Genome-Wide Identification and Expression Analysis of PP2C Gene Family in Eelgrass
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Characteristics of PP2C Family Members in Eelgrass
2.2. Phylogenetic and Conserved Motif Analysis
2.3. Chromosomal Localization and Synteny Analysis
2.4. Expression Analysis of PP2C Members
2.5. qRT-PCR Experiment
2.6. Cis-Acting Regulatory Elements Analysis
2.7. Prediction of PYL-PP2C-SnRK2 Module in Eelgrass
3. Results
3.1. Genome-Wide Identification of Eelgrass PP2Cs
3.2. Phylogenetic Analysis and Classification of Eelgrass PP2Cs
3.3. The Gene Structure of Eelgrass PP2Cs
3.4. Protein Domain and Conserved Motifs of Eelgrass PP2Cs
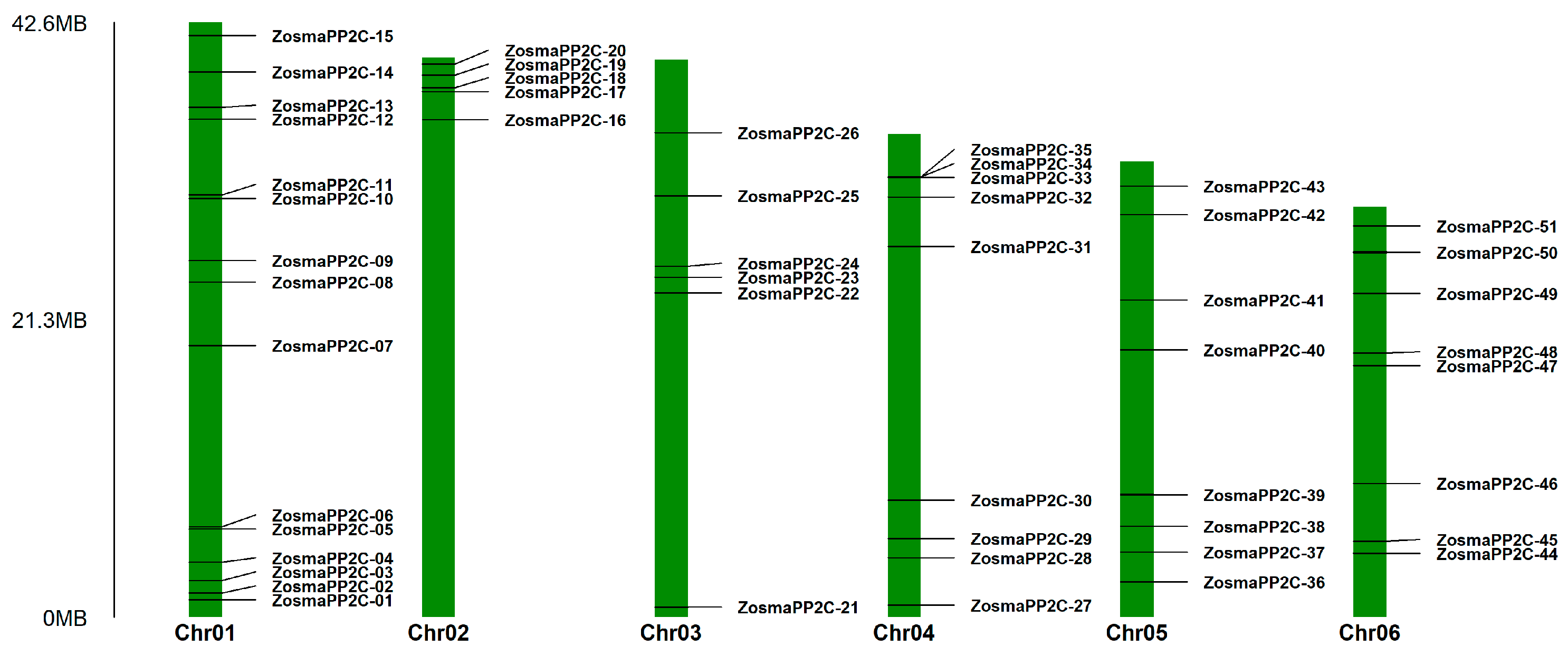
3.5. Chromosomal Localization and Synteny Analysis of Eelgrass PP2Cs
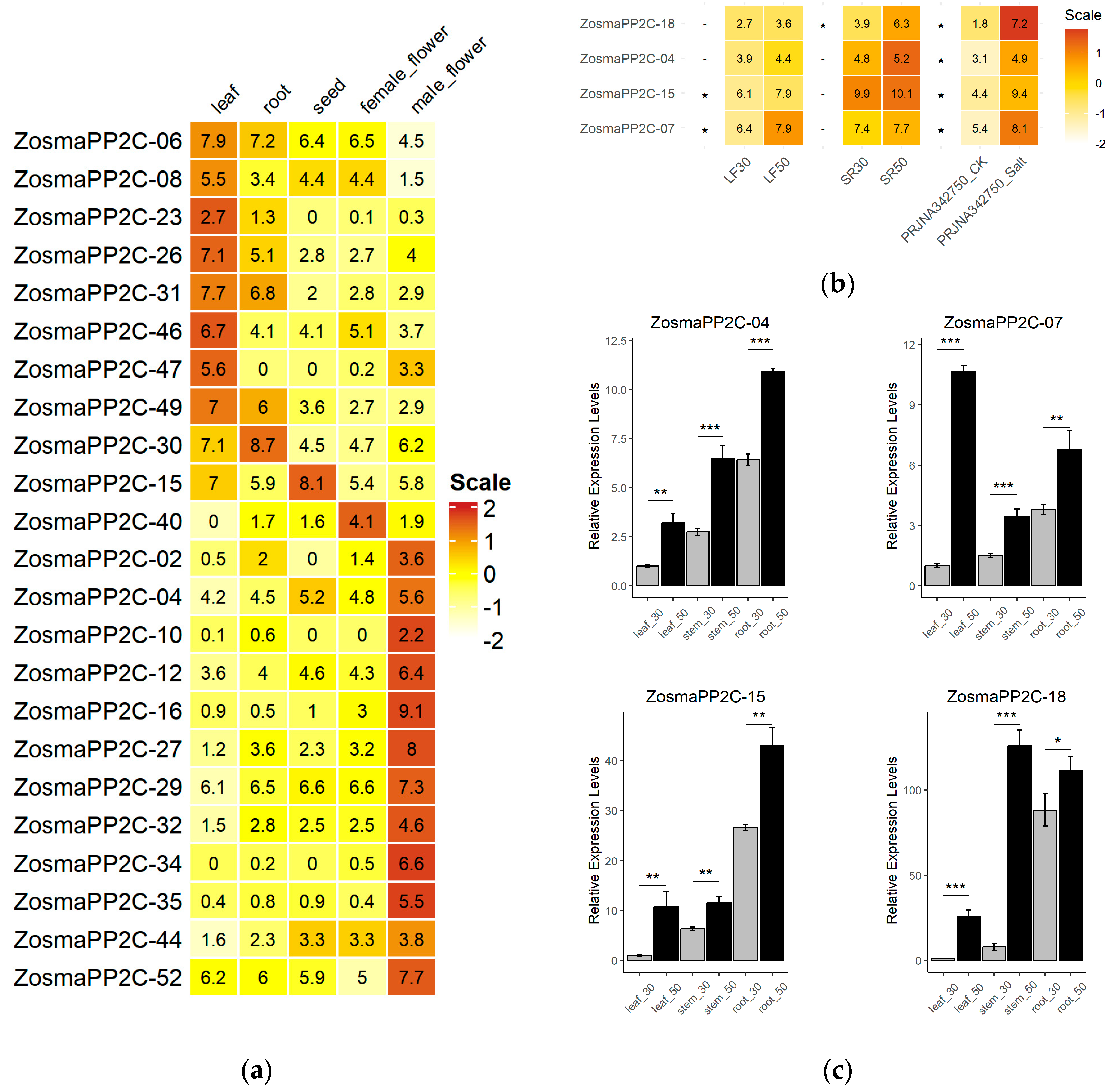
3.6. The Expression of Eelgrass PP2Cs in Different Tissues and in Response to Salt Stress
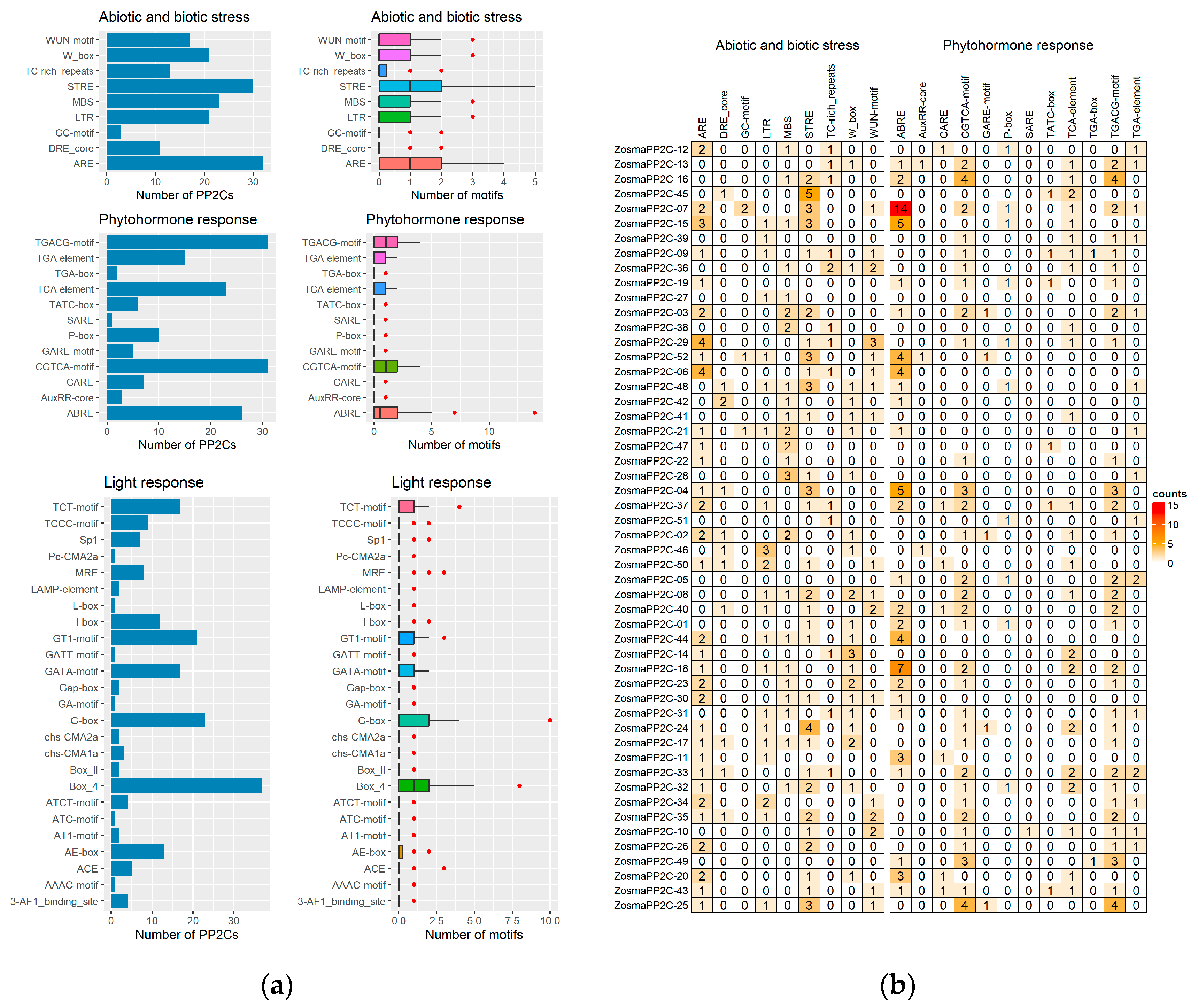
3.7. Cis-Acting Regulatory Element of Eelgrass PP2Cs on Promoter Region
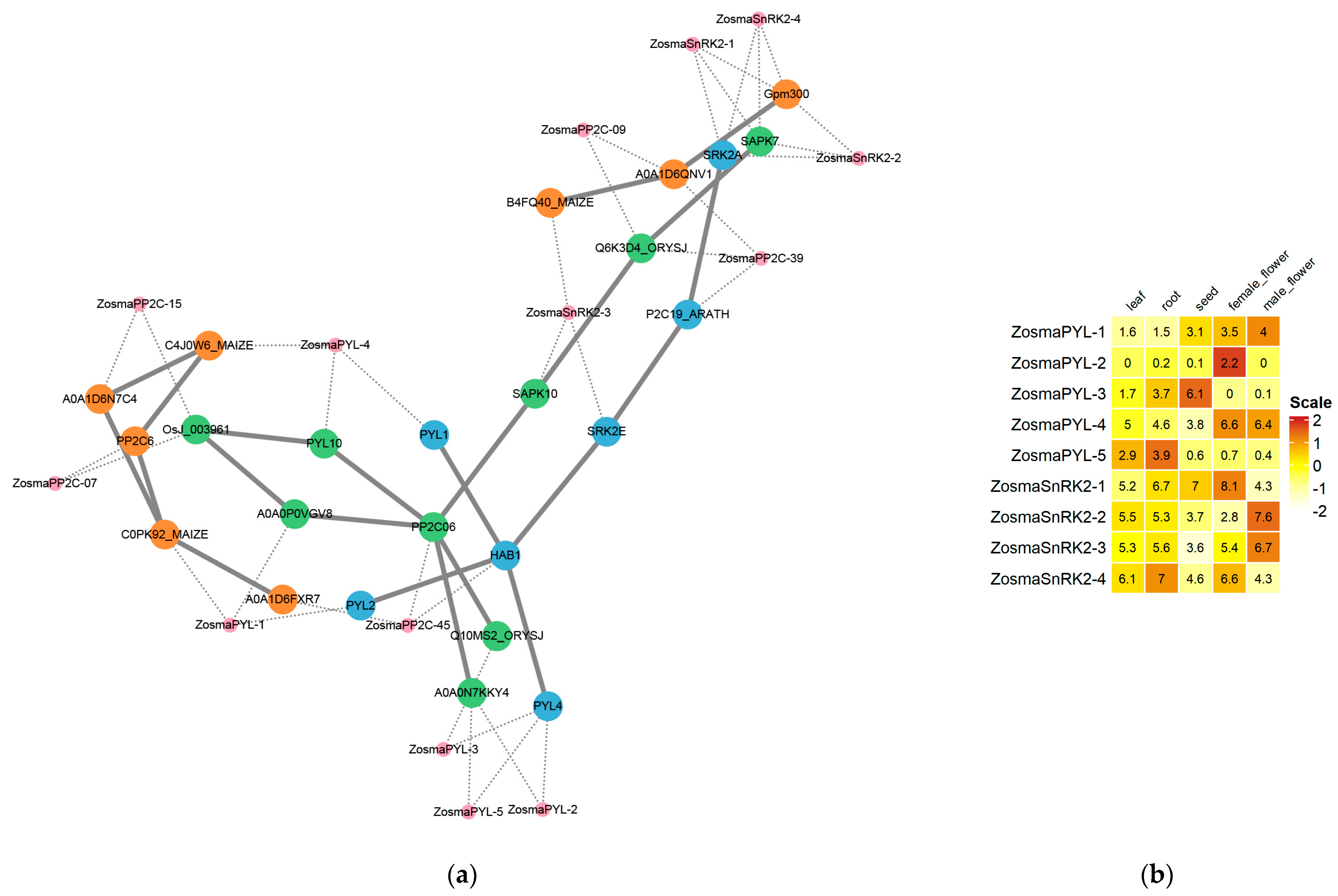
3.8. Prediction of PP2C-Interacting Proteins in Eelgrass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PP2C | Protein Phosphatase 2C |
| ABA | abscisic acid |
| ABRE | Abscisic Acid Response Element |
| PYL | Pyrabactin resistance 1-like |
| SnRK2 | SNF1-related protein kinase 2 |
| nucl | nucleus |
| cyto | cytoplasm |
| chlo | chloroplast |
| mito | mitochondria |
| cysk | cytoskeleton |
| extr | extracellular |
References
- Das, A.K.; Helps, N.R.; Cohen, P.T.; Barford, D. Crystal Structure of the Protein Serine/Threonine Phosphatase 2C at 2.0 A Resolution. EMBO J. 1996, 15, 6798–6809. [Google Scholar] [CrossRef] [PubMed]
- Chuong, N.N.; Hoang, X.L.T.; Nghia, D.H.T.; Dai, T.N.T.; Thi, V.-A.L.; Thao, N.P. Protein Phosphatase Type 2C Functions in Phytohormone-Dependent Pathways and in Plant Responses to Abiotic Stresses. Curr. Protein Pept. Sci. 2021, 22, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Mo, W.; Zheng, X.; Shi, Q.; Zhao, X.; Chen, X.; Yang, Z.; Zuo, Z. Unveiling the Crucial Roles of Abscisic Acid in Plant Physiology: Implications for Enhancing Stress Tolerance and Productivity. Front. Plant Sci. 2024, 15, 1437184. [Google Scholar] [CrossRef]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 Protein Phosphatases 2C Act in a Negative Feedback Regulatory Loop of the Abscisic Acid Signalling Pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C Protein Phosphatases Directly Regulate Abscisic Acid-Activated Protein Kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Miao, J.; Bu, L.; Tan, W.; Wang, P.; Li, X.; Li, X.; Chen, C.; Zhang, K.; Shen, W.; Gong, Z.; et al. OsPP2C49, a Negative Regulatory Factor in the Abscisic Acid Signaling Pathway, Positively Regulates Grain Yield in Rice. Rice 2024, 17, 65. