Resequencing and Transcriptome Analyses Reveal Variations and Expression Patterns of the RR Gene Family in Cucumber
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of CsRR Genes in Cucumber
2.2. Variant Identification and Annotation
2.3. Genetic Diversity Analysis
2.4. RNA-Seq Data Analysis
2.5. Plant Materials, qRT-PCR Analysis of CsRR Genes After Foc Inoculation
3. Results
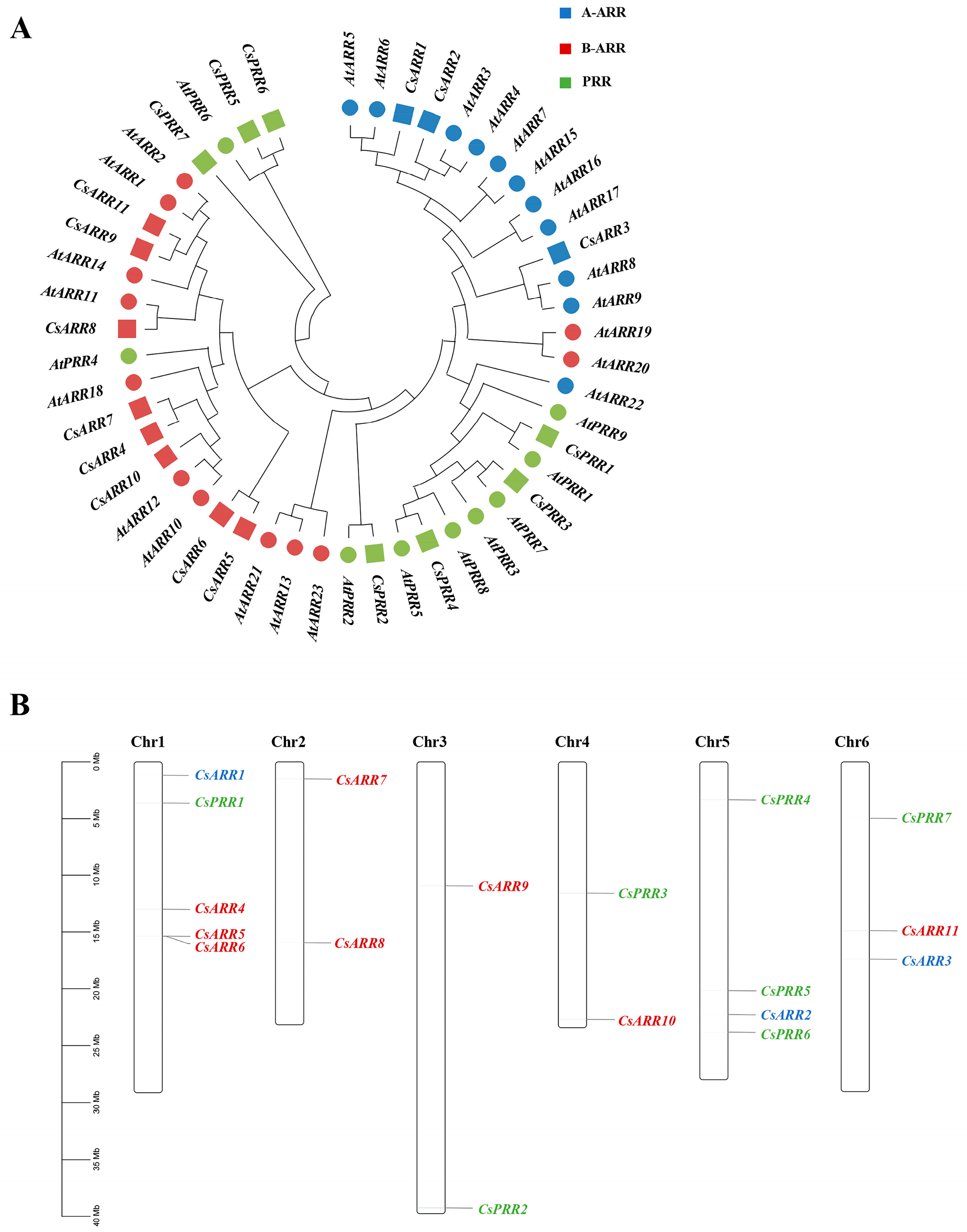
3.1. The Identification of CsRR Genes
