Genetic Variations and Haplotype Diversity of the Wheat FRIZZY PANICLE (WFZP) Gene in 98 Aegilops tauschii Accessions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Primer Design, Plant DNA Amplification and Sequencing
2.3. Protein 3D Modeling Prediction
2.4. Sequence Alignments and Phylogenetic Study
3. Results
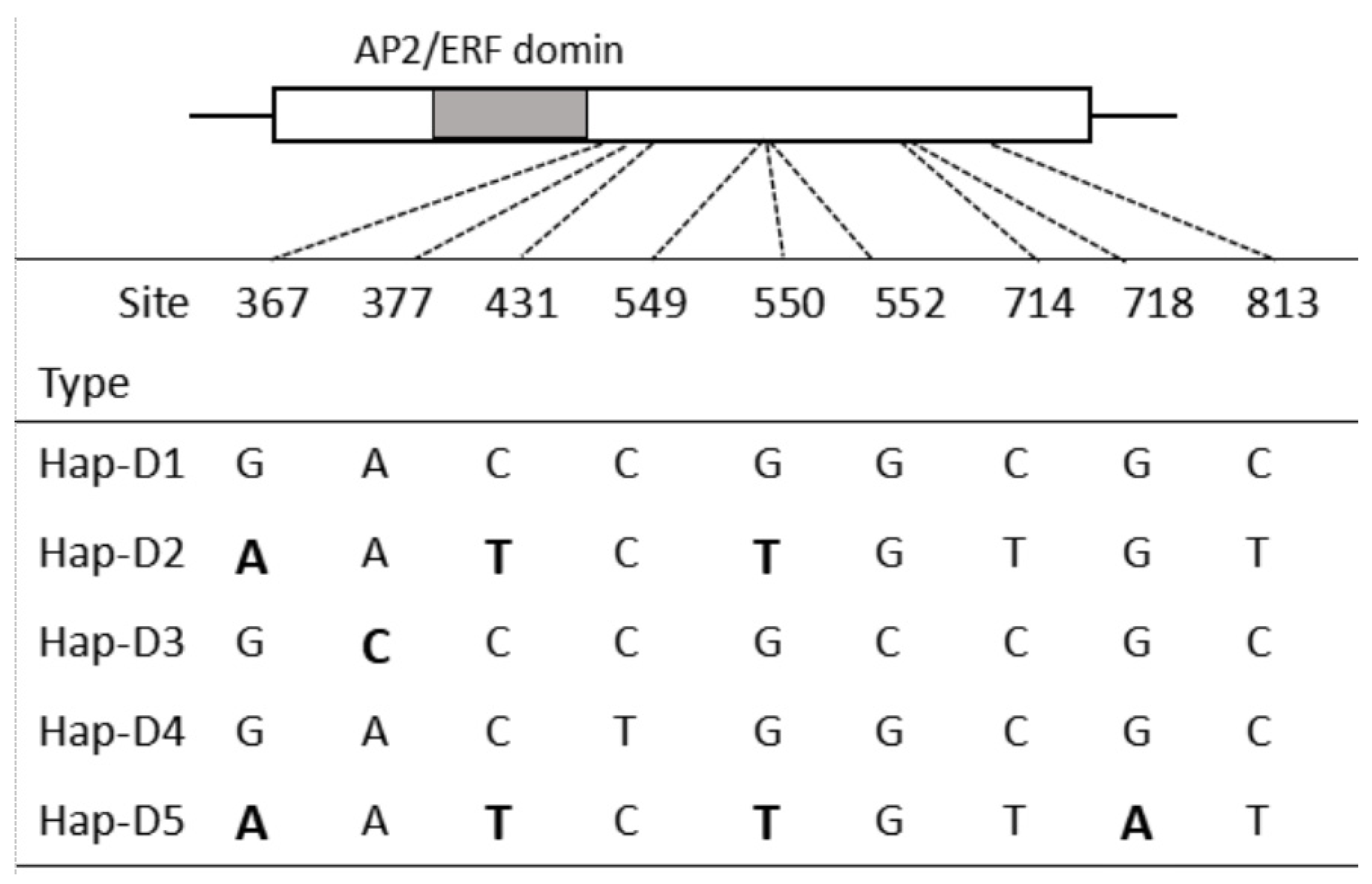
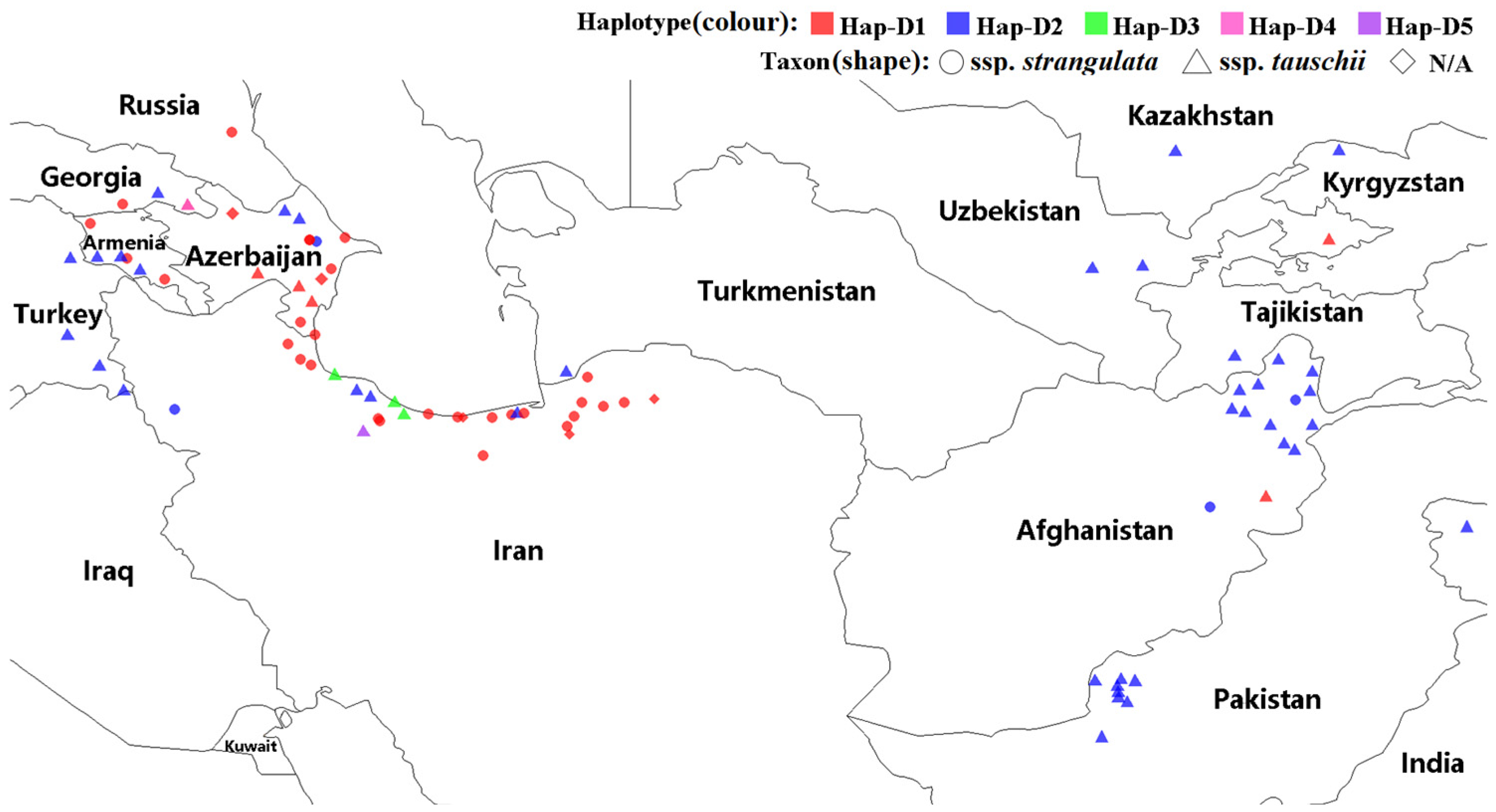
3.1. Ae. tauschii WFZP-D: Nucleotide Sequence Polymorphism
3.2. Ae. tauschii WFZP-D: Peptide Sequences
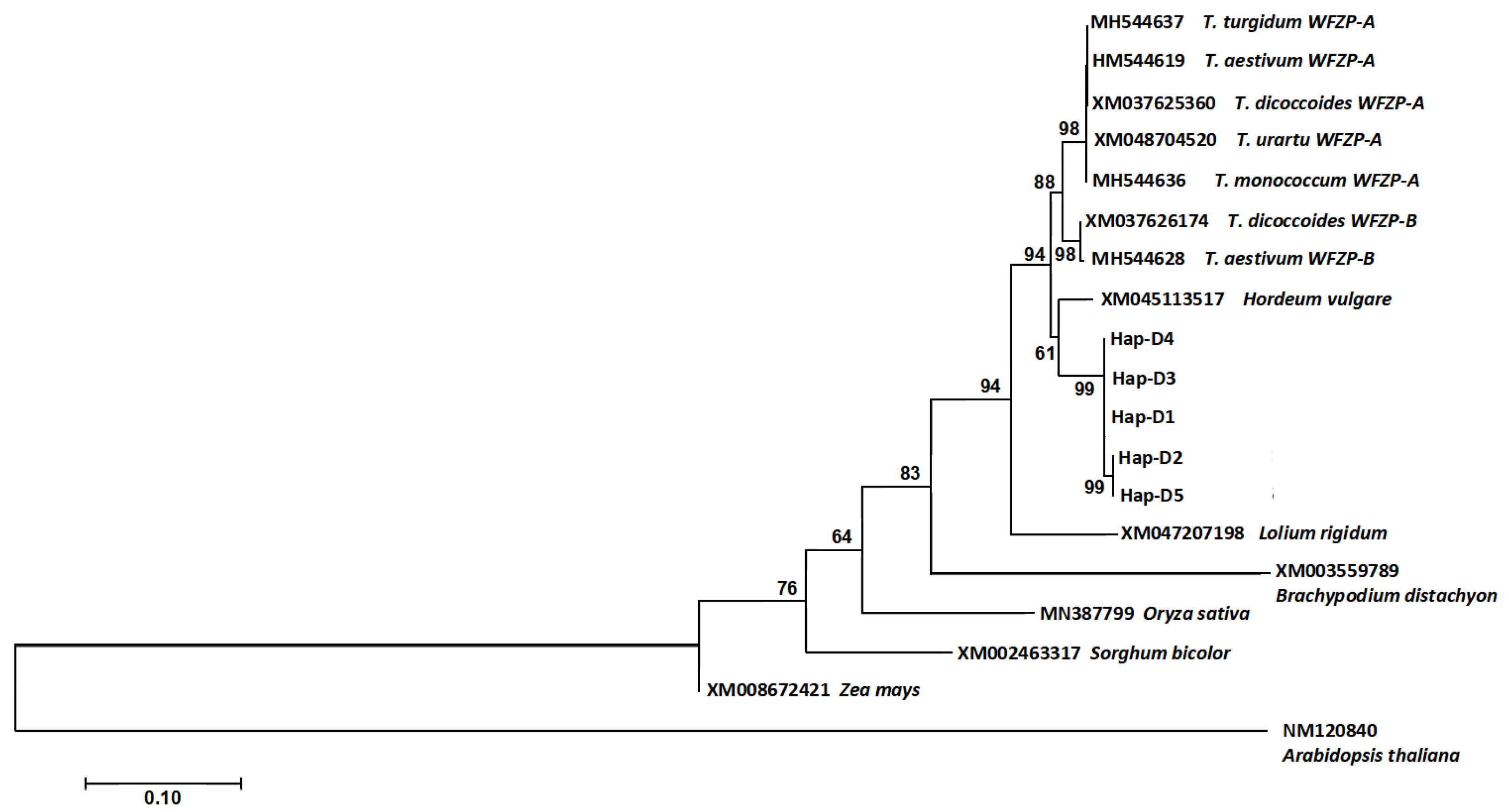
3.3. Phylogeny of the WFZP Homoeologs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakuma, S.; Schnurbusch, T. Of foral fortune: Tinkering with the grain yield potential of cereal crops. New Phytol. 2020, 225, 1873–1882. [Google Scholar]
- Zhang, J.; Gizaw, S.; Bossolini, E.; Hegarty, J.; Howell, T.; Carter, A.; Akhunov, E.; Dubcovsky, J. Identification and validation of QTL for grain yield and plant water status under contrasting water treatments in fall-sown spring wheats. Theor. Appl. Genet. 2018, 131, 1741–1759. [Google Scholar]
- Boden, S.A.; Cavanagh, C.; Cullis, B.R.; Ramm, K.; Greenwood, J.; Finnegan, E.J.; Trevaskis, B.; Swain, S.M. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plants 2015, 1, 14016. [Google Scholar]
- Zhang, J.; Burguener, G.F.; Paraiso, F.; Dubcovsky, J. Natural alleles of LEAFY and WAPO1 interact to regulate spikelet number per spike in wheat. Theor. Appl. Genet. 2024, 137, 257. [Google Scholar]
- Poursarebani, N.; Seidensticker, T.; Koppolu, R.; Trautewig, C.; Gawroński, P.; Bini, F.; Govind, G.; Rutten, T.; Sakuma, S.; Tagiri, A.; et al. The genetic basis of composite spike form in barley and ‘Miracle-wheat’. Genetics 2015, 201, 155–165. [Google Scholar]
- Poursarebani, N.; Trautewig, C.; Melzer, M.; Nussbaumer, T.; Lundqvist, U.; Rutten, T.; Schmutzer, T.; Brandt, R.; Himmelbach, A.; Altschmied, L.; et al. COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 2020, 11, 5138. [Google Scholar] [CrossRef]