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.-W.; Lee, Y.-N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.-D.; Lee, S.; Lee, S.C.; Kim, B.-G. A Rice Orthologue of the ABA Receptor, OsPYL/RCAR5, Is a Positive Regulator of the ABA Signal Transduction Pathway in Seed Germination and Early Seedling Growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Gao, S.; Qin, F.; Dai, M. Deletion of an Endoplasmic Reticulum Stress Response Element in a ZmPP2C-A Gene Facilitates Drought Tolerance of Maize Seedlings. Mol. Plant 2017, 10, 456–469. [Google Scholar] [CrossRef]
- Ni, L.; Fu, X.; Zhang, H.; Li, X.; Cai, X.; Zhang, P.; Liu, L.; Wang, Q.; Sun, M.; Wang, Q.-W.; et al. Abscisic Acid Inhibits Rice Protein Phosphatase PP45 via H2O2 and Relieves Repression of the Ca2+/CaM-Dependent Protein Kinase DMI3. Plant Cell 2019, 31, 128–152. [Google Scholar] [CrossRef]
- Guo, Y.; Shi, Y.; Wang, Y.; Liu, F.; Li, Z.; Qi, J.; Wang, Y.; Zhang, J.; Yang, S.; Wang, Y.; et al. The Clade F PP2C Phosphatase ZmPP84 Negatively Regulates Drought Tolerance by Repressing Stomatal Closure in Maize. New Phytol. 2023, 237, 1728–1744. [Google Scholar] [CrossRef]
- Lu, F.; Li, W.; Peng, Y.; Cao, Y.; Qu, J.; Sun, F.; Yang, Q.; Lu, Y.; Zhang, X.; Zheng, L.; et al. ZmPP2C26 Alternative Splicing Variants Negatively Regulate Drought Tolerance in Maize. Front. Plant Sci. 2022, 13, 851531. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Jeong, S.; Baek, W.; Choi, H.; Lee, S.C. A Positive Role for CaMEKK17 in Response to Drought Stress, Modulated by Clade A PP2Cs. Plant Cell Environ. 2025, 48, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sun, J. SlbHLH1 Mediates ABA Treatment-Retarded Chilling Injury by Repressing SlPP2C29 in Tomato Fruit. Plant Sci. 2024, 344, 112086. [Google Scholar] [CrossRef]
- Urrea Castellanos, R.; Friedrich, T.; Petrovic, N.; Altmann, S.; Brzezinka, K.; Gorka, M.; Graf, A.; Bäurle, I. FORGETTER2 Protein Phosphatase and Phospholipase D Modulate Heat Stress Memory in Arabidopsis. Plant J. 2020, 104, 7–17. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.-B.; Wei, N.; Liu, Z.-H.; Li, Y.; Zheng, Y.; Li, X.-B. A Type-2C Protein Phosphatase (GhDRP1) Participates in Cotton (Gossypium hirsutum) Response to Drought Stress. Plant Mol. Biol. 2021, 107, 499–517. [Google Scholar] [CrossRef]