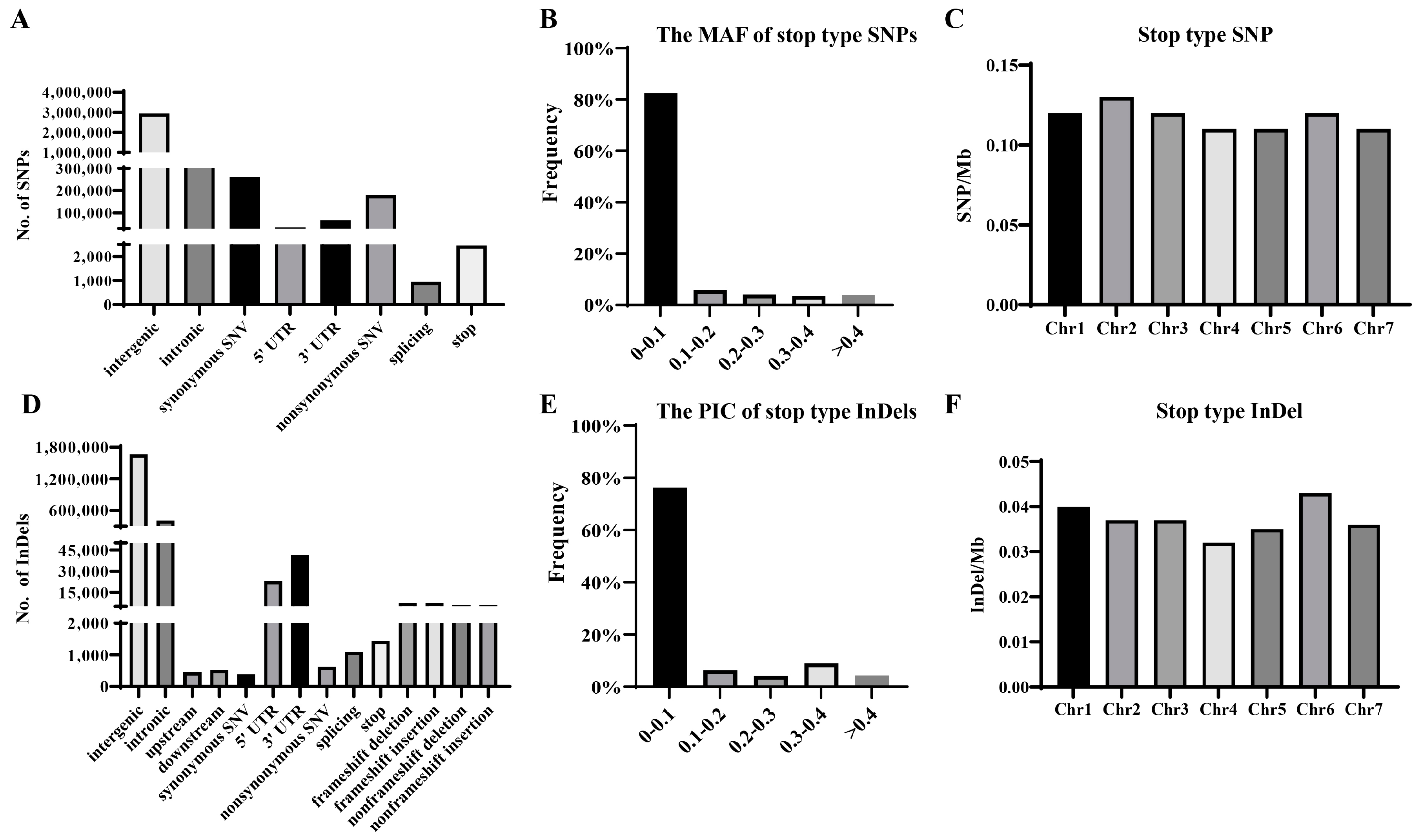
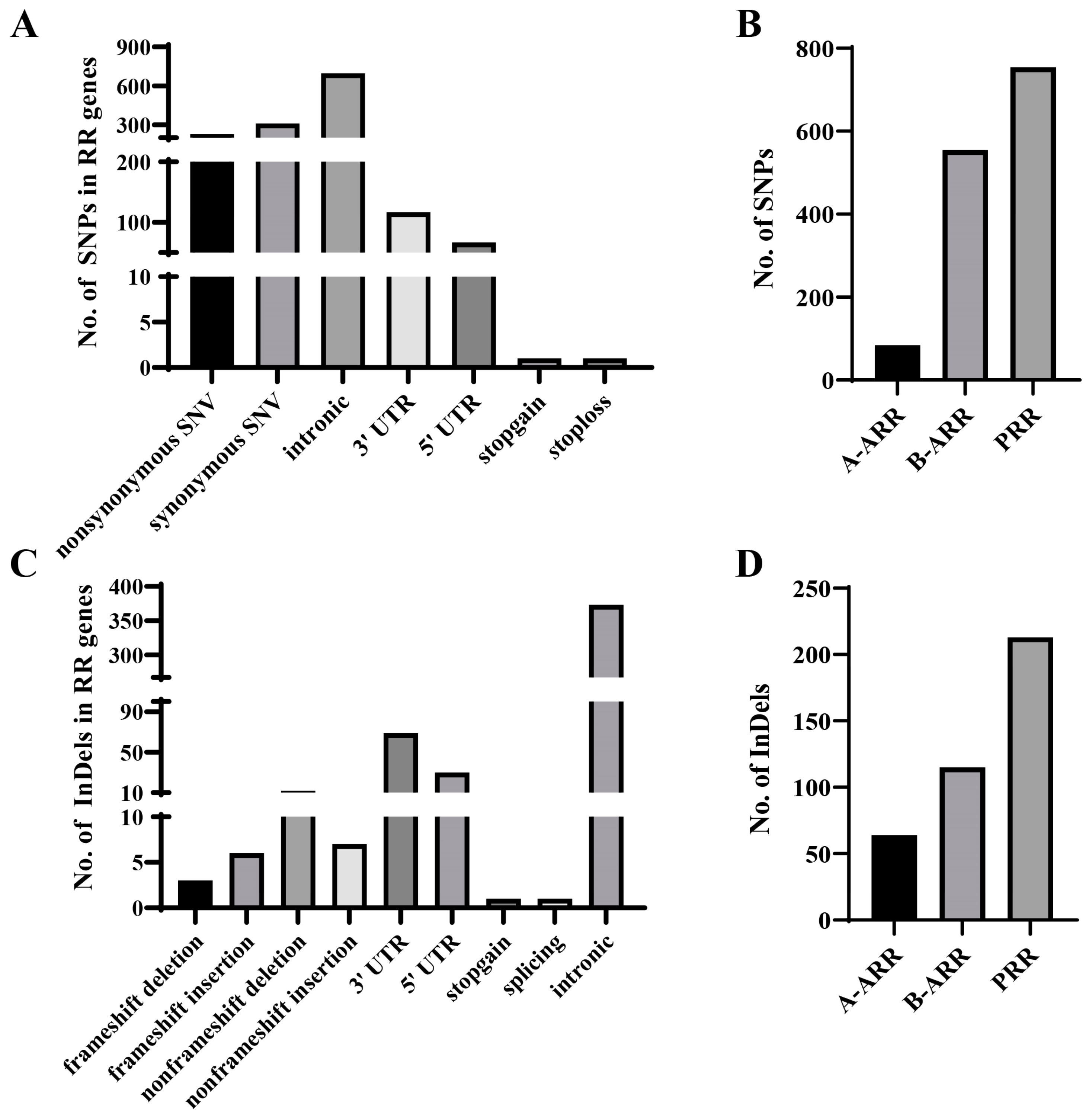
3.2. SNP and InDel Variations in 182 Cucumber Germplasms
