Non-Invasive Preimplantation Genetic Testing
Abstract
1. The Fundamentals of Embryo Chromosomal Abnormalities
2. Identifying Chromosomal Abnormalities in Preimplantation Embryos: The Old, the New, and the Future
3. Embryonic Cell-Free DNA Analysis: How Close Are We to an niPGT Solution?
3.1. Cell-Free DNA Analysis for niPGT-A: Concordance Studies
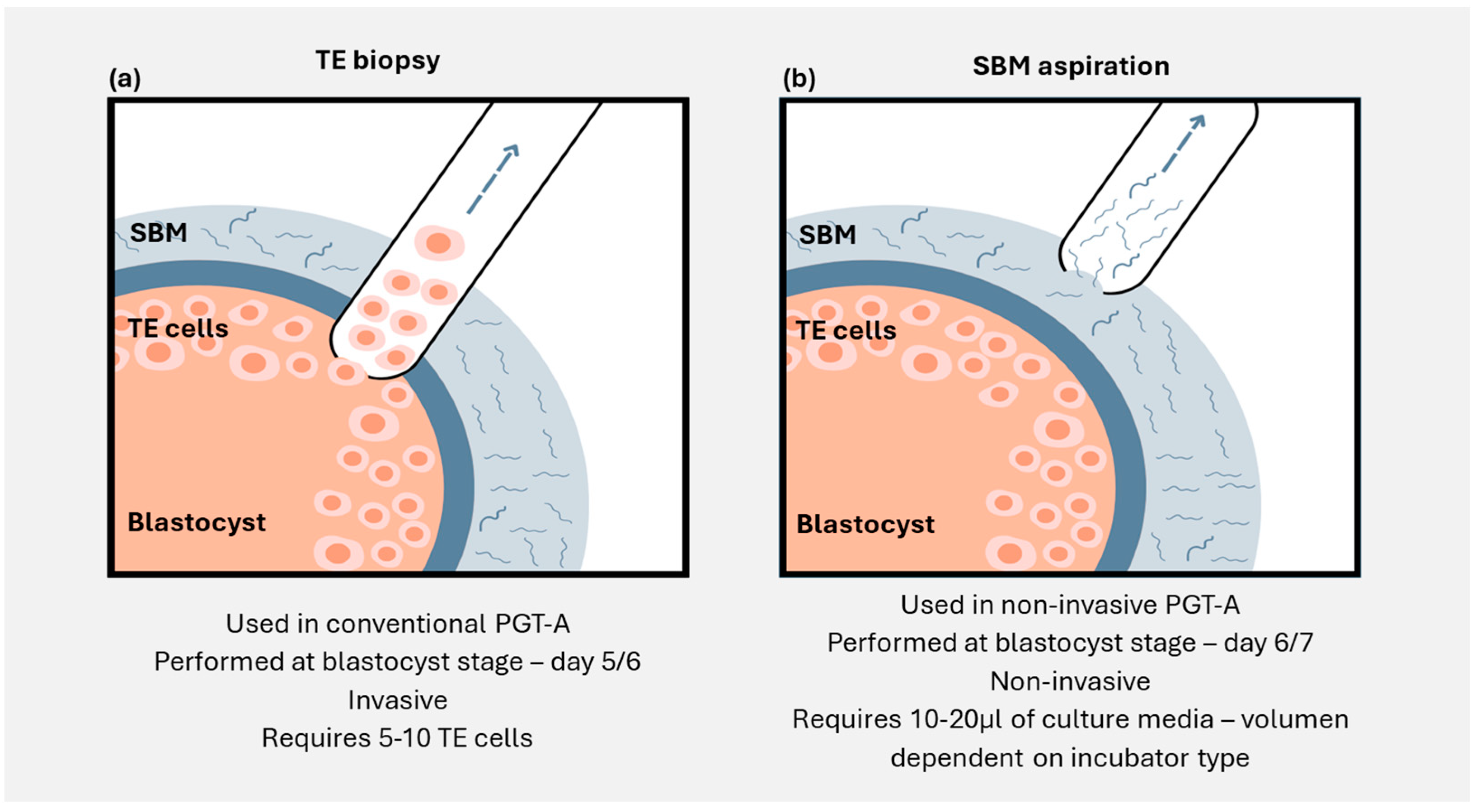
3.2. Minimally Invasive Embryo Analysis
4. Origin of Embryonic Cell-Free DNA in Spent Blastocyst Media
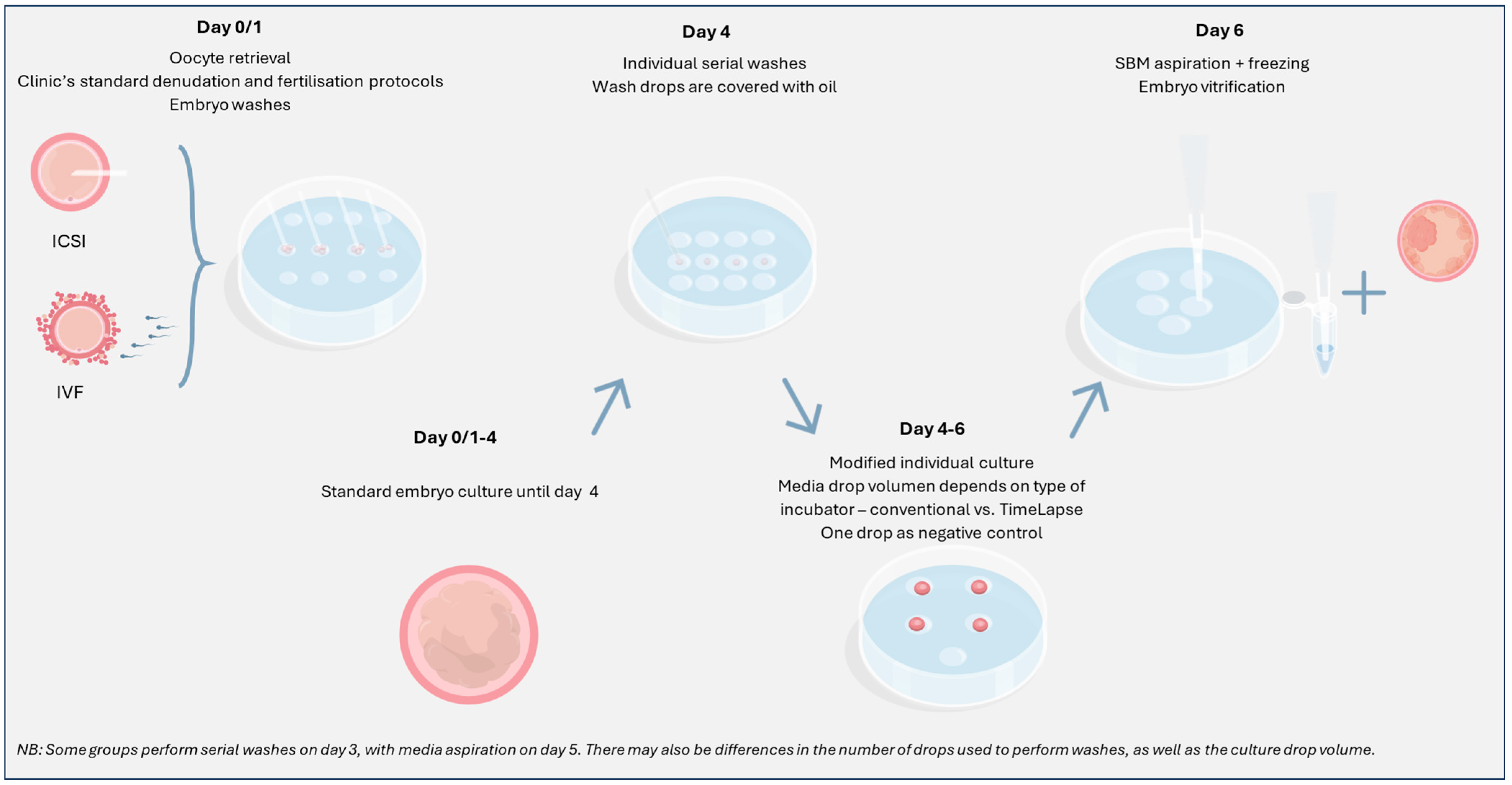
5. Methodology: Protocol Modifications
5.1. Managing Contamination
5.2. Extended Embryo Culture
5.3. Previously Vitrified Blastocysts
6. Benefits of Implementing niPGT-A
7. Barriers to Implementation of Non-Invasive PGT-A
7.1. Concordance: How High Is High Enough?
7.2. Maternal and Exogenous DNA Contamination
7.3. Result Interpretation and Clinical Guidance
8. Clinical Outcomes
9. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef]
- Cimadomo, D.; Fabozzi, G.; Vaiarelli, A.; Ubaldi, N.; Ubaldi, F.M.; Rienzi, L. Impact of maternal age on oocyte and embryo competence. Front. Endocrinol. 2018, 9, 327. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Hoffmann, E.; Cimadomo, D.; Ubaldi, F.; Rienzi, L. Human female meiosis revised: New insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum. Reprod. Update 2017, 23, 706–722. [Google Scholar] [CrossRef]
- Gabriel, A.S.; Hassold, T.; Thornhill, A.; Affara, N.; Handyside, A.; Griffin, D. An algorithm for determining the origin of trisomy and the positions of chiasmata from SNP genotype data. Chromosome Res. 2011, 19, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zong, C.; Fan, W.; Yang, M.; Li, J.; Chapman, A.; Zhu, P.; Hu, X.; Xu, L.; Yan, L.; et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 2012, 338, 1627–1630. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Behr, B.; Quake, S. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 2012, 150, 402–412. [Google Scholar] [CrossRef]
- Jimbo, M.; Kunisaki, J.; Ghaed, M.; Yu, V.; Flores, H.; Hotaling, J. Fertility in the aging male: A systematic review. Fertil. Steril. 2022, 118, 1022–1034. [Google Scholar] [CrossRef] [PubMed]
- Mazzilli, R.; Rucci, C.; Vaiarelli, A.; Cimadomo, D.; Ubaldi, F.; Foresta, C.; Ferlin, A. Male factor infertility and assisted reproductive technologies: Indications, minimum access criteria and outcomes. J. Endocrinol. Investig. 2023, 46, 1079–1085. [Google Scholar] [CrossRef]
- Franasiak, J.; Forman, E.; Hong, K.; Werner, M.; Upham, K.; Treff, N.; Scott, R. Aneuploidy across individual chromosomes at the embryonic level in trophectoderm biopsies: Changes with patient age and chromosome structure. J. Assist. Reprod. Genet. 2014, 31, 1501–1509. [Google Scholar] [CrossRef]
- Rodrigo, L.; Mateu, E.; Mercader, A.; Cobo, A.; Peinado, V.; Milán, M.; Al-Asmar, N.; Campos-Galindo, I.; García-Herrero, S.; Mir, P.; et al. New tools for embryo selection: Comprehensive chromosome screening by array comparative genomic hybridization. BioMed Res. Int. 2014, 2014, 517125. [Google Scholar] [CrossRef]
- Capalbo, A.; Ubaldi, F.; Cimadomo, D.; Maggiulli, R.; Patassini, C.; Dusi, L.; Sanges, F.; Buffo, L.; Venturella, R.; Rienzi, L. Consistent and reproducible outcomes of blastocyst biopsy and aneuploidy screening across different biopsy practitioners: A multicentre study involving 2586 embryo biopsies. Hum. Reprod. 2016, 31, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Rubio, A.; Rodrigo, L.; Simón, C. Chromosome abnormalities in human embryos. Reproduction 2020, 160, A33–A44. [Google Scholar] [CrossRef]
