Abstract
Background: Geranylgeranyl pyrophosphate synthase (GGPS) is a pivotal enzyme in terpene biosynthesis, influencing the production of carotenoids, chlorophylls, and diverse phytohormones. This study aimed to identify and characterize the StGGPS gene family in potato (Solanum tuberosum) to elucidate its involvement in carotenoid synthesis and responses to abiotic stresses. Methods: Employing bioinformatics approaches, including HMMER, SMART, and Pfam, we conducted a genome-wide identification of StGGPS genes. Subsequent phylogenetic analysis, gene structure characterization, conserved motif detection, and synteny analysis were performed to investigate evolutionary relationships within the family. The expression patterns of StGGPS genes were then analyzed using RNA-seq data and quantitative real-time PCR (qRT-PCR) in potato tubers exhibiting different pigmentation and under drought and salt stress conditions. Results: Eleven StGGPS genes were identified, unevenly distributed across seven chromosomes, and classified into three subfamilies based on phylogenetic and structural analyses. Synteny analysis revealed one intra-genomic duplicate pair (StGGPS1/StGGPS4) and conserved orthologs with other Solanaceae species. Promoter analysis identified cis-elements related to light response and abiotic stress (e.g., ABRE and CGTCA-motif). Expression data showed differential regulation of StGGPS genes in colored tubers, with yellow and red tubers exhibiting higher expression of carotenoid-related genes. Under drought stress, StGGPS10 was significantly upregulated (5.2-fold, p < 0.001), while StGGPS6 showed salt-responsive induction (3.8-fold, p < 0.001), linking them to ABA signaling and cytoskeletal dynamics, respectively. Conclusions: This study provides a comprehensive overview of the StGGPS gene family, highlighting their roles in carotenoid biosynthesis and abiotic stress responses. The stress-specific expression patterns of StGGPS10 and StGGPS6 offer potential targets for genetic improvement of potato stress resilience.
1. Introduction
Carotenoids, a diverse class of terpenoid pigments, are ubiquitous in bacteria, fungi, algae, and terrestrial plants, where they fulfill critical functions in photosynthesis and photoprotection [1,2,3,4,5,6]. Within photosynthetic membranes, carotenoids associate with proteins to form pigment–protein complexes, acting as integral light-harvesting components [7]. Beyond their direct roles in photosynthesis, carotenoids contribute to plant development and stress resistance by serving as precursors for retrograde signaling molecules [8]. For example, carotenoids are metabolized via the methylerythritol phosphate (MEP) pathway to synthesize abscisic acid [9], a phytohormone that enhances plant resilience to abiotic stresses such as drought and salinity [10]. Furthermore, strigolactones, another group of carotenoid-derived plant hormones, regulate plant architecture by inhibiting shoot branching, modulating root development, and promoting leaf expansion, thereby influencing plant aging and secondary growth processes [11].
Geranylgeranyl pyrophosphate synthase (GGPS) is a pivotal enzyme in terpenoid biosynthesis, catalyzing the formation of geranylgeranyl pyrophosphate (GGPP) from farnesyl pyrophosphate (FPP) and isopentenyl pyrophosphate (IPP) [12]. GGPP functions as a precursor for the synthesis of crucial plant pigments and hormones, including carotenoids, chlorophyll, gibberellin, and abscisic acid [13]. Accumulating evidence from diverse plant species underscores the multifaceted role of GGPS in both primary metabolism and stress adaptation. For instance, in sweet potato (Ipomoea batatas), IbGGPS overexpression enhances osmotic stress tolerance by modulating carotenoid biosynthesis [14], while, in cotton (Gossypium hirsutum), GGPS isoforms regulate chlorophyll synthesis under abiotic stress [15]. Analogously, Liriodendron tulipifera GGPS controls terpenoid accumulation with tissue-specific expression patterns [16], and Arabidopsis AtGGPS11 is essential for photosynthesis-related isoprenoid production [17]. Taken together, these findings suggest that GGPS gene families exhibit functional diversification, likely shaped by evolutionary pressures to fulfill species-specific metabolic and environmental requirements.
Potato (S. tuberosum), the most productive noncereal food crop, is a vital source of carbohydrates, dietary fiber, vitamins, and antioxidants [18]. Despite its nutritional importance, potato cultivation is highly vulnerable to abiotic stresses such as drought, salinity, and heat [19]. Although GGPS-mediated terpenoid metabolism is recognized as a critical component of stress resilience in other plants [14,15,16,17], the functional roles of the potato GGPS gene family remain unexplored. This study was guided by the hypothesis that the potato GGPS family has evolved functional divergence to regulate carotenoid biosynthesis and abiotic stress adaptation, with distinct members contributing to stress-specific responses.