- Baek, W.; Lim, C.W.; Lee, S.C. Functional Analysis of the Pepper Protein Phosphatase, CaAIPP1, and Its Interacting Partner CaAIRF1: Modulation of ABA Signalling and the Drought Stress Response. Plant Cell Environ. 2017, 40, 2359–2368. [Google Scholar] [CrossRef]
- Jeong, S.; Lim, C.W.; Lee, S.C. CaADIP1-Dependent CaADIK1-Kinase Activation Is Required for Abscisic Acid Signalling and Drought Stress Response in Capsicum annuum. New Phytol. 2021, 231, 2247–2261. [Google Scholar] [CrossRef]
- Liu, Q.; Ding, J.; Huang, W.; Yu, H.; Wu, S.; Li, W.; Mao, X.; Chen, W.; Xing, J.; Li, C.; et al. OsPP65 Negatively Regulates Osmotic and Salt Stress Responses Through Regulating Phytohormone and Raffinose Family Oligosaccharide Metabolic Pathways in Rice. Rice 2022, 15, 34. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.-I.; Kwon, H.; Cho, M.H.; Kim, B.-G.; Chung, J.H.; Nam, M.H.; Song, J.S.; Kim, K.-H.; Yoon, I.S. The Rice Abscisic Acid-Responsive RING Finger E3 Ligase OsRF1 Targets OsPP2C09 for Degradation and Confers Drought and Salinity Tolerance in Rice. Front. Plant Sci. 2021, 12, 797940. [Google Scholar] [CrossRef]
- Xing, B.; Gu, C.; Zhang, T.; Zhang, Q.; Yu, Q.; Jiang, J.; Liu, G. Functional Study of BpPP2C1 Revealed Its Role in Salt Stress in Betula platyphylla. Front. Plant Sci. 2020, 11, 617635. [Google Scholar] [CrossRef]
- Chu, M.; Chen, P.; Meng, S.; Xu, P.; Lan, W. The Arabidopsis Phosphatase PP2C49 Negatively Regulates Salt Tolerance through Inhibition of AtHKT1;1. J. Integr. Plant Biol. 2021, 63, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wei, Q.; Shi, J.; Jin, X.; He, Y.; Zhang, Y.; Luo, Q.; Wang, Y.; Chang, J.; Yang, G.; et al. Brachypodium distachyon BdPP2CA6 Interacts with BdPYLs and BdSnRK2 and Positively Regulates Salt Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Krzywińska, E.; Kulik, A.; Bucholc, M.; Fernandez, M.A.; Rodriguez, P.L.; Dobrowolska, G. Protein Phosphatase Type 2C PP2CA Together with ABI1 Inhibits SnRK2.4 Activity and Regulates Plant Responses to Salinity. Plant Signal Behav. 2016, 11, e1253647. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Olsen, J.L.; Reusch, T.B.H.; Procaccini, G.; Kudrna, D.; Williams, M.; Grimwood, J.; Rajasekar, S.; Jenkins, J.; Schmutz, J.; et al. Improved Chromosome-Level Genome Assembly and Annotation of the Seagrass, Zostera marina (Eelgrass). F1000Res 2021, 10, 289. [Google Scholar] [CrossRef]