- Konstantinidis, M.; Prates, R.; Goodall, N.; Fischer, J.; Tecson, V.; Lemma, T.; Chu, B.; Jordan, A.; Armenti, E.; Wells, D.; et al. Live births following Karyomapping of human blastocysts: Experience from clinical application of the method. Reprod. Biomed. Online 2015, 31, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Asim, A.; Kumar, A.; Muthuswamy, S.; Jain, S.; Agarwal, S. Down syndrome: An insight of the disease. J. Biomed. Sci. 2015, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- Alfarawati, S.; Fragouli, E.; Colls, P.; Stevens, J.; Gutiérrez-Mateo, C.; Schoolcraft, W.; Katz-Jaffe, M.; Wells, D. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil. Steril. 2011, 95, 520–524. [Google Scholar] [CrossRef]
- Capalbo, A.; Rienzi, L.; Cimadomo, D.; Maggiulli, R.; Elliott, T.; Wright, G.; Nagy, Z.; Ubaldi, F. Correlation between standard blastocyst morphology, euploidy and implantation: An observational study in two centers involving 956 screened blastocysts. Hum. Reprod. 2014, 29, 1173–1181. [Google Scholar] [CrossRef]
- Swain, J. Could time-lapse embryo imaging reduce the need for biopsy and PGS? J. Assist. Reprod. Genet. 2013, 30, 1081–1090. [Google Scholar] [CrossRef]
- Melzer, K.; McCaffrey, C.; Adler, A.; Colls, P.; Munne, S.; Grifo, J. Developmental morphology and continuous time-lapse microscopy (TLM) of human embryos: Can we predict euploidy? Fertil. Steril. 2012, 98, S136. [Google Scholar] [CrossRef]
- Campbell, A.; Fishel, S.; Bowman, N.; Duffy, S.; Sedler, M.; Hickman, C. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod. Biomed. Online 2013, 26, 477–485. [Google Scholar] [CrossRef]
- Minasi, M.; Colasante, A.; Riccio, T.; Ruberti, A.; Casciani, V.; Scarselli, F.; Spinella, F.; Fiorentino, F.; Varricchio, M.; Greco, E. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: A consecutive case series study. Hum. Reprod. 2016, 31, 2245–2254. [Google Scholar] [CrossRef]
- Desai, N.; Ploskonka, S.; Goodman, L.; Attaran, M.; Goldberg, J.; Austin, C.; Falcone, T. Delayed blastulation, multinucleation, and expansion grade are independently associated with live-birth rates in frozen blastocyst transfer cycles. Fertil. Steril. 2016, 106, 1370–1378. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, J.; Salem, S.; Liu, X.; Kuang, Y.; Salem, R.; Liu, J. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: A prospective study with sibling oocytes. BMC Med. Genom. 2014, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Rienzi, L.; Capalbo, A.; Stoppa, M.; Romano, S.; Maggiulli, R.; Albricci, L.; Scarica, C.; Farcomeni, A.; Vajta, G.; Ubaldi, F. No evidence of association between blastocyst aneuploidy and morphokinetic assessment in a selected population of poor-prognosis patients: A longitudinal cohort study. Reprod. Biomed. Online 2015, 30, 57–66. [Google Scholar] [CrossRef]
- Rubio, C.; Bellver, J.; Rodrigo, L. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: A randomized, controlled study. Fertil. Steril. 2017, 107, 30254–30256. [Google Scholar] [CrossRef]
- Munne, S.; Kaplan, B.; Frattarelli, J. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: A multicenter randomized clinical trial. Fertil. Steril. 2019, 112, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Chambers, G.; Hale, L. Assisted reproductive technology (ART) cumulative live birth rates following preimplantation genetic diagnosis for aneuploidy (PGD-A) or morphological assessment of embryos: A cohort analysis. Aust. N. Z. J. Obstet. Gynaecol. 2018, 58, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Mastenbroek, S.; Twisk, M.; van der Veen, F. Preimplantation genetic screening: A systematic review and meta-analysis of RCTs. Hum. Reprod. Update 2011, 17, 454–466. [Google Scholar] [CrossRef]
- Somigliana, E.; Busnelli, A.; Paoni, A.; Vigano, P.; Riccaboni, A.; Rubio, C.; Capalbo, A. Cost-effectiveness of preimplantation genetic testing for aneuploidies. Fertil. Steril. 2019, 111, 1169–1176. [Google Scholar] [CrossRef]
- Neal, S.; Morin, S.; Franasiak, J.; Goodman, L.; Juneau, C.; Forman, E.; Werner, M.; Scott, R. Preimplantation genetic testing for aneuploidy is cost-effective, shortens treatment time, and reduces the risk of failed embryo transfer and clinical miscarriage. Fertil. Steril. 2018, 110, 896–904. [Google Scholar] [CrossRef]