To investigate this hypothesis, we conducted a comprehensive analysis of the GGPS family in the potato genome, encompassing characterization of physicochemical properties, phylogenetic relationships, and subcellular localization. Furthermore, we explored the evolutionary mechanisms underlying gene family expansion, including duplication events and collinearity analyses with homologs in model species such as Arabidopsis, tomato, and pepper. Finally, we validated the functional association of candidate genes with abiotic stress responses through transcriptomic profiling and quantitative PCR, with a specific focus on drought and salt stress due to their significant impact on potato productivity. By integrating evolutionary insights with expression dynamics, this study elucidates the role of functional specialization within the StGGPS gene family in contributing to stress resilience and identifies promising targets, such as StGGPS6 and StGGPS10, for breeding programs aimed at developing stress-tolerant cultivars.
2. Materials and Methods
2.1. RNA Extraction and qRT-PCR Analysis
The potato variety Jinong Shu 8511, bred by the Agricultural University of Hebei, served as the experimental material for this study. The plants, characterized by yellow skin and flesh, were grown for 21 days before being subjected to stress conditions. To simulate drought stress, seedlings were treated with 25 mmol/L PEG6000 [18] and, to simulate salt stress, 70 mmol/L of NaCl was used [20]. Samples were collected at 0, 12, and 24 h post-stress, flash-frozen in liquid nitrogen, and stored at −80 °C.
Total RNA was extracted using the Promega RNA Extraction kit (LS1040, Promega Corporation, Madison, WI, USA), and RNA integrity and concentration were assessed through agarose gel electrophoresis and a Nanodrop ND-2000 spectrophotometer (Nanodrop Technologies, now part of Thermo Fisher Scientific, Waltham, MA, USA). First-strand cDNA synthesis was performed using the Kangwei Century RT gDNA kit (CW2020M, Kangwei Century Biotechnology Co., Ltd., Beijing, China). qRT-PCR was carried out on a CFX96 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the US EVERBRIGHT AugeGreen qPCR Master Mix (S2008L, US EVERBRIGHT Biotechnology, Santa Clara, CA, USA) The reaction conditions were based on the AugeGreen qRT-PCR Master Mix instructions. qRT-PCR assays were performed with three biological replicates and three technical replicates. Significant differences were determined by one-way ANOVA using SPSS 26.0. StEF-1α was used as the reference gene [21], and relative expression was calculated using the 2−ΔΔct methods [22].
2.2. Identification and Sequence Analysis of Potato GGPS Genes
The genome annotation files and nucleotide sequences for S. tuberosum were downloaded from the Ensembl Plant database (http://plants.ensembl.org/index.html, on 1 May 2024). The GGPS structural domain (PF00348) was identified using HMMER 3.1 (http://hmmer.org/download.html, on 5 May 2024) within the potato genome protein sequences. Sequences lacking the GGPS structural domain were excluded, and the remaining sequences were verified using SMART (http://smart.embl-heidelberg.de, on 11 May 2024) and Pfam (v.36, https://www.ebi.ac.uk/interpro/, on 11 May 2024) tools. The physical and chemical properties of the identified GGPS family members, including amino acid count, molecular weight, and theoretical isoelectric points, were analyzed using BioPerl (v1.5.1) [23].
2.3. Phylogenetic Analysis
Multiple sequence alignments of potato GGPS, along with GGPS sequences from Arabidopsis, tobacco, tomato (SlGGPS), and pepper, were performed using the MUSCLE algorithm. A phylogenetic tree was constructed using IQ-TREE, with 1000 bootstrap replicates [24].
2.4. Gene Structure, Conserved Motifs, and 3D Structural Analysis
Gene structure information for potato GGPS family members was extracted from genome annotation files, and intron/exon structures were visualized using GSDS 2.0 (http://gsds.gao-lab.org/, on 25 May 2024) [25]. Conserved motifs were identified using the MEME 5.5.5 online tool (http://meme-suite.org/, on 25 May 2024) with the following parameters: a maximum number of 10 motifs, a motif width of 6–50, amino acids, and default settings for other parameters [26]. Subcellular location predictions were conducted using WoLF PSORT [27]. The three-dimensional structural models of potato GGPS genes were retrieved from the RCSB PDB database (https://www.rcsb.org/, on 27 May 2024) and refined using PSI-BLAST, SWISS-MODEL (https://swissmodel.expasy.org/, on 25 May 2024), and SAVES (https://saves.mbi.ucla.edu/, on 25 May 2024) for quality evaluation [28,29].
2.5. Analysis of Cis-Acting Elements in Promoter Regions
The cis-acting elements within 2 kb upstream of the potato GGPS gene promoters [30] were analyzed using the PlantCARE database, and the elements were visualized using the GGPS 2.0 website (http://gsds.gao-lab.org/, on 27 May 2024) [25].