- Lamb, J.B.; van de Water, J.A.J.M.; Bourne, D.G.; Altier, C.; Hein, M.Y.; Fiorenza, E.A.; Abu, N.; Jompa, J.; Harvell, C.D. Seagrass Ecosystems Reduce Exposure to Bacterial Pathogens of Humans, Fishes, and Invertebrates. Science 2017, 355, 731–733. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Li, J.-D.; Yan, W.-J.; Luo, F.-S.; Wang, L.; Zuo, L.-M.; Xu, J.-G.; Li, W.; Peidong, Z. The Combined Effect of Seawater Salinity and Duration on the Survival and Growth of Eelgrass Zostera Marina. Aquat. Bot. 2023, 187, 103652. [Google Scholar] [CrossRef]
- Cullen-Unsworth, L.C.; Unsworth, R. A Call for Seagrass Protection. Science 2018, 361, 446–448. [Google Scholar] [CrossRef]
- York, P.H.; Gruber, R.K.; Hill, R.; Ralph, P.J.; Booth, D.J.; Macreadie, P.I. Physiological and Morphological Responses of the Temperate Seagrass Zostera muelleri to Multiple Stressors: Investigating the Interactive Effects of Light and Temperature. PLoS ONE 2013, 8, e76377. [Google Scholar] [CrossRef]
- Maxwell, P.S.; Pitt, K.A.; Burfeind, D.D.; Olds, A.D.; Babcock, R.C.; Connolly, R.M. Phenotypic Plasticity Promotes Persistence Following Severe Events: Physiological and Morphological Responses of Seagrass to Flooding. J. Ecol. 2014, 102, 54–64. [Google Scholar] [CrossRef]
- Ontoria, Y.; Webster, C.; Said, N.; Ruiz, J.M.; Pérez, M.; Romero, J.; McMahon, K. Positive Effects of High Salinity Can Buffer the Negative Effects of Experimental Warming on Functional Traits of the Seagrass Halophila Ovalis. Mar. Pollut. Bull. 2020, 158, 111404. [Google Scholar] [CrossRef]
- Zhao, H.; Dong, X.; Yang, D.; Ge, Q.; Lu, P.; Liu, C. New Insights into the Salt-Responsive Regulation in Eelgrass at Transcriptional and Post-Transcriptional Levels. Front. Plant Sci. 2025, 16, 1497064. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Toriyama, K. DCW11, down-Regulated Gene 11 in CW-Type Cytoplasmic Male Sterile Rice, Encoding Mitochondrial Protein Phosphatase 2c Is Related to Cytoplasmic Male Sterility. Plant Cell Physiol. 2008, 49, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 Can Positively Regulate Leaf Senescence by Suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2020, 61, 644–658. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, Z.; Liu, W.; Qi, M.; Xu, T.; Wu, Y.; Li, T. Silencing SlPP2C Expression Delayed Plant Senescence and Fruit Ripening in Tomato. Physiol. Plant 2023, 175, e13925. [Google Scholar] [CrossRef]
- Liang, B.; Sun, Y.; Wang, J.; Zheng, Y.; Zhang, W.; Xu, Y.; Li, Q.; Leng, P. Tomato Protein Phosphatase 2C Influences the Onset of Fruit Ripening and Fruit Glossiness. J. Exp. Bot. 2021, 72, 2403–2418. [Google Scholar] [CrossRef]