- Fiorentino, F.; Biricik, A.; Bono, S.; Spizzichino, L.; Cotroneo, E.; Cottone, G.; Kokocinski, F.; Michel, C. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil. Steril. 2014, 101, 1375–1382. [Google Scholar] [CrossRef]
- Huang, J.; Yan, L.; Lu, S.; Zhao, N.; Xie, X.; Qiao, J. Validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of blastocysts. Fertil. Steril. 2016, 105, 1532–1536. [Google Scholar] [CrossRef] [PubMed]
- Friedenthal, J.; Maxwell, S.; Munné, S.; Kramer, Y.; McCulloh, D.; McCaffrey, C.; Grifo, J. Next generation sequencing for preimplantation genetic screening improves pregnancy outcomes compared with array comparative genomic hybridization in single thawed euploid embryo transfer. Fertil. Steril. 2018, 109, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Viotti, M.; Victor, A.; Barnes, F.; Zouves, C.; Besser, A.; Grifo, J.; Cheng, E.; Lee, M.; Horcajadas, J.; Corti, L.; et al. Using outcome data from one thousand mosaic embryo transfers to formulate an embryo ranking system for clinical use. Fertil. Steril. 2021, 115, 1212–1224. [Google Scholar] [CrossRef]
- Palini, S.; Galluzzi, L.; De Stefani, S.; Bianchi, M.; Wells, D.; Magnani, M.; Bulletti, C. Genomic DNA in human blastocoele fluid. Reprod. Biomed. Online 2013, 26, 603–610. [Google Scholar] [CrossRef]
- Hammond, E.; Shelling, A.; Cree, L. Nuclear and mitochondrial DNA in blastocoele fluid and embryo culture medium: Evidence and potential clinical use. Hum. Reprod. 2016, 31, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Rule, K.; Chosed, R.; Chang, A.; Wininger, D.; Roudebush, W. Relationship between blastocoel cell-free DNA and day-5 blastocyst morphology. J. Assist. Reprod. Genet. 2018, 35, 1497–1501. [Google Scholar] [CrossRef]
- Gianaroli, L.; Magli, M.; Pomante, A.; Crivello, A.; Cafueri, G.; Valerio, M.; Ferraretti, A. Blastocentesis: A source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil. Steril. 2014, 102, 1692–1699. [Google Scholar] [CrossRef]
- Tobler, K.; Zhao, Y.; Ross, R.; Benner, A.; Xu, X.; Du, L.; Broman, K.; Thrift, K.; Brezina, P.; Kearns, W. Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertil. Steril. 2015, 104, 418–425. [Google Scholar] [CrossRef]
- Magli, M.; Pomante, A.; Cafueri, G.; Valerio, M.; Crippa, A.; Ferraretti, A.; Gianaroli, L. Preimplantation genetic testing: Polar bodies, blastomeres, trophectoderm cells, or blastocoelic fluid? Fertil. Steril. 2016, 105, 676–683. [Google Scholar] [CrossRef]
- Tšuiko, O.; Zhigalina, D.; Jatsenko, J.; Skryabin, N.; Kanbekova, O.; Artyukhova, V.; Svetlakov, A.; Teearu, K.; Trošin, A.; Salumets, A.; et al. Karyotype of the blastocoel fluid demonstrates low concordance with both trophectoderm and inner cell mass. Fertil. Steril. 2018, 109, 1127–1134. [Google Scholar] [CrossRef]
- Magli, M.; Albanese, C.; Crippa, A.; Tabanelli, C.; Ferraretti, A.; Gianaroli, L. Deoxyribonucleic acid detection in blastocoelic fluid: A new predictor of embryo ploidy and viable pregnancy. Fertil. Steril. 2019, 111, 77–85. [Google Scholar] [CrossRef]
- Zhang, W.; von Versen-Höynck, F.; Kapphahn, K.; Fleischmann, R.; Zhao, Q.; Baker, V. Maternal and neonatal outcomes associated with trophectoderm biopsy. Fertil. Steril. 2019, 112, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kort, J.; Baker, V. Embryo biopsy and perinatal outcomes of singleton pregnancies: An analysis of 16,246 frozen embryo transfer cycles reported in the Society for Assisted Reproductive Technology Clinical Outcomes Reporting System. Am. J. Obstet. Gynecol. 2021, 224, 500.e1–500.e18. [Google Scholar] [CrossRef] [PubMed]
- Makhijani, R.; Bartels, C.; Godiwala, P.; Bartolucci, A.; DiLuigi, A.; Nulsen, J.; Grow, D.; Benadiva, C.; Engmann, L. Impact of trophectoderm biopsy on obstetric and perinatal outcomes following frozen-thawed embryo transfer cycles. Hum. Reprod. 2021, 36, 340–348. [Google Scholar] [CrossRef]