2.6. Chromosomal Location, Gene Distribution, and Synteny Analysis
The chromosomal locations of StGGPS family members were extracted from genome annotation files and mapped using TBtools II (TBtools v1.120) [31]. Duplication events among potato GGPS genes were analyzed with MscanX (MscanX v1.0) software [32], and synteny relationships were visualized using Circos (Circos v0.69-14) [33]. Synonymous substitution (Ks) and nonsynonymous substitution (Ka) rates for duplicated gene pairs were calculated using KaKs Calculator 2.0 [34]. Furthermore, MScanX software was used to analyze the collinearity between potato GGPS genes and multiple species.
2.7. Expression Analysis of RNA Data
RNA-seq raw counts were normalized using the DESeq2 package (v1.30.1) with median-of-ratios method. RNA-seq data were obtained from NCBI to assess the expression pattern of GGPS genes in potato tubers of various colors (white, yellow, red, and purple) and under abiotic stresses, including salt stress (200 mM NaCl for 12 and 24 h) and simulated drought stress (150 mM mannitol for 12 and 24 h). A heat map of gene expression was generated using TBtools II [31].
3. Results
3.1. Identification of GGPS Gene Family Members and Analysis of Protein Physicochemical Properties in Potatoes
Eleven GGPS (StGGPS) gene family members were identified in potatoes using HMMER 3.1 in conjunction with the SMART, Pfam, and NCBI CDD databases. The StGGPS genes were sequentially named (StGGPS1–StGGPS11). BioPerl analysis showed that the amino acid length of StGGPS family members ranges from 294 to 398 residues, with an average length of 351 residues. The molecular weight varies from 32,784.6 to 43,490.4 Da, with an average molecular weight of 38,973.8 Da. The theoretical isoelectric point ranges from 4.87 to 8.38, with an average of 6.41. Among the StGGPS genes, six exhibit predominant localization in the chloroplast. Two genes show subcellular distribution across the cytoplasm, nucleus, and mitochondria, while one gene is associated with the cytoskeleton (Table 1).

Table 1.
Analysis of potato GGPS gene family members and physicochemical properties.
3.2. Potato GGPS Family Evolution Analysis, Gene Structure, and Conserved Motifs
To explore the evolution of the GGPS gene family, a phylogenic tree was constructed using genes from Arabidopsis (Arabidopsis thaliana), potato (S. tuberosum), tobacco (Nicotiana attenuata), tomato (Solanum lycopersicum), and hot pepper (Capsicum annuum) (Figure 1). The GGPS genes were divided into three subfamilies: Group 3, with seven potato StGGPS members; Group 2, with three potato StGGPS members; and Group 1, with one potato StGGPS member. Members within the same branch likely have similar functions and evolutionary relationships.
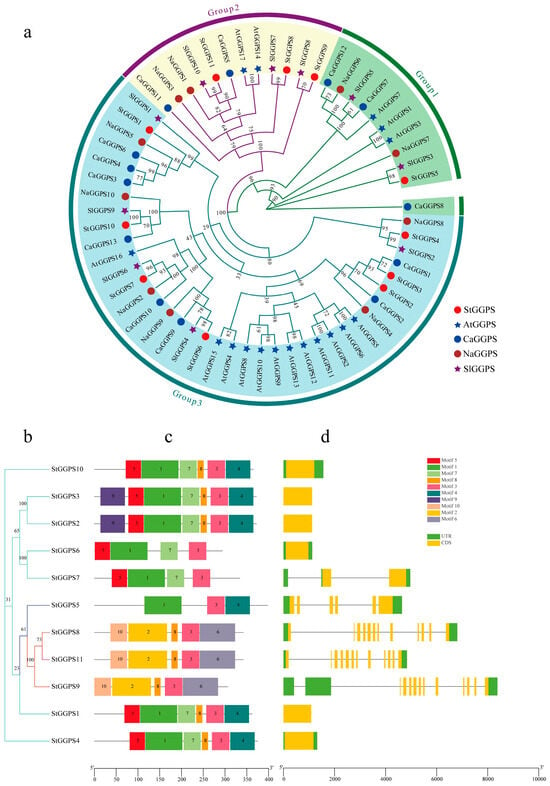
Figure 1.
Phylogenetic, gene structure, and conserved motif analysis of the GGPS gene family. (a): Phylogenetic tree of the GGPS gene family in A. thaliana, S. tuberosum, N. attenuata, S. lycopersicum, and C. annuum. (b): Phylogenetic tree of the StGGPS gene family. (c): Conserved motif analysis of the StGGPS gene. (d): Analysis of StGGPS gene structure.