- Yu, X.; Han, J.; Li, L.; Zhang, Q.; Yang, G.; He, G. Wheat PP2C-A10 Regulates Seed Germination and Drought Tolerance in Transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 635–651. [Google Scholar] [CrossRef]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of Seed Dormancy and Germination by DOG1-AHG1 PP2C Phosphatase Complex via Binding to Heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 Requires PP2C Phosphatases of the ABA Signalling Pathway to Control Seed Dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J.J. Reduced Dormancy5 Encodes a Protein Phosphatase 2C That Is Required for Seed Dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R.; Chen, H.; Deng, X.W.; Wei, N. SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis. Plant Cell 2020, 32, 3792–3811. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-Wide and Expression Analysis of Protein Phosphatase 2C in Rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, Y.; Zhai, H.; Cai, H.; Ji, W.; Luo, X.; Li, J.; Bai, X. AtPP2CG1, a Protein Phosphatase 2C, Positively Regulates Salt Tolerance of Arabidopsis in Abscisic Acid-Dependent Manner. Biochem. Biophys. Res. Commun. 2012, 422, 710–715. [Google Scholar] [CrossRef]
- Arefian, M.; Antil, N.; Najar, M.A.; Behera, S.K.; Subba, P.; Prasad, T.S.K. Identifying Novel Genes and Proteins Involved in Salt Stress Perception and Signaling of Rice Seedlings. OMICS 2022, 26, 151–164. [Google Scholar] [CrossRef]
- Yu, X.; Han, J.; Wang, E.; Xiao, J.; Hu, R.; Yang, G.; He, G. Genome-Wide Identification and Homoeologous Expression Analysis of PP2C Genes in Wheat (Triticum aestivum L.). Front. Genet. 2019, 10, 561. [Google Scholar] [CrossRef]
- Shazadee, H.; Khan, N.; Wang, J.; Wang, C.; Zeng, J.; Huang, Z.; Wang, X. Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L. Int. J. Mol. Sci. 2019, 20, 1395. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique Drought Resistance Functions of the Highly ABA-Induced Clade A Protein Phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef]
- Guo, X.-H.; Deng, K.-Q.; Wang, J.; Yu, D.-S.; Zhao, Q.; Liu, X.-M. Mutational Analysis of Arabidopsis PP2CA2 Involved in Abscisic Acid Signal Transduction. Mol. Biol. Rep. 2010, 37, 763–769. [Google Scholar] [CrossRef]
- Brock, A.K.; Willmann, R.; Kolb, D.; Grefen, L.; Lajunen, H.M.; Bethke, G.; Lee, J.; Nürnberger, T.; Gust, A.A. The Arabidopsis Mitogen-Activated Protein Kinase Phosphatase PP2C5 Affects Seed Germination, Stomatal Aperture, and Abscisic Acid-Inducible Gene Expression. Plant Physiol. 2010, 153, 1098–1111. [Google Scholar] [CrossRef]
- Zha, D.; He, Y.; Song, J. Regulatory Role of ABA-Responsive Element Binding Factors in Plant Abiotic Stress Response. Physiol. Plant 2025, 177, e70233. [Google Scholar] [CrossRef]