- Alteri, A.; Cermisoni, G.C.; Pozzoni, M.; Gaeta, G.; Cavoretto, P.; Viganò, P. Obstetric, neonatal, and child health outcomes following embryo biopsy for preimplantation genetic testing. Hum. Reprod. Update 2023, 29, 291–306. [Google Scholar] [CrossRef]
- Scott, R.; Upham, K.; Forman, E.; Zhao, T.; Treff, N. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: A randomized and paired clinical trial. Fertil. Steril. 2013, 100, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Forman, E.; Hong, K.; Franasiak, J.; Scott, R.T., Jr. Obstetrical and neonatal outcomes from the BEST Trial: Single embryo transfer with aneuploidy screening improves outcomes after in vitro fertilization without compromising delivery rates. Am. J. Obstet. Gynecol. 2014, 69, 742–744. [Google Scholar]
- Awadalla, M.; Park, K.; Latack, K.; McGinnis, L.; Ahmady, A.; Paulson, R. Influence of trophectoderm biopsy prior to frozen blastocyst transfer on obstetrical outcomes. Reprod. Sci. 2021, 28, 3459–3465. [Google Scholar] [CrossRef]
- Stigliani, S.; Anserini, P.; Venturini, P.; Scaruffi, P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum. Reprod. 2013, 28, 2652–2660. [Google Scholar] [CrossRef]
- Stigliani, S.; Persico, L.; Lagazio, C.; Anserini, P.; Venturini, P.; Scaruffi, P. Mitochondrial DNA in Day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Mol. Hum. Reprod. 2014, 20, 1238–1246. [Google Scholar] [CrossRef]
- Galluzzi, L.; Palini, S.; Stefani, S.; de Andreoni, F.; Primiterra, M.; Diotallevi, A. Extracellular embryo genomic DNA and its potential for genotyping applications. Future Sci. OA 2015, 1, FSO62. [Google Scholar] [CrossRef]
- Wu, H.; Ding, C.; Shen, X.; Wang, J.; Li, R.; Cai, B. Medium-based noninvasive preimplantation genetic diagnosis for human α-thalassemia-SEA. Medicine 2015, 94, e669. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Du, H.; Ling, J.; Sun, X.; Chen, D. Non-invasive pre-implantation aneuploidy screening and diagnosis of beta thalassemia IVSII654 mutation using spent embryo culture medium. Ann. Med. 2017, 49, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Romanelli, V.; Patassini, C.; Poli, M.; Girardi, L.; Giancani, A. Diagnostic efficacy of blastocoele fluid and spent media as sources of DNA for preimplantation genetic testing in standard clinical conditions. Fertil. Steril. 2018, 110, 870–879. [Google Scholar] [CrossRef]
- Xu, J.; Fang, R.; Chen, L.; Chen, D.; Xiao, J.; Yang, W. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilisation. Proc. Natl. Acad. Sci. USA 2016, 113, 11907–11912. [Google Scholar] [CrossRef] [PubMed]
- Shamonki, M.; Jin, H.; Haimowitz, Z.; Liu, L. Proof of concept: Preimplantation genetic screening without embryo biopsy through analysis of cell-free DNA in spent embryo culture media. Fertil. Steril. 2016, 106, 1312–1318. [Google Scholar] [CrossRef]
- Vera-Rodriguez, M.; Diez-Juan, A.; Jimenez-Almazan, J.; Martinez, S.; Navarro, R.; Peinado, V.; Mercader, A.; Meseguer, M.; Blesa, D.; Moreno, I.; et al. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum. Reprod. 2018, 33, 745–756. [Google Scholar] [CrossRef]
- Ho, J.; Arrach, N.; Rhodes-Long, K.; Ahmady, A.; Ingles, S.; Chung, K.; Bendikson, K.; Paulson, R.; McGinnis, L. Pushing the limits of detection: Investigation of cell-free DNA for aneuploidy screening in embryos. Fertil. Steril. 2018, 110, 467–475. [Google Scholar] [CrossRef]
- Huang, L.; Bogale, B.; Tang, Y.; Lu, S.; Xie, X.; Racowsky, C. Noninvasive preimplantation genetic testing for aneuploidy in spent medium may be more reliable than trophectoderm biopsy. Proc. Natl. Acad. Sci. USA 2019, 116, 14105–14112. [Google Scholar] [CrossRef]
- Yeung, Q.; Zhang, Y.; Chung, J.; Lui, W.; Kwok, Y.; Gui, B.; Kong, G.; Cao, Y.; Li, T.; Choy, K. A prospective study of non-invasive preimplantation genetic testing for aneuploidies (NiPGT-A) using next-generation sequencing (NGS) on spent culture media (SCM). J. Assist. Reprod. Genet. 2019, 36, 1609–1621. [Google Scholar] [CrossRef]