- González, F.G.; Slafer, G.A.; Miralles, D.J. Pre-anthesis development and number of fertile florets in wheat as affected by photoperiod sensitivity genes Ppd-D1 and Ppd-B1. Euphytica 2005, 146, 253–269. [Google Scholar] [CrossRef]
- Ochagavía, H.; Prieto, P.; Savin, R.; Griffiths, S.; Slafer, G.A. Dynamics of leaf and spikelet primordia initiation in wheat as affected by Ppd-1a alleles under field conditions. J. Exp. Bot. 2018, 69, 2621–2631. [Google Scholar] [CrossRef]
- Ning, S.; Zhao, L.; Li, S.; Li, S.; Zang, T.; Liu, Y.E.; Yang, H.; Chen, X.; Chen, X.; Yi, Y.; et al. Delays in heading and improvements in both spikelet number and spike length are associated with the Aegilops tausschii photoperiod-sensitive ppd-D1b allele. Cereal Res. Commun. 2023, 51, 593–601. [Google Scholar]
- Chen, Z.; Ke, W.; He, F.; Chai, L.; Cheng, X.; Xu, H.; Wang, X.; Du, D.; Zhao, Y.; Chen, X.; et al. A single nucleotide deletion in the third exon of FT-D1 increases the spikelet number and delays heading date in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2022, 20, 920–933. [Google Scholar]
- Li, Y.; Xiong, H.; Guo, H.; Xie, Y.; Zhao, L.; Gu, J.; Li, H.; Zhao, S.; Ding, Y.; Zhou, C.; et al. A gain-of-function mutation at the C-terminus of FT-D1 promotes heading by interacting with 14-3-3A and FDL6 in wheat. Plant Biotechnol. J. 2025, 23, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Wu, F.; Zhou, W.; Li, J.; Shi, H.; Wang, Z.; Lin, Y.; Yang, X.; Wei, Y.; Zheng, Y.; et al. Mapping of QTL for total spikelet number per spike on chromosome 2D in wheat using a high-density genetic map. Genet. Mol. Biol. 2019, 42, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ding, P.; Liu, J.; Li, T.; Zou, Y.; Habib, A.; Mu, Y.; Tang, H.; Jiang, Q.; Liu, Y.; et al. Identification and validation of a major and stably expressed QTL for spikelet number per spike in bread wheat. Theor. Appl. Genet. 2019, 132, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, H.; Zhou, X.; Mou, Y.; Zhang, Y.; Yu, L.; Chen, X.; Wu, F.; Zhou, H.; Lin, Y.; et al. Identification, validation and candidate gene analysis of major QTL for Supernumerary spikelets in wheat. BMC Genom. 2024, 25, 675. [Google Scholar] [CrossRef]
