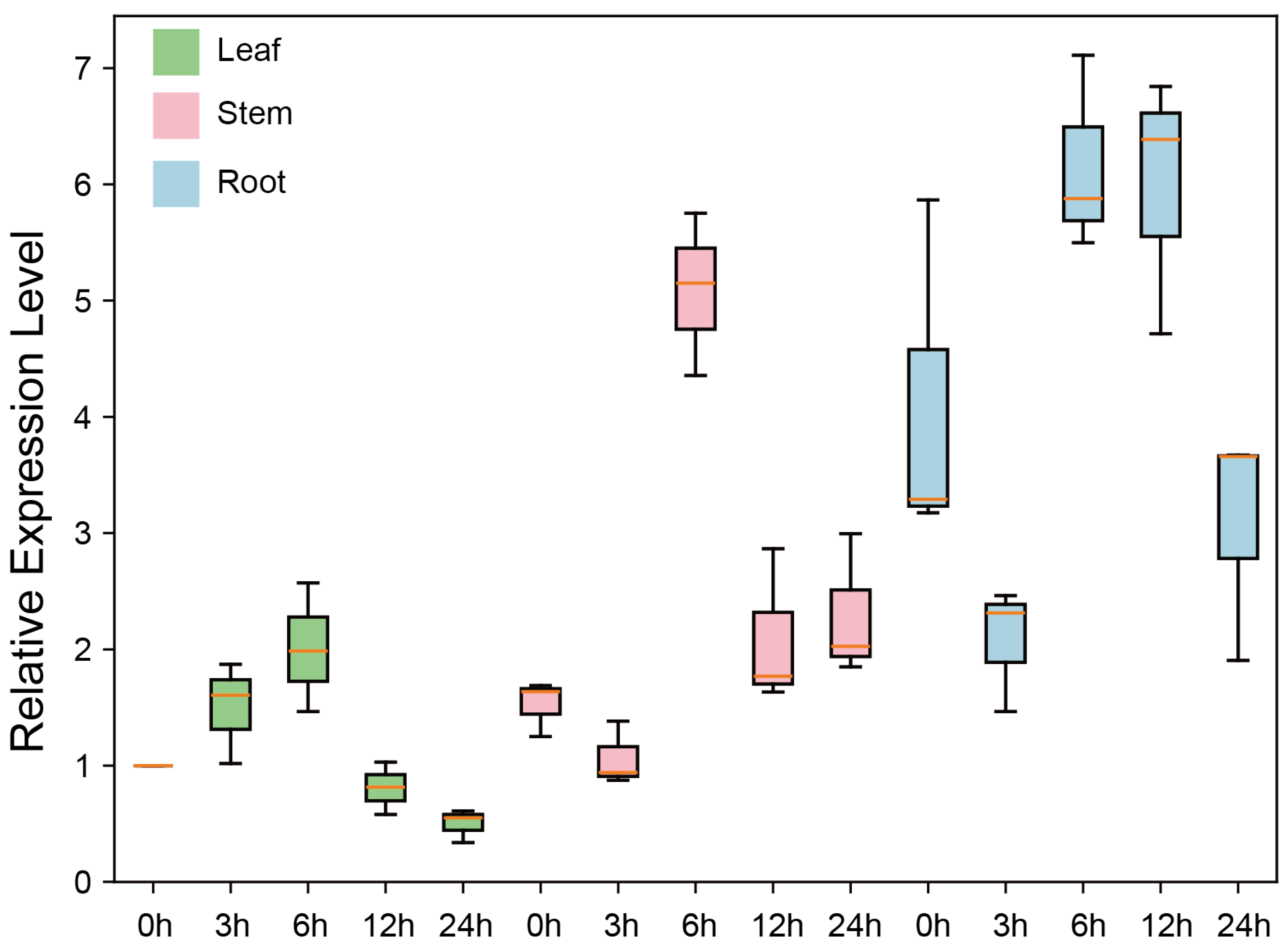
3.8. Tissue Expression Patterns of GhSTARD45
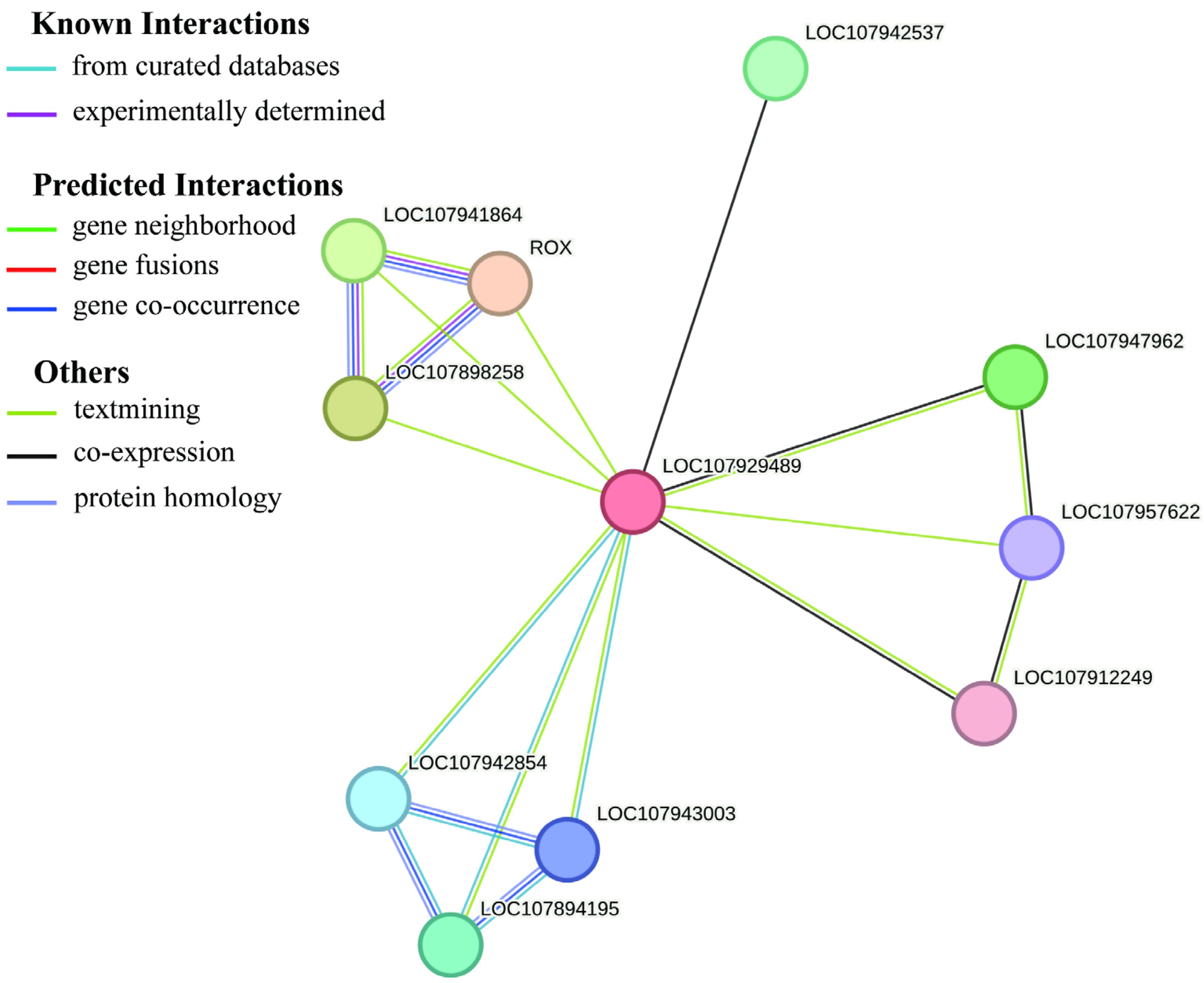
3.9. Gene Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roderick, S.L.; Chan, W.W.; Agate, D.S.; Olsen, L.R.; Vetting, M.W.; Rajashankar, K.R.; Cohen, D.E. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 2002, 9, 507–511. [Google Scholar] [CrossRef]
- Schrick, K.; Nguyen, D.; Karlowski, W.M.; Mayer, K.F.X. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004, 5, R41. [Google Scholar] [CrossRef]
- Clark, B.J. The mammalian START domain protein family in lipid transport in health and disease. J. Endocrinol. 2012, 212, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Tillman, M.C.; Imai, N.; Li, Y.; Khadka, M.; Okafor, C.D.; Juneja, P.; Adhiyaman, A.; Hagen, S.J.; Cohen, D.E.; Ortlund, E.A. Allosteric regulation of thioesterase superfamily member 1 by lipid sensor domain binding fatty acids and lysophosphatidylcholine. Proc. Natl. Acad. Sci. USA 2020, 117, 22080–22089. [Google Scholar] [CrossRef]
- Alpy, F.; Legueux, F.; Bianchetti, L.; Tomasetto, C. Les protéines à domaine START, des trafiquants intracellulaires de lipides. Médecine/Sci. 2009, 25, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.F.; Kishida, T.; Christenson, L.K.; Fujimoto, T.; Hiroi, H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol. Cell. Endocrinol. 2003, 202, 59–65. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Schrick, K.; Bruno, M.; Khosla, A.; Cox, P.N.; Marlatt, S.A.; Roque, R.A.; Nguyen, H.C.; He, C.; Snyder, M.P.; Singh, D.; et al. Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol. 2014, 12, 70. [Google Scholar] [CrossRef]
- Mukherjee, T.; Subedi, B.; Khosla, A.; Begler, E.M.; Stephens, P.M.; Warner, A.L.; Lerma-Reyes, R.; Thompson, K.A.; Gunewardena, S.; Schrick, K. The START domain mediates Arabidopsis GLABRA2 dimerization and turnover independently of homeodomain DNA binding. Plant Physiol. 2022, 190, 2315–2334. [Google Scholar] [CrossRef]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Wang, Z.; Wang, S.; Wang, Y.; Zhu, Q.; Li, S.; Xiang, C. Arabidopsis enhanced drought tolerance1/homeodomain glabrous11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013, 162, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-H.; Wu, S.-J.; Peng, Y.-S.; Liu, R.-N.; Chen, X.; Zhao, P.; Xu, P.; Zhu, J.-B.; Jiao, G.-L.; Pei, Y.; et al. Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 2016, 14, 72–84. [Google Scholar] [CrossRef]