- Rubio, C.; Rienzi, L.; Navarro-Sánchez, L.; Cimadomo, D.; García-Pascual, C.; Albricci, L.; Soscia, D.; Valbuena, D.; Capalbo, A.; Ubaldi, F.; et al. Embryonic cell-free DNA versus trophectoderm biopsy for aneuploidy testing: Concordance rate and clinical implications. Fertil. Steril. 2019, 112, 510–519. [Google Scholar] [CrossRef]
- Rubio, C.; Navarro-Sánchez, L.; García-Pascual, C.; Ocali, O.; Cimadomo, D.; Venier, W.; Barroso, G.; Kopcow, L.; Bahçeci, M.; Kulmann, M.; et al. Multicenter prospective study of concordance between embryo cell-free DNA and trophectoderm biopsies from 1,301 human blastocysts. Am. J. Obstet. Gynecol. 2020, 223, 751.e1–751.e13. [Google Scholar] [CrossRef] [PubMed]
- Lledo, B.; Morales, R.; Ortiz, J.; Rodriguez-Arnedo, A.; Ten, J.; Castillo, J.; Bernabeu, A.; Llacer, J.; Bernabeu, R. Consistent results of non-invasive PGT-A of human embryos using two different techniques for chromosomal analysis. Reprod. Biomed. Online 2021, 42, 555–563. [Google Scholar] [CrossRef]
- Shitara, A.; Takahashi, K.; Goto, M.; Takahashi, H.; Iwasawa, T.; Onodera, Y.; Makino, K.; Miura, H.; Shirasawa, H.; Sato, W.; et al. Cell-free DNA in spent culture medium effectively reflects the chromosomal status of embryos following culturing beyond implantation compared to trophectoderm biopsy. PLoS ONE 2021, 16, e0246438. [Google Scholar] [CrossRef]
- Hanson, A.; Tao, X.; Hong, K.; Comito, C.; Pangasnan, R.; Seli, E.; Jalas, C.; Scott, R. Noninvasive preimplantation genetic testing for aneuploidy exhibits high rates of deoxyribonucleic acid amplification failure and poor correlation with results obtained using trophectoderm biopsy. Fertil. Steril. 2021, 115, 1461–1470. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Jia, J.; Chang, L.; Liu, P.; Qiao, J.; Tang, F.; Wen, L. DNA methylome reveals cellular origin of cell-free DNA in spent medium of human preimplantation embryos. J. Clin. Investig. 2021, 131, e146051. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Fu, J.; Li, X.; Zhou, J.; Xiao, M.; Zhang, S.; Sun, Y.; Sun, X. Re-denudation of residual cumulus cells on day 3 increases the accuracy of cell-free DNA detection in spent embryo culture medium. J. Assist. Reprod. Genet. 2022, 39, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Zhang, S.; Gu, Y.; Jiang, B.; Hu, L.; Tan, Y.; Yao, Y.; Tang, Y.; Wan, A.; Cai, S.; et al. Non-invasive preimplantation genetic testing for conventional IVF blastocysts. J. Transl. Med. 2022, 20, 396. [Google Scholar] [CrossRef]
- Handayani, N.; Aubry, D.; Boedino, A.; Bowolaksono, A.; Sini, I.; Haq, N.; Sirait, B.; Periastiningrum, G.; Mutia, K.; Wiwko, B. Non-invasive pre-implantation genetic testing’s reliability for aneuploidy using cell-free DNA in embryo culture media. J. Gynecol. Obstet. Hum. Reprod. 2024, 53, 102808. [Google Scholar] [CrossRef]
- Feichtinger, M. Non-invasive preimplantation genetic screening using array comparative genomic hybridization on spent culture media: A proof-of-concept pilot study. Reprod. Biomed. Online 2017, 34, 583–589. [Google Scholar] [CrossRef]
- Sonehara, H.; Matsumoto, R.; Nakayama, N.; Kobanawa, M.; Numata, K.; Kawasaki, A.; Shozu, M. Aneuploidy and sex concordance rate between cell-free DNA analysis from spent culture media of preimplantation embryo and DNA from whole embryo with respect to different morphological grading. Reprod. Med. Biol. 2022, 21, e12493. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Zhang, H.; Xie, J.; Wei, Y.; Zhang, C.; Meng, L. Validation of preimplantation genetic tests for aneuploidy (PGT-A) with DNA from spent culture media (SCM): Concordance assessment and implication. Reprod. Biol. Endocrinol. 2021, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wei, Y.; Tan, Q.; Huang, Y.; Wu, J.; Li, C.; Ma, Y.; Zhou, L.; Liang, B.; Kong, L.; et al. Concordance of PGT for aneuploidies between blastocyst biopsies and spent blastocyst culture medium. Reprod. Biomed. Online 2023, 46, 483–490. [Google Scholar] [CrossRef]