- Dobrovolskaya, O.; Pont, C.; Sibout, R.; Martinek, P.; Badaeva, E.; Murat, F.; Chosson, A.; Watanabe, N.; Prat, E.; Gautier, N.; et al. FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol. 2015, 167, 189–199. [Google Scholar] [CrossRef]
- Du, D.; Zhang, D.; Yuan, J.; Feng, M.; Li, Z.; Wang, Z.; Zhang, Z.; Li, X.; Ke, W.; Li, R.; et al. FRIZZY PANICLE defines a regulatory hub for simultaneously controlling spikelet formation and awn elongation in bread wheat. New Phytol. 2021, 231, 814–833. [Google Scholar] [CrossRef]
- Kihara, H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Biol. Agric. Hortic. 1944, 19, 889–890. [Google Scholar]
- McFadden, E.S.; Sears, E.R. The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 1944, 13, 26–27. [Google Scholar]
- McFadden, E.S.; Sears, E.R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 1946, 37, 81–89. [Google Scholar] [CrossRef]
- Yen, C.; Yang, J.; Yuan, Z.; Ning, S.; Zhang, L.; Hao, M.; Liu, D. Biosystematics of Triticeae: Volume I. Triticum-Aegilops Complex; China Agriculture Press: Beijing, China, 2020. [Google Scholar]
- Kimber, G.; Yen, Y. Analysis of pivotal-differential evolutionary patterns. Proc. Natl. Acad. Sci. USA 1988, 85, 9106–9108. [Google Scholar] [CrossRef]
- Van Slageren, M.W. Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae); International Center for Agricultural Research in Dry Areas, Agricultural University, Wageningen: Aleppo, Syria, 1994. [Google Scholar]
- Zhang, F.; Shi, Y.; Ali, J.; Xu, J.; Li, Z. Breeding by selective introgression: Theory, practices, and lessons learned from rice. Crop J. 2021, 9, 646–657. [Google Scholar] [CrossRef]
- Liu, D. Towards cultivar-oriented gene discovery for better crops. Crop J. 2024, 12, 670–675. [Google Scholar]