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Chi, Y.; Lin, D.; Tyler, B.M.; Liu, X. Mutations in ORP1 conferring oxathiapiprolin resistance confirmed by genome editing using CRISPR/Cas9 in Phytophthora capsici and P sojae. Phytopathology 2018, 108, 1412–1419. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y. Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 2018, 46, 7–13. [Google Scholar] [CrossRef]
- Lai, S.-H.; Chye, M.-L. Plant Acyl-CoA-Binding Proteins—Their Lipid and Protein Interactors in Abiotic and Biotic Stresses. Cells 2021, 10, 1064. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Tian, T.; Feng, L.; Yang, X.; Li, L.; Tan, X.; Wu, W.; Li, Z.; Treves, H.; Serneels, F.; et al. Pathogenesis-related protein 1 suppresses oomycete pathogen by targeting against AMPK kinase complex. J. Adv. Res. 2023, 43, 13–26. [Google Scholar] [CrossRef]
- Kamata, M.; Yamashita, T.; Kina, A.; Funata, M.; Mizukami, A.; Sasaki, M.; Tani, A.; Funami, M.; Amano, N.; Fukatsu, K. Design, synthesis, and structure–activity relationships of novel spiro-piperidines as acetyl-CoA carboxylase inhibitors. Bioorganic Med. Chem. Lett. 2012, 22, 3643–3647. [Google Scholar] [CrossRef]
- Lu, S.; He, H.; Wang, P.; Gou, H.; Cao, X.; Ma, Z.; Chen, B.; Mao, J. Evolutionary relationship analysis of STARD gene family and VvSTARD5 improves tolerance of salt stress in transgenic tomatoes. Physiol. Plant. 2022, 174, e13772. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, S.; Chen, J.; Zhu, S.; Zhu, Q.; Zhao, T. Advances in cotton genomics, genetics and breeding. Plants 2024, 13, 2579. [Google Scholar] [CrossRef]
- Jareczek, J.J.; Grover, C.E.; Wendel, J.F. Cotton fiber as a model for understanding shifts in cell development under domestication. Front. Plant Sci. 2023, 14, 1146802. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, M.T.; Majeed, S.; Rana, I.A.; Ali, Z.; Jia, Y.; Du, X.; Hinze, L.; Azhar, M.T. Impact of salinity stress on cotton and opportunities for improvement through conventional and biotechnological approaches. BMC Plant Biol. 2024, 24, 20. [Google Scholar] [CrossRef] [PubMed]