- Wang, L.; Ju, C.; Han, C.; Yu, Z.; Bai, M.-Y.; Wang, C. The Interaction of Nutrient Uptake with Biotic and Abiotic Stresses in Plants. J. Integr. Plant Biol. 2025, 67, 455–487. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Ho, T.H. Functional Dissection of an Abscisic Acid (ABA)-Inducible Gene Reveals Two Independent ABA-Responsive Complexes Each Containing a G-Box and a Novel Cis-Acting Element. Plant Cell 1995, 7, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Porras, J.L.; Riaño-Pachón, D.M.; Dreyer, I.; Mayer, J.E.; Mueller-Roeber, B. Genome-Wide Analysis of ABA-Responsive Elements ABRE and CE3 Reveals Divergent Patterns in Arabidopsis and Rice. BMC Genom. 2007, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Nguyen, N.H.; Cheong, J.-J. Transcriptional Regulation of Protein Phosphatase 2C Genes to Modulate Abscisic Acid Signaling. Int. J. Mol. Sci. 2020, 21, 9517. [Google Scholar] [CrossRef]
- Waheed, A.; Zhuo, L.; Wang, M.; Hailiang, X.; Tong, Z.; Wang, C.; Aili, A. Integrative Mechanisms of Plant Salt Tolerance: Biological Pathways, Phytohormonal Regulation, and Technological Innovations. Plant Stress. 2024, 14, 100652. [Google Scholar] [CrossRef]



| Gene_ID | Gene_Name | Genomic Position | Exon Number | Amino Acid | Mw | pI | Hydropathicity | Location |
|---|---|---|---|---|---|---|---|---|
| Zosma02g21130 | ZosmaPP2C-17 | Chr02:37645131-37646558(+) | 1 | 476 | 52,968.61 | 5.22 | −0.345 | nucl |
| Zosma02g23620 | ZosmaPP2C-19 | Chr02:38842882-38844276(+) | 4 | 338 | 37,679.66 | 6.21 | −0.367 | nucl |
| Zosma02g18050 | ZosmaPP2C-16 | Chr02:35640155-35642265(+) | 9 | 402 | 43,424.27 | 4.74 | −0.211 | cyto |
| Zosma02g25420 | ZosmaPP2C-20 | Chr02:39635934-39637992(+) | 4 | 378 | 41,661.17 | 6.25 | −0.273 | cyto |
| Zosma02g21670 | ZosmaPP2C-18 | Chr02:37935610-37936910(+) | 2 | 388 | 42,198.18 | 5.63 | −0.339 | chlo |
| Zosma01g02300 | ZosmaPP2C-02 | Chr01:1723172-1724206(+) | 1 | 345 | 38,260.02 | 5.07 | −0.365 | nucl |
| Zosma01g03800 | ZosmaPP2C-03 | Chr01:2596913-2597182(−) | 1 | 90 | 9874.10 | 4.56 | 0.046 | cyto |
| Zosma01g18970 | ZosmaPP2C-08 | Chr01:24010873-24012921(−) | 10 | 323 | 34,955.8 | 5.65 | −0.116 | chlo |
| Zosma01g36660 | ZosmaPP2C-14 | Chr01:39054616-39056106(−) | 3 | 374 | 40,638.99 | 5.98 | −0.317 | chlo |
| Zosma01g25210 | ZosmaPP2C-11 | Chr01:30232032-30234173(−) | 3 | 657 | 72,553.42 | 5.6 | −0.432 | nucl |
| Zosma01g33240 | ZosmaPP2C-13 | Chr01:36512517-36515120(+) | 7 | 443 | 47,269.78 | 5.86 | −0.185 | nucl |
| Zosma01g06030 | ZosmaPP2C-04 | Chr01:3891751-3894444(−) | 4 | 401 | 43,330.71 | 4.72 | −0.154 | chlo |