To further elucidate the evolutionary relationships among StGGPS genes, we analyzed the intron–exon substructure and conserved motifs of potato StGGPS members (Figure 1). The genetic structure of different StGGPS members exhibits considerable variation, ranging from intronless genes to those containing up to 11 introns. This diversity in gene structure suggests that StGGPS genes may have been subjected to divergent selection pressures during evolution. Notably, StGGPS genes within the same subfamily generally displayed similar gene structures, a pattern consistent with the phylogenetic tree, indicating a strong correlation between the intron–exon gene structure of StGGPS genes and their phylogenetic relationships. Given the association of conserved sequence elements with protein function, we employed MEME to identify 10 distinct motifs (Motif 1 through Motif 10). The number of motifs present in each StGGPS member ranged from three to seven; for instance, StGGPS5 contains a minimum of three motifs, while StGGPS2 and StGGPS3 possess a maximum of seven. Although certain motifs are absent in some StGGPS members, a conserved pattern, particularly Motif 3, is consistently observed across all StGGPS genes. Furthermore, analysis of the predicted three-dimensional structures revealed a higher degree of structural similarity among proteins within the same subfamily compared to those from different subfamilies. For example, StGGPS8, StGGPS9, and StGGPS11 from Group 2 exhibit similar structures, as do StGGPS1, StGGPS2, StGGPS3, StGGPS4, and StGGPS10 from Group 3 (Figure 2). Collectively, this integrated analysis of the phylogenetic tree, gene structure, and conserved motifs suggests that StGGPS members within the same subfamily share similar gene functions and protein structures, implying analogous functional roles. We hypothesize that the observed similarities in gene structures and conserved motifs among StGGPS genes are indicative of functional convergence.
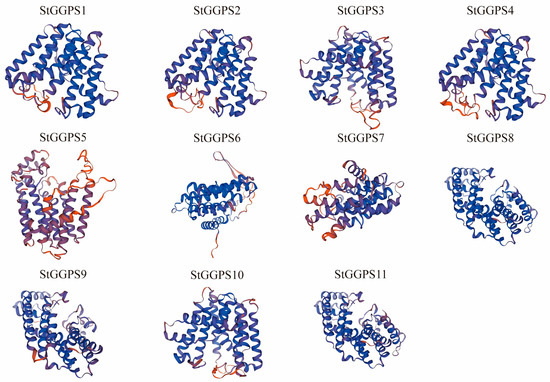
Figure 2.
Three-dimensional structural modeling of proteins of the StGGPS gene family.
3.3. Chromosome Localization of Genes and Analysis of Gene Duplication Events
Chromosomal positions of StGGPS family members were extracted from the genome annotation file using Tbtools II. We found that chromosome 2 contained three StGGPS genes; chromosomes 7 and 10 contained two StGGPS genes; and chromosomes 4, 9, 11, and 12 contained only one StGGPS gene (Figure 3a).
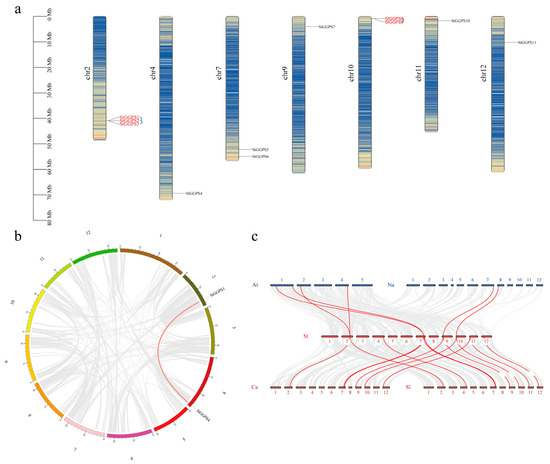
Figure 3.
Gene duplication events in the GGPS family of S. tuberosum. (a): Chromosomal localization map of StGGPS gene. (b): Covariance analysis of StGGPS gene. (c): Collinearity analysis of GGPS genes in multiple species.
To explore the structural and functional characteristics of the StGGPS genes, we examined gene duplications through tandem repeats and segmental duplications. We identified StGGPS gene pairs resulting from duplication events, specifically StGGPS1/StGGPS4 (Figure 3b). We found three pairs of tandem repeat genes: StGGPS1, StGGPS2, StGGPS2, StGGPS3, StGGPS8, and StGGPS9. Gene duplication and fragment duplication likely drove the evolution of StGGPS genes. Further analysis of the Ka/Ks ratios for each pair indicated that the ratios ranged from 0.0634 (StGGPS1/StGGPS4) to 0.6052 (StGGPS8/StGGPS9), suggesting selective pressure on these genes.
To understand the evolutionary mechanisms of the potato GGPS family, we performed linear analysis using Arabidopsis and Solanaceae plants (tobacco, tomatoes, peppers, and potatoes) (Figure 3c). Four pairs of homologous genes were identified between potato and A. thaliana. Two homologous pairs were found between potatoes and tobacco, eight homologous genes between potatoes and tomatoes, and seven between potatoes and peppers. These homologous genes likely existed before the divergence of ancestral lineages.