- Sharif, I.; Aleem, S.; Farooq, J.; Rizwan, M.; Younas, A.; Sarwar, G.; Chohan, S.M. Salinity stress in cotton: Effects, mechanism of tolerance and its management strategies. Physiol. Mol. Biol. Plants 2019, 25, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Hu, G.; Yu, J.; Thu, S.W.; Grover, C.E.; Zhu, S.; Wendel, J.F. Salt-tolerance diversity in diploid and polyploid cotton (Gossypium) species. Plant J. 2020, 101, 1135–1151. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Stahlberg, A.; Hakansson, J.; Xian, X.; Semb, H.; Kubista, M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004, 50, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Di, Y.; Liu, Y.; Huang, G.; Zheng, Y.; Zhang, Y.; Fang, W. Development and application of a novel reverse transcription real-time PCR method for miR-499 quantification. Clin. Biochem. 2013, 46, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of appropriate reference genes for quantitative real-time PCR in oxytropis ochrocephala bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef] [PubMed]
- Thorsell, A.-G.; Lee, W.H.; Persson, C.; Siponen, M.I.; Nilsson, M.; Busam, R.D.; Kotenyova, T.; Schüler, H.; Lehtiö, L. Comparative structural analysis of lipid binding START domains. PLoS ONE 2011, 6, e19521. [Google Scholar] [CrossRef]
- Dong, Y.; Hu, G.; Grover, C.E.; Miller, E.R.; Zhu, S.; Wendel, J.F. Parental legacy versus regulatory innovation in salt stress responsiveness of allopolyploid cotton (Gossypium) species. Plant J. 2022, 111, 872–887. [Google Scholar] [CrossRef]
- Mahtha, S.K.; Purama, R.K.; Yadav, G. StAR-related lipid transfer (START) domains across the rice pangenome reveal how ontogeny recapitulated selection pressures during rice domestication. Front. Genet. 2021, 12, 737194. [Google Scholar] [CrossRef]
- Chew, W.; Hrmova, M.; Lopato, S. Role of homeodomain leucine zipper (HD-Zip) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 2013, 14, 8122–8147. [Google Scholar] [CrossRef]
- Roodbarkelari, F.; Groot, E.P. Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol. 2017, 213, 95–104. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple links between HD-Zip proteins and hormone networks. Int. J. Mol. Sci. 2018, 19, 4047. [Google Scholar] [CrossRef]
- Satheesh, V.; Parameswaran, C.; Tejkumar, J.P.; Vajinder, K.; Jain, P.K.; Viswanathan, C.; Shripad, R.B.; Srinivasan, R. Transmembrane START domain proteins: In silico identification, characterization and expression analysis under stress conditions in chickpea (Cicer arietinum L.). Plant Signal. Behav. 2016, 11, e992698. [Google Scholar] [CrossRef] [PubMed]