- Pritchard, D.J.; Hollington, P.A.; Davies, W.P.; Gorham, J.; de Diaz Leon, J.L.; Mujeeb-Kazi, A. K+/Na+ discrimination in synthetic hexaploid wheat lines: Transfer of the trait for K+/Na+ discrimination from Aegilops tauschii into a Triticum turgidum background. Cereal Res. Commun. 2002, 30, 261–267. [Google Scholar]
- Yang, W.; Liu, D.; Li, J.; Zhang, L.; Wei, H.; Hu, X.; Zheng, Y.; He, Z.; Zou, Y. Synthetic hexaploid wheat and its utilization for wheat genetic improvement in China. J. Genet. Genom. 2009, 36, 539–546. [Google Scholar] [CrossRef]
- Hao, M.; Zhang, L.; Zhao, L.; Dai, S.; Li, A.; Yang, W.; Xie, D.; Li, Q.; Ning, S.; Yan, Z.; et al. A breeding strategy targeting the secondary gene pool of bread wheat: Introgression from a synthetic hexaploid wheat. Theor. Appl. Genet. 2019, 132, 2285–2294. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar]
- Brancourt-Hulmel, M.; Doussinault, G.; Lecomte, C.; Bérard, P.; Le Buanec, B.; Trottet, M. Genetic improvement of agronomic traits of winter wheat cultivars released in France from 1946 to 1992. Crop Sci. 2003, 43, 37–45. [Google Scholar]
- Sreenivasulu, N.; Schnurbusch, T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 2012, 17, 91–101. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Ford, B.; Wallace, X.; Pettolino, F.; Griffin, P.T.; Schmitz, R.J.; Zhang, P.; Barrero, J.M.; Hayden, M.J.; Boden, S.A.; et al. Zebularine treatment is associated with deletion of FT-B1 leading to an increase in spikelet number in bread wheat. Plant Cell Environ. 2018, 41, 1346–1360. [Google Scholar]
- Shaw, L.M.; Turner, A.S.; Laurie, D.A. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 2012, 71, 71–84. [Google Scholar] [PubMed]
- Bentley, A.R.; Horsnell, R.; Werner, C.P.; Turner, A.S.; Rose, G.A.; Bedard, C.; Howell, P.; Wilhelm, E.P.; Mackay, I.J.; Howells, R.M.; et al. Short, natural, and extended photoperiod response in BC2F4 lines of bread wheat with different photoperiod-1 (Ppd-1) alleles. J. Exp. Bot. 2013, 64, 1783–1793. [Google Scholar] [PubMed]
- Scarth, R.; Kirby, E.J.M.; Law, C.N. Effects of the photoperiod genes Ppd1 and Ppd2 on growth and development of the shoot apex in wheat. Ann. Bot. 1985, 55, 351–359. [Google Scholar] [CrossRef]