3.4. Analysis of Cis-Acting Elements in StGGPS Gene Promoters
To explore the potential functions of StGGPS genes, we analyzed the 2000 bp upstream promoter sequence. We identified 10 cis-acting elements (Figure 4) associated with various physiological processes. Notably, elements such as Box4, G-box, GATA-motif, GT1-motif, and TCT-motif, which are related to light sensing, were prevalent. This suggests that the StGGPS gene family is involved in potato photosynthesis. Additionally, StGGPS genes associated with abiotic stress responses contained cis-elements such as ABRE (involved in abscisic acid response), ARE (anaerobic response), CGTCA-motif and TGACG-motif (jasmonic acid response), and TGA (auxin response). Overall, StGGPS genes likely play significant roles in light and abiotic stress responses in potatoes.
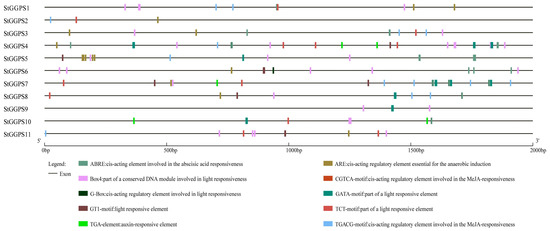
Figure 4.
Original analysis of the cis-acting 2000 bp promoter of the StGGPS gene.
3.5. Expression Patterns of Genes in Tubers of Different Colors
Given the established correlation between tuber color and carotenoid content [17], RNA-sequencing data were employed to investigate the expression patterns of StGGPS genes across potato tubers exhibiting four distinct colors (Figure 5). The analysis revealed differential expression of StGGPS genes dependent on tuber color. Specifically, StGGPS1, StGGPS3, StGGPS4, and StGGPS7 were upregulated in yellow tubers, whereas StGGPS2, StGGPS5, StGGPS8, and StGGPS9 showed increased expression in white tubers. Red tubers exhibited upregulation of StGGPS6, StGGPS10, and StGGPS11, while StGGPS gene expression was generally downregulated in purple tubers. These findings suggest a potentially higher carotenoid content in yellow and red tubers and a correspondingly lower carotenoid content in purple tubers. The observed downregulation of StGGPS genes in purple tubers likely represents a metabolic and transcriptional shift, prioritizing anthocyanin biosynthesis over carotenoid production. This adaptive reallocation of resources may serve to optimize stress tolerance or mediate ecological interactions.
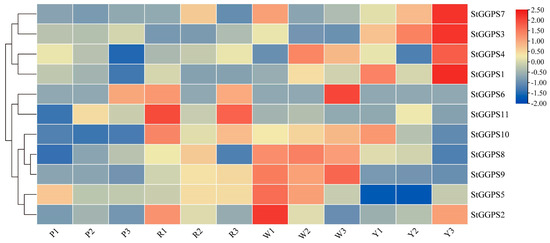
Figure 5.
Expression of StGGPS gene in different colored tubers (P: purple potato; R: red potato; W: white potato; Y: yellow potato).
3.6. Gene Expression Patterns in Response to Abiotic Stress
Abiotic stresses, such as drought and salt stress, significantly impact potato production. To investigate the response of the StGGPS family to these stresses, we analyzed RNA-seq data from NCBI, validated by RT-qPCR. Under drought stress, the RNA-seq data revealed differential expression of StGGPS genes. Specifically, StGGPS9 and StGGPS10 were upregulated, whereas other StGGPS members were downregulated. Reverse transcription quantitative PCR validation confirmed the drought-responsive nature of StGGPS10, demonstrating a significant 5.2-fold increase in transcript abundance under drought stress (p < 0.001). This substantial induction suggests a critical role for StGGPS10 in mediating drought resistance in potato (Figure 6). Under salt stress conditions, RNA-seq analysis revealed differential expression of multiple StGGP family members, including StGGPS3, StGGPS4, StGGPS9, StGGPS10, StGGPS2, StGGPS5, StGGPS6, and StGGPS11. Subsequent reverse transcription quantitative real-time PCR (RT-qPCR) validation confirmed the salt-responsive transcription of StGGPS6, which exhibited a significant 3.8-fold upregulation of transcript abundance under salt stress (p < 0.001) (Figure 7). Based on these findings, we hypothesize that StGGPS6 and StGGPS10 play important roles in the abiotic stress response of potatoes.
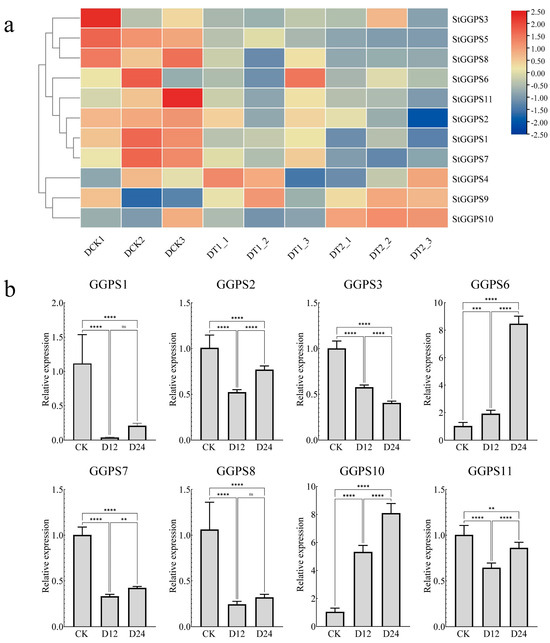
Figure 6.