- Takeuchi, H.; Morishita, M.; Uemura, M.; Maezawa, T.; Shibahara, T.; Takayama, E.; Nishioka, M.; Kondo, E.; Minoura, H.; Ikeda, T. Conditions for improved accuracy of noninvasive preimplantation genetic testing for aneuploidy: Focusing on the zona pellucida and early blastocysts. Reprod. Med. Biol. 2024, 23, e12604. [Google Scholar] [CrossRef]
- Ardestani, G.; Banti, M.; Garcia-Pascual, C.; Navarro-Sanchez, L.; Zyl, E.; Castellon, J.; Simon, C.; Sakkas, D.; Rubio, C. Culture time to optimise embryo cell-free DNA analysis for frozen-thawed blastocysts undergoing noninvasive preimplantation genetic testing for aneuploidy. Fertil. Steril. 2024, 122, 465–473. [Google Scholar] [CrossRef]
- Kuznyetsov, V.; Madjunkova, S.; Antes, R.; Abramov, R.; Motamedi, G.; Ibarrientos, Z.; Librach, C. Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS ONE 2018, 13, e0197262. [Google Scholar] [CrossRef]
- Li, P.; Song, Z.; Yao, Y.; Huang, T.; Mao, R.; Huang, J.; Ma, Y.; Dong, X.; Huang, W.; Huang, J.; et al. Preimplantation genetic screening with spent culture medium/blastocoel fluid for in vitro fertilization. Sci. Rep. 2018, 8, 9275. [Google Scholar]
- Jiao, J.; Shi, B.; Sagnelli, M.; Yang, D.; Yao, Y.; Li, W.; Shao, L.; Lu, S.; Li, D.; Wang, X. Minimally invasive preimplantation genetic testing using blastocyst culture medium. Hum. Reprod. 2019, 34, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xia, H.; Chen, H.; Yao, C.; Feng, L.; Song, X.; Bai, X. Less-invasive chromosome screening of embryos and embryo assessment by genetic studies of DNA in embryo culture medium. J. Assist. Reprod. Genet. 2019, 36, 2505–2513. [Google Scholar] [CrossRef]
- Kuznyetsov, V.; Madjunkova, S.; Abramov, R.; Antes, R.; Ibarrientos, Z.; Motamedi, G.; Zaman, A.; Kuznyetsova, I.; Librach, C. Minimally invasive cell-free human embryo aneuploidy testing (miPGT-A) utilizing combined spent embryo culture medium and blastocoel fluid -towards development of a clinical assay. Sci. Rep. 2020, 10, 7244. [Google Scholar] [CrossRef]
- Chen, J.; Jia, L.; Li, T.; Guo, Y.; He, S.; Zhang, Z.; Su, W.; Zhang, S.; Fang, C. Diagnostic efficiency of blastocyst culture medium in noninvasive preimplantation genetic testing. Fertil. Steril. 2020, 2, 88–94. [Google Scholar] [CrossRef]
- Li, X.; Hao, Y.; Chen, D.; Ji, D.; Zhu, W.; Zhu, X.; Wei, Z.; Cao, Y.; Zhang, Z.; Zhou, P. Non-invasive preimplantation genetic testing for putative mosaic blastocysts: A pilot study. Hum. Reprod. 2021, 36, 2020–2034. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.R.; McGillivray, B.; Wicker, S.; Peek, J.; Shelling, A.; Stone, P.; Chamley, L.; Cree, L. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: Genetic contamination identified. Fertil. Steril. 2017, 107, 220–228. [Google Scholar] [CrossRef]
- Hardy, K.; Spanos, S.; Becker, D.; Iannelli, P.; Winston, R.; Stark, J. From cell death to embryo arrest: Mathematical models of human preimplantation embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Vyas, P.; Balakier, H.; Librach, C. Ultrastructural identification of CD9 positive extracellular vesicles released from human embryos and transported through the zona pellucida. Syst. Biol. Reprod. Med. 2019, 65, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Veraguas, D.; Aguilera, C.; Henriquez, C. Evaluation of extracellular vesicles and gDNA from culture medium as a possible indicator of developmental competence in human embryos. Zygote 2021, 29, 138–149. [Google Scholar] [CrossRef]
- Domingo-Muelas, A.; Skory, R.; Moverley, A. Human embryo live imaging reveals nuclear DNA shedding during blastocyst expansion and biopsy. Cell 2023, 186, 3166–3181. [Google Scholar] [CrossRef]
- Navarro-Sánchez, L.; Ocali, O.; García-Pascual, C.; Andrade, G.M.; Salom, D.C.; Lai, F.; Dutra, C.G.; Rubio, C.; Simon, C.; Frantz, N.; et al. High concordance of the embryonic cell-free DNA with the inner cell mass: Impact of blastocyst quality, patient age and mode of fertilization. Hum. Reprod. 2022, 37, 551. [Google Scholar] [CrossRef]