| Zosma01g16060 | ZosmaPP2C-07 | Chr01:19447211-19448485(−) | 2 | 398 | 43,151.96 | 5.6 | −0.27 | nucl |
| Zosma01g41250 | ZosmaPP2C-15 | Chr01:41671572-41673020(+) | 4 | 402 | 43,831.35 | 7.2 | −0.414 | cyto |
| Zosma01g09130 | ZosmaPP2C-05 | Chr01:6297640-6298838(−) | 6 | 271 | 29,309.02 | 5.04 | −0.167 | cyto |
| Zosma01g24910 | ZosmaPP2C-10 | Chr01:30001865-30003225(+) | 4 | 375 | 42,879.38 | 8.61 | −0.281 | nucl |
| Zosma01g20530 | ZosmaPP2C-09 | Chr01:25525319-25526706(−) | 3 | 408 | 44,707.5 | 4.47 | −0.335 | cyto |
| Zosma01g01360 | ZosmaPP2C-01 | Chr01:1226328-1228062(+) | 4 | 415 | 44,827.16 | 7.19 | −0.259 | chlo |
| Zosma01g32210 | ZosmaPP2C-12 | Chr01:35669026-35687172(−) | 14 | 645 | 71,138.92 | 5.88 | −0.214 | chlo |
| Zosma01g09260 | ZosmaPP2C-06 | Chr01:6435541-6436897(−) | 5 | 280 | 30,367.56 | 9.23 | −0.431 | cyto |
| Zosma04g06830 | ZosmaPP2C-29 | Chr04:5613604-5614375(−) | 4 | 162 | 17,801.05 | 4.61 | −0.254 | cyto |
| Zosma04g16720 | ZosmaPP2C-31 | Chr04:26553016-26554963(−) | 4 | 567 | 62,401.61 | 6.68 | −0.493 | cyto |
| Zosma04g22800 | ZosmaPP2C-34 | Chr04:31522865-31524049(+) | 1 | 395 | 45,150.45 | 6.31 | −0.536 | nucl |
| Zosma04g22790 | ZosmaPP2C-33 | Chr04:31505790-31522743(+) | 24 | 913 | 103,337.17 | 6.99 | −0.146 | chlo |
| Zosma04g21550 | ZosmaPP2C-32 | Chr04:30094279-30095776(−) | 4 | 412 | 46,772.25 | 6.26 | −0.276 | chlo |
| Zosma04g09260 | ZosmaPP2C-30 | Chr04:8351211-8352819(−) | 3 | 462 | 50,534.86 | 4.67 | -0.214 | nucl |
| Zosma04g05430 | ZosmaPP2C-28 | Chr04:4230755-4234336(+) | 10 | 378 | 41,317.95 | 8.04 | −0.365 | cyto |
| Zosma04g00980 | ZosmaPP2C-27 | Chr04:863428-864629(−) | 3 | 225 | 25,499.52 | 6.25 | −0.622 | chlo |
| Zosma04g22810 | ZosmaPP2C-35 | Chr04:31524979-31526648(−) | 4 | 481 | 54,856.74 | 9.2 | −0.236 | cysk |
| Zosma06g09020 | ZosmaPP2C-45 | Chr06:5412181-5413730(−) | 4 | 431 | 47,071.05 | 5.03 | −0.206 | chlo |
| Zosma06g12670 | ZosmaPP2C-46 | Chr06:9544529-9548632(−) | 11 | 416 | 45,739.09 | 7.73 | −0.11 | chlo |
| Zosma06g07830 | ZosmaPP2C-44 | Chr06:4547808-4551544(+) | 4 | 404 | 43,622.19 | 8.05 | −0.141 | chlo |
| Zosma06g28610 | ZosmaPP2C-51 | Chr06:28014762-28016144(+) | 4 | 379 | 41,245.42 | 5.97 | −0.351 | chlo |
| Zosma06g22190 | ZosmaPP2C-49 | Chr06:23169549-23170715(+) | 2 | 360 | 40,348.93 | 7.77 | −0.358 | chlo |
| Zosma06g16930 | ZosmaPP2C-48 | Chr06:18926155-18927551(+) | 5 | 299 | 32,721.32 | 7.85 | −0.325 | cyto |
| Zosma06g26550 | ZosmaPP2C-50 | Chr06:26097002-26170395(+) | 20 | 1062 | 121,353.82 | 6.98 | −0.373 | chlo |
| Zosma06g15960 | ZosmaPP2C-47 | Chr06:18007315-18009251(+) | 10 | 357 | 39,322.92 | 8.01 | −0.322 | cysk |