Expression of StGGPS gene under drought stress. (a): Expression of StGGPS under drought stress. (b): Relative expression of StGGPS under drought stress (**: p < 0.01, ***: p < 0.001, and ****: p < 0.0001).
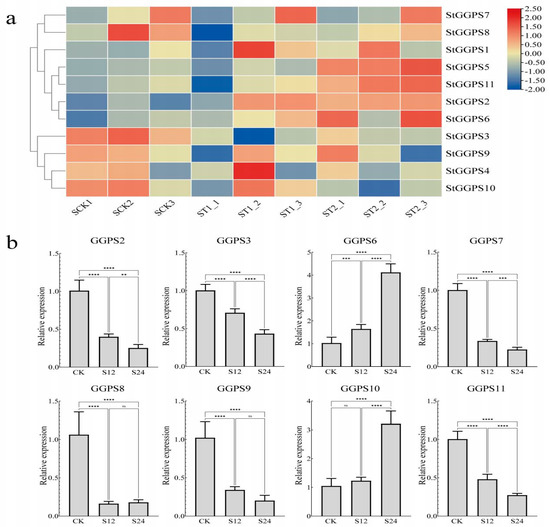
Figure 7.
Expression of StGGPS gene under salt stress. (a): Expression of StGGPS under salt stress. (b): Relative expression of StGGPS under salt stress (**: p < 0.01, ***: p < 0.001, and ****: p < 0.0001).
4. Discussion
The StGGPS gene family in potato comprises three phylogenetically distinct subfamilies: Group 3 (seven members), Group 2 (two members), and Group 1 (a single member). Ancient duplication events with high retention rates of gene pairs have driven the proliferation of duplicated genes in plant genomes [35]. Notably, we identified a conserved duplicate pair (StGGPS1/StGGPS4) within Group 3, suggesting this subfamily’s pivotal role in StGGPS expansion. RNA-seq analyses revealed co-expression patterns of StGGPS1 and StGGPS4 across differentially pigmented tubers, implicating their co-ordinated regulation in carotenoid biosynthesis pathways.
Furthermore, RNA-seq and RT-qPCR analyses confirmed that StGGPS1 was downregulated under drought stress conditions, whereas StGGPS4 was upregulated. In contrast, under salt stress conditions, StGGPS1 was upregulated and StGGPS4 was downregulated. These findings suggest that StGGPS1 and StGGPS4 may have distinct roles in the potato plant’s response to abiotic stress. Additionally, our collinearity across species revealed that certain StGGPS genes are evolutionarily conserved. For instance, StGGPS7 (PGSC0003DMG400002687) in potatoes is homologous to NaGGPS2 (A4A49_09152) in tobacco, SlGGPS6 (Solyc09g008920.3) in tomato, and CaGGPS10 (T459_22463) in pepper. Additionally, StGGPS9 (PGSC0003DMG400014369) in potatoes was homologous to NaGGPS3 (A4A49_24015) in tobacco, SlGGPS8 (Solyc10g005840.3) in tomatoes, and CaGGPS11 (T459_24642) in pepper plants. These genes may have been present before the divergence of the ancestral lineages of Solanaceae and, at the same time, these orthologous genes may play similar functions and roles in different Solanaceae species.
As a key enzyme in the synthesis of terpenoids, geranyl geranyl pyrophosphate synthase (GGPS) catalyzes the production of geranyl geranyl pyrophosphate (GGPP), a crucial compound in plant photosynthesis and biotic/abiotic stress responses [15,16,36]. Previous studies have demonstrated that NtGGPPS1 and NtPSY1 in tobacco form a multi-enzyme complex that regulates carotenoid biosynthesis and the co-expression of genes in the carotenoid biosynthesis pathway [36]. Similarly, ApGGPS is the key enzyme in andrographolide synthesis and plays a role in photosynthesis, growth, and other physiological processes in plants by participating in GGPP synthesis [37].