- Leaver, M.; Wells, D. Non-invasive preimplantation genetic testing (niPGT): The next revolution in reproductive genetics? Hum. Reprod. Update 2020, 26, 16–42. [Google Scholar] [CrossRef]
- Chow, J.; Lam, K.; Cheng, H.; Lai, S.; Yeung, W.; Ng, E. Optimising non-invasive preimplantation genetic testing: Investigating culture conditions, sample collection, and IVF treatment for improved non-invasive PGT-A results. J. Assist. Reprod. Genet. 2024, 41, 465–472. [Google Scholar] [CrossRef]
- Sakkas, D.; Navarro-Sánchez, L.; Ardestani, G. The impact of implementing a non-invasive preimplantation genetic testing for aneuploidies (niPGT-A) embryo culture protocol on embryo viability and clinical outcomes. Hum. Reprod. 2024, 39, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Kaye, L.; Will, E.; Bartolucci, A.; Nulsen, J.; Benadiva, C.; Engmann, L. Pregnancy rates for single embryo transfer (SET) of day 5 and day 6 blastocysts after cryopreservation by vitrification and slow freeze. J. Assist. Reprod. Genet. 2017, 34, 913–919. [Google Scholar] [CrossRef]
- Tiegs, A.; Sun, L.; Patounakis, G.; Scott, R. Worth the wait? Day 7 blastocysts have lower euploidy rates but similar sustained implantation rates as day 5 and day 6 blastocysts. Hum. Reprod. 2019, 34, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, X.V.; Odia, R.; Naja, R.; Serhal, P.; Saab, W.; Seshadri, S.; Ben-Nagi, J. Euploid blastocysts implant irrespective of their morphology after NGS-(PGT-A) testing in advanced maternal age patients. J. Assist. Reprod. Genet. 2019, 36, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Shear, M.; Vaughan, D.; Modest, A.; Seidler, E.; Leung, A.; Hacker, M.; Sakkas, D.; Penzias, A. Blasts from the past: Is morphology useful in PGT-A tested and untested frozen embryo transfers? Reprod. Biomed. Online 2020, 41, 981–989. [Google Scholar] [CrossRef]
- Franco, J.; de Albornoz-Riaza, E.C.; Villa-Milla, A.; Gay-Fernandez-Vegue, R.; Sotos-Borras, F.; de Albornoz, A.V.-C.; Martinez-Acera, A.; Bueno-Olalla, B.; Iniesta-Perez, S.; Melia-Fullana, E.; et al. Comparative analysis of non-invasive preimplantation genetic testing of aneuploidies (niPGT-A), PGT-A and IVF cycles without aneuploidy testing: Preliminary results. Hum. Reprod. 2021, 36, 560. [Google Scholar] [CrossRef]
- Maggiulli, R.; Cimadomo, D.; Giancani, A.; Soscia, D.; Albricci, L.; Rubio, C.; Pascual, C.G.; Sanchez, L.N.; Capalbo, A.; Simon, C.; et al. VF culture media refresh in a reduced volume on day 4 aimed at improving non-invasive embryo selection does not affect embryo competence: A prospective analysis of 2605 embryos. Reprod. Biomed. Online 2022, 45, e30–e31. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, Y.; Yan, J.; Wu, Y.; Zhuang, X.; Lin, S.; Zhu, J.; Lian, Y.; Qiao, J.; Liu, P. Effect of repeated cryopreservation on human embryo developmental potential. Reprod. Biomed. Online 2017, 35, 627–632. [Google Scholar] [CrossRef]
- Aluko, A.; Vaughan, D.A.; Modest, A.M.; Penzias, A.; Hacker, M.R.; Thornton, K.; Sakkas, D. Multiple cryopreservation-warming cycles, coupled with blastocyst biopsy, negatively affect IVF outcomes. Reprod. Biomed. Online 2021, 42, 572–578. [Google Scholar] [CrossRef]
- Al Hashimi, A.; Linara-Demakakou, E.; Harvey, S.; Harvey, K.; Griffin, D.; Ahuja, K.; Macklon, N. Double vitrification and warming of blastocysts does not affect pregnancy, miscarriage, or live birth rates. Reprod. Biomed. Online 2024, 49, 104103. [Google Scholar] [CrossRef]
- Theodorou, E.; Jones, B.; Armas, D.; Heath, C.; Serhal, P.; Ben-Nagi, J. Live birth rate following a euploid blastocyst transfer is not affected by double vitrification and warming at cleavage or blastocyst stage. J. Assist. Reprod. Genet. 2022, 39, 987–993. [Google Scholar] [CrossRef] [PubMed]