- Worland, A.J.; Börner, A.; Korzun, V.; Li, W.M.; Petrovíc, S.; Sayers, E.J. The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 1998, 100, 385–394. [Google Scholar]
- Snape, J.W.; Butterworth, K.; Whitechurch, E.; Worland, A.J. Waiting for fine times: Genetics of flowering time in wheat. Euphytica 2001, 119, 185–190. [Google Scholar]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy number variation affecting the photoperiod-B1 and vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS ONE 2012, 7, e33234. [Google Scholar] [CrossRef]
- Royo, C.; Dreisigacker, S.; Alfaro, C.; Ammar, K.; Villegas, D. Effect of Ppd-1 genes on durum wheat flowering time and grain filling duration in a wide range of latitudes. J. Agric. Sci. 2015, 154, 612–631. [Google Scholar]
- Pérez-Gianmarco, T.I.; Slafer, G.A.; González, F.G. Wheat pre-anthesis development as affected by photoperiod sensitivity genes (Ppd-1) under contrasting photoperiods. Funct. Plant Biol. 2018, 45, 645–657. [Google Scholar]
- Pérez-Gianmarco, T.I.; Slafer, G.A.; González, F.G. Photoperiod-sensitivity genes shape floret development in wheat. J. Exp. Bot. 2019, 70, 1339–1348. [Google Scholar]
- Guo, Z.; Song, Y.; Zhou, R.; Ren, Z.; Jia, J. Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 2010, 185, 841–851. [Google Scholar]
- Chuck, G.; Muszynski, M.; Kellogg, E.; Hake, S.; Schmidt, R.J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 2002, 298, 1238–1241. [Google Scholar]
- Komatsu, M.; Chujo, A.; Nagato, Y.; Shimamoto, K.; Kyozuka, J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 2003, 130, 3841–3850. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhao, S.; Fu, Y.; Sun, H.; Ma, X.; Tan, L.; Liu, F.; Sun, X.; Sun, H.; Gu, P.; et al. Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Crop J. 2018, 96, 716–733. [Google Scholar]