Through collinearity analysis, we identified four pairs of homologous genes in potato and Arabidopsis, including StGGPS2 (PGSC0003DMG400041508), AtGGPS5 (AT2G18640), AtGGPS15 (AT4G36810), StGGPS5 (PGSC0003DMG400007081), AtGGPS1 (AT1G17050), and AtGGPS3 (AT1G78510). AtGGPS15 (AT4G36810) is a key isoenzyme required for the production of most isoprene compounds involved in photosynthesis [17]. The conserved gene structures and motifs between StGGPS2 and AtGGPS15 suggest functional conservation, potentially underpinning their shared roles in photosynthetic processes of potato. In addition, AtGGPS1 (AT1G17050) and AtGGPS3 (AT1G78510) are paralogous genes involved in the synthesis of [20] the REDOX cofactor plastoquinine-9 in photosynthesis [38], a key antioxidant in plant leaves that plays a critical role in photoprotection [39]. Higher plastoquinin-9 activity in potatoes could enhance resistance to the effects of salt stress on photosynthesis. StGGPS5 is homologous to AtGGPS1 (AT1G17050) and AtGGPS3 (AT1G78510), and RNA-seq data show significant upregulation of StGGPS5 under salt stress; we propose that StGGPS5 may be closely related to plastoquinone-9 synthesis in potatoes and participate in their response to salt stress, thereby mitigating the adverse effects of salt on potato growth and development.
Future research should integrate metabolomic profiling with transgenic approaches, such as the overexpression of StMYB1, to rigorously validate the proposed metabolic trade-off between carotenoid and anthocyanin biosynthesis in potato tubers. Further elucidation of the transcriptional cross-regulation and resource allocation mechanisms governing these pathways is crucial for a more complete understanding of their competitive interaction. Furthermore, characterizing the ecological advantages conferred by anthocyanin dominance may inform strategies aimed at engineering stress-resilient cultivars with tailored nutritional profiles. This will allow for a more informed balancing of pigmentation and stress adaptation traits for the improvement of agricultural outcomes.
5. Conclusions
Eleven StGGPS family genes were identified and classified into three subfamilies based on gene structure and conserved motif analysis. Bioinformatics analysis and gene expression verification revealed that these genes contain numerous photo-responsive and cis-acting elements related to abiotic stress responses. Combined with transcriptome data and q-PCR verification, our findings suggest that StGGPS family members play a significant role in helping potatoes resist damage caused by abiotic stress, thereby supporting the plant’s growth and development under adverse conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16060646/s1, Figure S1: High resolution images used in this article; Table S1: The primer sequences used in this study.
Author Contributions
Conceptualization, C.F., W.L., S.Z. and J.C.; Methodology, S.G.; Formal analysis, C.F.; Investigation, S.G.; Writing—original draft, C.F. and W.L.; Writing—review and editing, X.C., M.J., S.Z. and J.C.; Visualization, W.L., X.C. and M.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Hebei Province modern agricultural industrial technology system tuber industry innovation team special and Hebei province detoxigenic potato breeding technology innovation center (HBCT2023060201 and SG2012016).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Avalos, J.; Carmen Limón, M. Biological Roles of Fungal Carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef] [PubMed]
- López, G.-D.; Álvarez-Rivera, G.; Carazzone, C.; Ibáñez, E.; Leidy, C.; Cifuentes, A. Bacterial Carotenoids: Extraction, Characterization, and Applications. Crit. Rev. Anal. Chem. 2023, 53, 1239–1262. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Carotenoids and Their Biosynthesis in Fungi. Molecules 2022, 27, 1431. [Google Scholar] [CrossRef]
- Pereira, A.G.; Otero, P.; Echave, J.; Carreira-Casais, A.; Chamorro, F.; Collazo, N.; Jaboui, A.; Lourenço-Lopes, C.; Simal-Gandara, J.; Prieto, M.A. Xanthophylls from the Sea: Algae as Source of Bioactive Carotenoids. Mar. Drugs 2021, 19, 188. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef]
- Xu, P.; Chukhutsina, V.U.; Nawrocki, W.J.; Schansker, G.; Bielczynski, L.W.; Lu, Y.; Karcher, D.; Bock, R.; Croce, R. Photosynthesis without β-Carotene. eLife 2020, 9, e58984. [Google Scholar] [CrossRef]
- Sierra, J.; McQuinn, R.P.; Leon, P. The Role of Carotenoids as a Source of Retrograde Signals: Impact on Plant Development and Stress Responses. J. Exp. Bot. 2022, 73, 7139–7154. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Jia, K.-P.; Baz, L.; Al-Babili, S. From Carotenoids to Strigolactones. J. Exp. Bot. 2018, 69, 2189–2204. [Google Scholar] [CrossRef]