- von Wülfingen, A.B. Contested change: How Germany came to allow PGD. Reprod. Biomed. Soc. Online 2016, 13, 60–67. [Google Scholar] [CrossRef]
- Hreinsson, J.; Lundin, K.; Iwarsson, E.; Hausken, J.; Einarsson, S.; Grøndahl, M.; Hydén-Granskog, C.; Ingerslev, H. Preimplantation genetic testing legislation and accessibility in the Nordic countries. Acta Obstet. Gynecol. Scand. 2020, 99, 716–721. [Google Scholar] [CrossRef]
- Fang, R.; Yang, W.; Zhao, X.; Xiong, F.; Guo, C.; Xiao, J.; Chen, L.; Song, X.; Wang, H.; Chen, J.; et al. Chromosome screening using culture medium of embryos fertilised in vitro: A pilot clinical study. J. Transl. Med. 2019, 17, 73. [Google Scholar] [CrossRef]
- Minasi, M.; Fabozzi, G.; Casciani, V.; Lobascio, A.; Colasante, A.; Scarselli, F.; Greco, E. Improved blastocyst formation with reduced culture volume: Comparison of three different culture conditions on 1128 sibling human zygotes. J. Assist. Reprod. Genet. 2015, 32, 215–220. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, P.; Lan, F.; Yao, Y.; Hu, L.; Tan, Y.; Jiang, B.; Wan, A.; Zhao, D.; Gong, F.; et al. Conventional IVF is feasible in preimplantation genetic testing for aneuploidy. J. Assist. Reprod. Genet. 2023, 40, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Poli, M.; Rienzi, L.; Girardi, L.; Patassini, C.; Fabiani, M.; Cimadomo, D.; Benini, F.; Farcomeni, A.; Cuzzi, J.; et al. Mosaic human preimplantation embryos and their developmental potential in a prospective, non-selection clinical trial. Am. J. Hum. Genet. 2021, 108, 2238–2247. [Google Scholar] [CrossRef] [PubMed]
- Chavli, E. Single-cell DNA sequencing reveals a high incidence of chromosomal abnormalities in human blastocysts. J. Clin. Investig. 2024, 134, e174483. [Google Scholar] [CrossRef]
- Papavassiliou, P.; Charalsawadi, C.; Rafferty, K.; Jackson-Cook, C. Mosaicism for trisomy 21: A review. J. Med. Genet. Part A 2015, 167, 26–39. [Google Scholar] [CrossRef]
- Spinillo, S. Pregnancy outcome of confined placental mosaicism: Meta-analysis of cohort studies. Am. J. Obstet. Gynecol. 2022, 227, 714–727. [Google Scholar] [CrossRef]
- Chen, R.; Tang, N.; Du, H.; Yso, Y.; Zou, Y.; Wang, J.; Zhao, D.; Zhou, X.; Luo, Y.; Li, L.; et al. Clinical application of noninvasive chromosomal screening for elective single-blastocyst transfer in frozen-thawed cycles. J. Transl. Med. 2022, 20, 553. [Google Scholar] [CrossRef] [PubMed]
- Nakhuda, G.; Rodriguez, S.; Tormasi, S.; Welch, C. A pilot study to investigate the clinically predictive values of copy number variations detected by next-generation sequencing of cell-free deoxyribonucleic acid in spent culture media. Fertil. Steril. 2024, 122, 0015–0282. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Xu, J.; Yao, Y.; Huang, X.; Zhao, D.; Lu, S.; Yao, B.; Chen, L. Efficacy of non-invasive chromosome screening, preimplantation genetic testing for aneuploidy, and morphological grading in selecting embryos of patients with advanced maternal age: A three-armed prospective cohort study. BMC Pregnancy Childbirth 2024, 24, 545. [Google Scholar] [CrossRef]
- Ocali, O. The impact of implementing a noninvasive preimplantation genetic testing for aneuploidy (niPGT-A) protocol on outcomes. Fertil. Steril. 2021, 116, e390. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakalova, D.N.; Navarro-Sánchez, L.; Rubio, C. Non-Invasive Preimplantation Genetic Testing. Genes 2025, 16, 552. https://doi.org/10.3390/genes16050552
Bakalova DN, Navarro-Sánchez L, Rubio C. Non-Invasive Preimplantation Genetic Testing. Genes. 2025; 16(5):552. https://doi.org/10.3390/genes16050552
Chicago/Turabian StyleBakalova, Daniela N., Luis Navarro-Sánchez, and Carmen Rubio. 2025. "Non-Invasive Preimplantation Genetic Testing" Genes 16, no. 5: 552. https://doi.org/10.3390/genes16050552
APA StyleBakalova, D. N., Navarro-Sánchez, L., & Rubio, C. (2025). Non-Invasive Preimplantation Genetic Testing. Genes, 16(5), 552. https://doi.org/10.3390/genes16050552