| Zosma154g00070 | ZosmaPP2C-52 | scaffold_154:65907-68395(−) | 8 | 288 | 30,882.72 | 5.36 | −0.189 | cyto |
| Zosma05g21020 | ZosmaPP2C-41 | Chr05:22707710-22709375(−) | 5 | 282 | 30,696.04 | 7.84 | −0.275 | cyto |
| Zosma05g31280 | ZosmaPP2C-43 | Chr05:30867806-30869625(+) | 4 | 397 | 43,802.91 | 8.94 | −0.291 | chlo |
| Zosma05g04480 | ZosmaPP2C-36 | Chr05:2491529-2498534(+) | 10 | 535 | 58,109.03 | 4.58 | −0.25 | cyto |
| Zosma05g18030 | ZosmaPP2C-40 | Chr05:19136954-19138522(−) | 4 | 429 | 46,362.18 | 6.59 | −0.176 | chlo |
| Zosma05g07410 | ZosmaPP2C-37 | Chr05:4635552-4637065(−) | 4 | 379 | 41,150.38 | 5.09 | −0.192 | cysk |
| Zosma05g28040 | ZosmaPP2C-42 | Chr05:28833135-28834253(+) | 3 | 332 | 36,036.37 | 9.26 | −0.489 | nucl |
| Zosma05g10930 | ZosmaPP2C-38 | Chr05:6485671-6486054(+) | 3 | 71 | 7833.00 | 4.41 | 0.176 | extr |
| Zosma05g13440 | ZosmaPP2C-39 | Chr05:8751625-8758541(−) | 15 | 1076 | 120,748.65 | 5.07 | −0.363 | nucl |
| Zosma03g12800 | ZosmaPP2C-22 | Chr03:23217337-23220806(+) | 10 | 389 | 42,278.27 | 5.03 | −0.406 | nucl |
| Zosma03g27860 | ZosmaPP2C-26 | Chr03:34707412-34709175(−) | 4 | 383 | 42,883.17 | 8.13 | −0.241 | chlo |
| Zosma03g14770 | ZosmaPP2C-24 | Chr03:25132638-25135189(−) | 4 | 772 | 85,688.82 | 5.7 | −0.504 | nucl |
| Zosma03g00750 | ZosmaPP2C-21 | Chr03:720520-724643(+) | 11 | 665 | 74,159.99 | 6.03 | −0.23 | mito |
| Zosma03g13840 | ZosmaPP2C-23 | Chr03:24352649-24353861(−) | 3 | 350 | 37,628.48 | 5.33 | −0.181 | extr |
| Zosma03g21390 | ZosmaPP2C-25 | Chr03:30186101-30189998(+) | 4 | 375 | 42,268.25 | 9.11 | −0.328 | chlo |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Dong, X.; Yang, D.; Ge, Q.; Dai, J.; Ma, Z.; Wang, R.; Zhao, H. Genome-Wide Identification and Expression Analysis of PP2C Gene Family in Eelgrass. Genes 2025, 16, 657. https://doi.org/10.3390/genes16060657
Liu C, Dong X, Yang D, Ge Q, Dai J, Ma Z, Wang R, Zhao H. Genome-Wide Identification and Expression Analysis of PP2C Gene Family in Eelgrass. Genes. 2025; 16(6):657. https://doi.org/10.3390/genes16060657
Chicago/Turabian StyleLiu, Chang, Xu Dong, Dazuo Yang, Qingchao Ge, Jiaxin Dai, Zhi Ma, Rongna Wang, and Huan Zhao. 2025. "Genome-Wide Identification and Expression Analysis of PP2C Gene Family in Eelgrass" Genes 16, no. 6: 657. https://doi.org/10.3390/genes16060657
APA StyleLiu, C., Dong, X., Yang, D., Ge, Q., Dai, J., Ma, Z., Wang, R., & Zhao, H. (2025). Genome-Wide Identification and Expression Analysis of PP2C Gene Family in Eelgrass. Genes, 16(6), 657. https://doi.org/10.3390/genes16060657