- Tata, S.K.; Jung, J.; Kim, Y.; Choi, J.Y.; Jung, J.; Lee, I.; Shin, J.S.; Ryu, S.B. Heterologous Expression of Chloroplast-localized Geranylgeranyl Pyrophosphate Synthase Confers Fast Plant Growth, Early Flowering and Increased Seed Yield. Plant Biotechnol. J. 2016, 14, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Hivert, G.; Davidovich-Rikanati, R.; Bar, E.; Sitrit, Y.; Schaffer, A.; Dudareva, N.; Lewinsohn, E. Prenyltransferases Catalyzing Geranyldiphosphate Formation in Tomato Fruit. Plant Sci. 2020, 296, 110504. [Google Scholar] [CrossRef]
- Chen, W.; He, S.; Liu, D.; Patil, G.B.; Zhai, H.; Wang, F.; Stephenson, T.J.; Wang, Y.; Wang, B.; Valliyodan, B.; et al. A Sweetpotato Geranylgeranyl Pyrophosphate Synthase Gene, IbGGPS, Increases Carotenoid Content and Enhances Osmotic Stress Tolerance in Arabidopsis Thaliana. PLoS ONE 2015, 10, e0137623. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Mehari, T.G.; Fang, H.; Ji, M.; Qu, Z.; Jia, M.; Wang, D.; Ditta, A.; Khan, M.K.R.; Cao, Y.; et al. Genome-Wide Identification of the Geranylgeranyl Pyrophosphate Synthase (GGPS) Gene Family Involved in Chlorophyll Synthesis in Cotton. BMC Genom. 2023, 24, 176. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, H.; Zong, Y.; Tu, Z.; Li, H. Isolation, Expression, and Functional Analysis of the Geranylgeranyl Pyrophosphate Synthase (GGPPS) Gene from Liriodendron tulipifera. Plant Physiol. Biochem. PPB 2021, 166, 700–711. [Google Scholar] [CrossRef]
- Ruiz-Sola, M.Á.; Coman, D.; Beck, G.; Barja, M.V.; Colinas, M.; Graf, A.; Welsch, R.; Rütimann, P.; Bühlmann, P.; Bigler, L.; et al. Arabidopsis GERANYLGERANYL DIPHOSPHATE SYNTHASE 11 Is a Hub Isozyme Required for the Production of Most Photosynthesis-Related Isoprenoids. New Phytol. 2016, 209, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Gervais, T.; Creelman, A.; Li, X.-Q.; Bizimungu, B.; Koeyer, D.D.; Dahal, K. Potato Response to Drought Stress: Physiological and Growth Basis. Front. Plant Sci. 2021, 12, 698060. [Google Scholar] [CrossRef]
- Lal, M.K.; Tiwari, R.K.; Kumar, A.; Dey, A.; Kumar, R.; Kumar, D.; Jaiswal, A.; Changan, S.S.; Raigond, P.; Dutt, S.; et al. Mechanistic Concept of Physiological, Biochemical, and Molecular Responses of the Potato Crop to Heat and Drought Stress. Plants 2022, 11, 2857. [Google Scholar] [CrossRef]
- Kolomeichuk, L.V.; Efimova, M.V.; Zlobin, I.E.; Kreslavski, V.D.; Murgan, O.K.; Kovtun, I.S.; Khripach, V.A.; Kuznetsov, V.V.; Allakhverdiev, S.I. 24-Epibrassinolide Alleviates the Toxic Effects of NaCl on Photosynthetic Processes in Potato Plants. Photosynth. Res. 2020, 146, 151–163. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and Validation of Reference Genes for RT-qPCR Analysis in Potato under Abiotic Stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise Optimization of Real-Time RT-PCR Analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E. An Introduction to BioPerl. Methods Mol. Biol. 2007, 406, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Duarte, J.M.; Dutta, S.; Fayazi, M.; Feng, Z.; et al. RCSB Protein Data Bank: Celebrating 50 Years of the PDB with New Tools for Understanding and Visualizing Biological Macromolecules in 3D. Protein Sci. A Publ. Protein Soc. 2022, 31, 187–208. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating γ-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants1 [OPEN]. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, M.; Song, S.; Wei, F.; Qin, L.; Fan, P.; Shi, Y.; Wang, X.; Wang, R. A Small Subunit of Geranylgeranyl Diphosphate Synthase Functions as an Active Regulator of Carotenoid Synthesis in Nicotiana Tabacum. Int. J. Mol. Sci. 2023, 24, 992. [Google Scholar] [CrossRef]
- Wang, J.; Lin, H.-X.; Su, P.; Chen, T.; Guo, J.; Gao, W.; Huang, L.-Q. Molecular Cloning and Functional Characterization of Multiple Geranylgeranyl Pyrophosphate Synthases (ApGGPPS) from Andrographis paniculata. Plant Cell Rep. 2019, 38, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Fristedt, R.; Rogers, S.; Kumar, J.; Barnes, B.; Barnes, J.; Elowsky, C.G.; Wamboldt, Y.; Mackenzie, S.A.; Redding, K.; et al. Functional Modeling Identifies Paralogous Solanesyl-Diphosphate Synthases That Assemble the Side Chain of Plastoquinone-9 in Plastids. J. Biol. Chem. 2013, 288, 27594–27606. [Google Scholar] [CrossRef]
- Ksas, B.; Becuwe, N.; Chevalier, A.; Havaux, M. Plant Tolerance to Excess Light Energy and Photooxidative Damage Relies on Plastoquinone Biosynthesis. Sci. Rep. 2015, 5, 10919. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).