- Luhua, S.; Hegie, A.; Suzuki, N.; Shulaev, E.; Luo, X.; Cenariu, D.; Ma, V.; Kao, S.; Lim, J.; Gunay, M.B.; et al. Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant. 2013, 148, 322–333. [Google Scholar] [CrossRef]
- Feng, L.; Chen, Y.; Ma, T.; Zhou, C.; Sang, S.; Li, J.; Ji, S. Integrative physiology and transcriptome sequencing reveal differences between G. hirsutum and G. barbadense in response to salt stress and the identification of key salt tolerance genes. BMC Plant Biol. 2024, 24, 787. [Google Scholar] [CrossRef] [PubMed]
- Abdullah-Zawawi, M.-R.; Ahmad-Nizammuddin, N.-F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.-A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, L.; Lan, Y.; Li, X.; Wang, J.; Dong, J.; Guo, W.; Jing, D.; Liu, Q.; Zhang, S.; et al. An aspartic protease 47 causes quantitative recessive resistance to rice black-streaked dwarf virus disease and southern rice black-streaked dwarf virus disease. New Phytol. 2022, 233, 2520–2533. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, J.; Zeng, X.; Guo, H.; Luo, Y.; Kear, P.; Zhang, S.; Zhu, G. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Hortic. Plant J. 2021, 7, 129–141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, R.; Sun, J.; Li, S.; Cui, Y.; Rui, C.; Sun, M.; Ye, W. Cotton STARD Gene Family: Characterization, Evolution, and Expression Profiles During Salt Stress. Genes 2025, 16, 813. https://doi.org/10.3390/genes16070813
Cui R, Sun J, Li S, Cui Y, Rui C, Sun M, Ye W. Cotton STARD Gene Family: Characterization, Evolution, and Expression Profiles During Salt Stress. Genes. 2025; 16(7):813. https://doi.org/10.3390/genes16070813
Chicago/Turabian StyleCui, Ruifeng, Jiuguang Sun, Shuyan Li, Yupeng Cui, Cun Rui, Minshan Sun, and Wuwei Ye. 2025. "Cotton STARD Gene Family: Characterization, Evolution, and Expression Profiles During Salt Stress" Genes 16, no. 7: 813. https://doi.org/10.3390/genes16070813
APA StyleCui, R., Sun, J., Li, S., Cui, Y., Rui, C., Sun, M., & Ye, W. (2025). Cotton STARD Gene Family: Characterization, Evolution, and Expression Profiles During Salt Stress. Genes, 16(7), 813. https://doi.org/10.3390/genes16070813