- Du, D.; Li, Z.; Jiang, Z.; Yuan, J.; Zhang, X.; Zhao, H.; Tian, L.; Liu, Y.; Li, R.; He, F.; et al. The Transcription Factor WFZP Interacts with the Chromatin Remodeler TaSYD to Regulate Root Architecture and Nitrogen Uptake Efficiency in Wheat. Adv. Sci. 2025; early view. [Google Scholar]
- Lagudah, E.S.; Appels, R.; Brown, A.D.H. The molecular-genetic analysis of Triticum tauschii, the D genome donor to hexaploid wheat. Genome 1991, 36, 913–918. [Google Scholar]
- Lubbers, E.L.; Gill, K.S.; Cox, T.S.; Gill, B.S. Variation of molecular markers among geographically diverse accessions of Triticum tauschii. Genome 1991, 34, 354–361. [Google Scholar]
- Dvorak, J.; Luo, M.; Yang, Z.; Zhang, H. The structure of Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar]
- Dvorak, J.; Deal, K.; Luo, M.; You, F.; Borstel, K.; Dehghani, H. The origin of spelt and free-threshing hexaploid wheat. J. Hered. 2012, 103, 426–441. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, R.; Li, S.; Liao, J.; Ye, F.; Yin, S.; Shen, J.; Cui, Q.; Wang, X.; Song, D.; Chen, W.; et al. Genetic Variations and Haplotype Diversity of the Wheat FRIZZY PANICLE (WFZP) Gene in 98 Aegilops tauschii Accessions. Genes 2025, 16, 414. https://doi.org/10.3390/genes16040414
Tao R, Li S, Liao J, Ye F, Yin S, Shen J, Cui Q, Wang X, Song D, Chen W, et al. Genetic Variations and Haplotype Diversity of the Wheat FRIZZY PANICLE (WFZP) Gene in 98 Aegilops tauschii Accessions. Genes. 2025; 16(4):414. https://doi.org/10.3390/genes16040414
Chicago/Turabian StyleTao, Ruilong, Shengke Li, Jia Liao, Fahui Ye, Shuxiang Yin, Jicheng Shen, Qingshan Cui, Xinfeng Wang, Deguo Song, Wenjie Chen, and et al. 2025. "Genetic Variations and Haplotype Diversity of the Wheat FRIZZY PANICLE (WFZP) Gene in 98 Aegilops tauschii Accessions" Genes 16, no. 4: 414. https://doi.org/10.3390/genes16040414
APA StyleTao, R., Li, S., Liao, J., Ye, F., Yin, S., Shen, J., Cui, Q., Wang, X., Song, D., Chen, W., & Ning, S. (2025). Genetic Variations and Haplotype Diversity of the Wheat FRIZZY PANICLE (WFZP) Gene in 98 Aegilops tauschii Accessions. Genes, 16(4), 414. https://doi.org/10.3390/genes16040414