3.3. CsRR Gene Variations Are Genetically Diverse
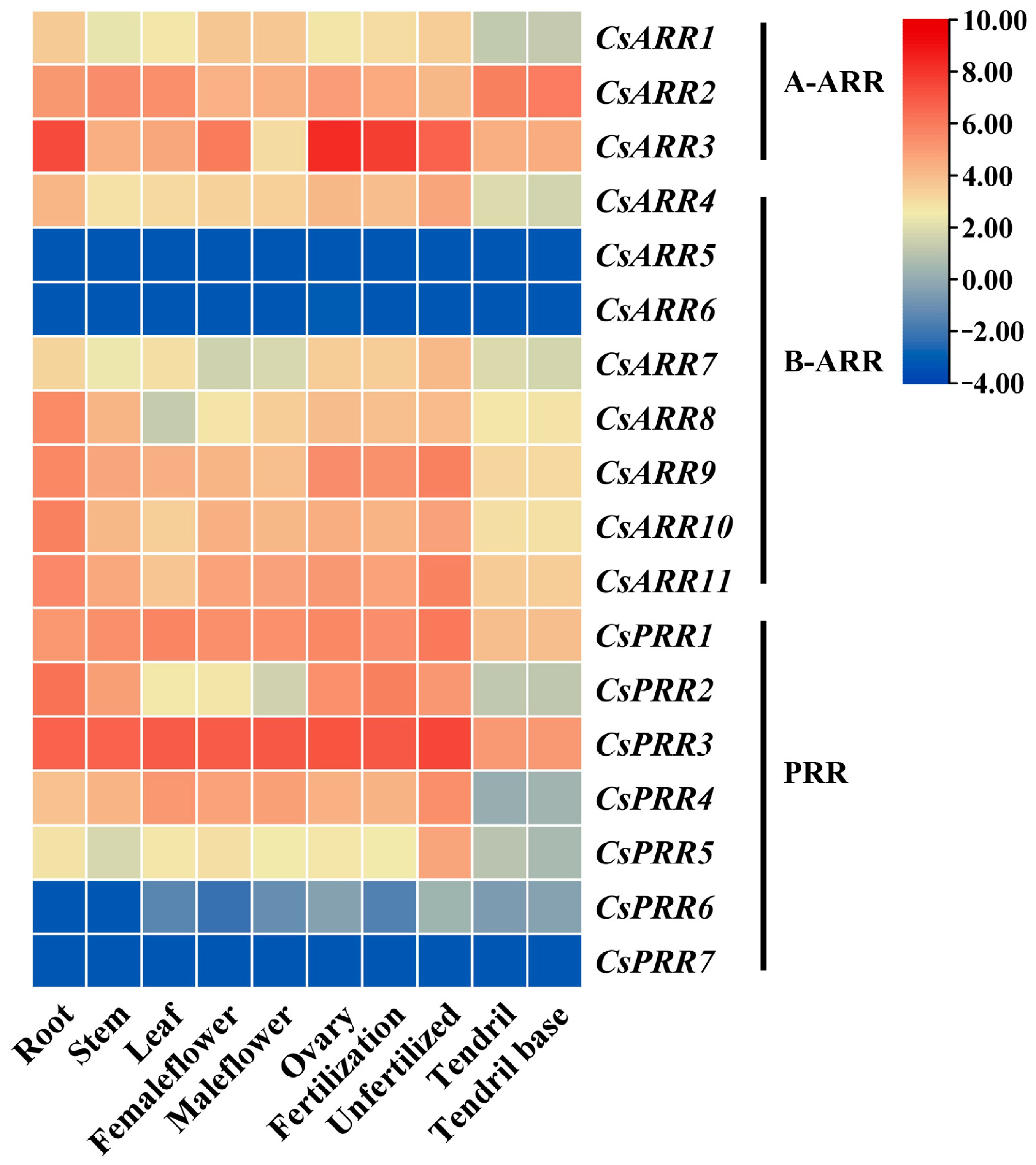
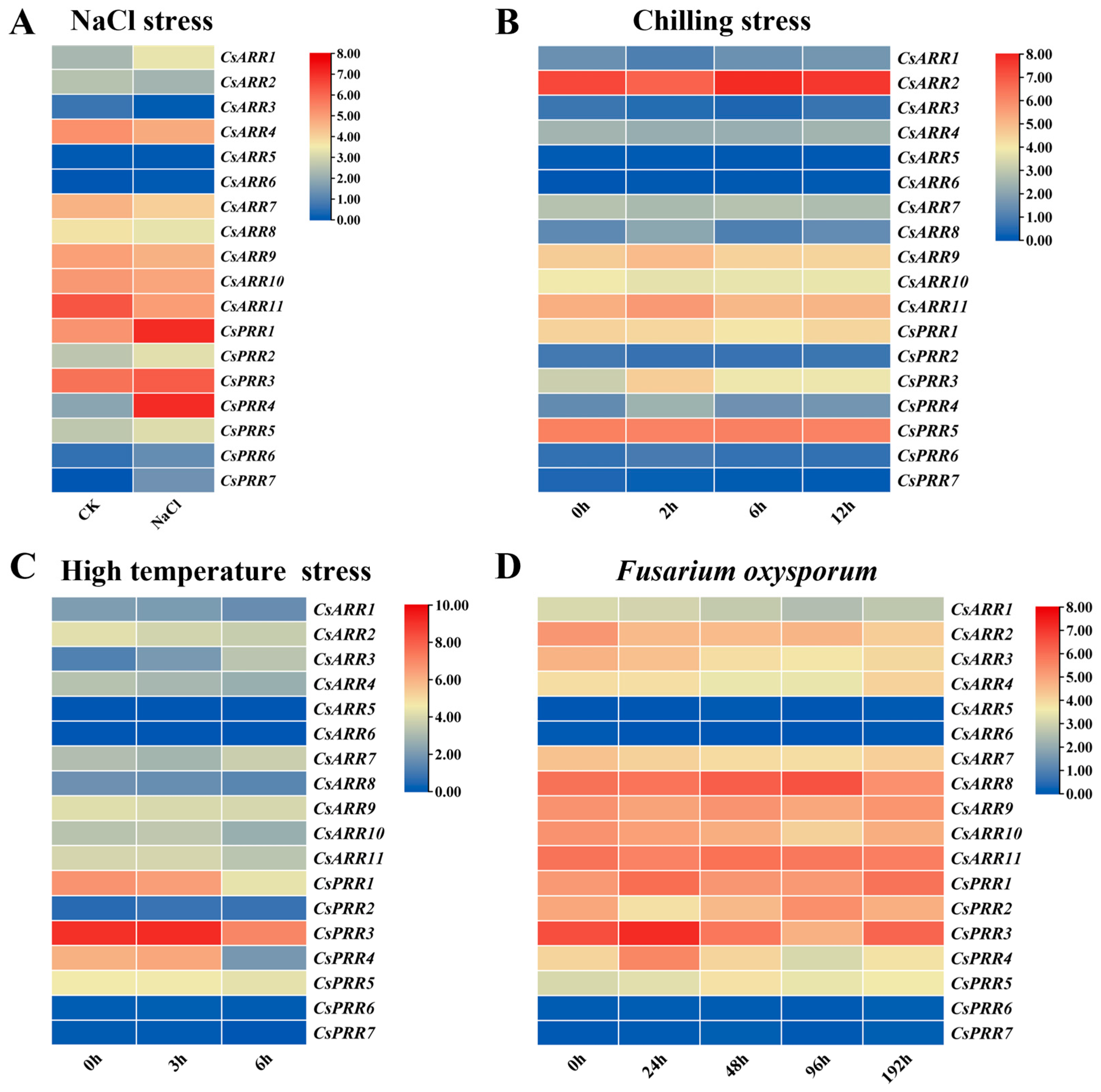
3.4. CsRR Gene Expression Patterns
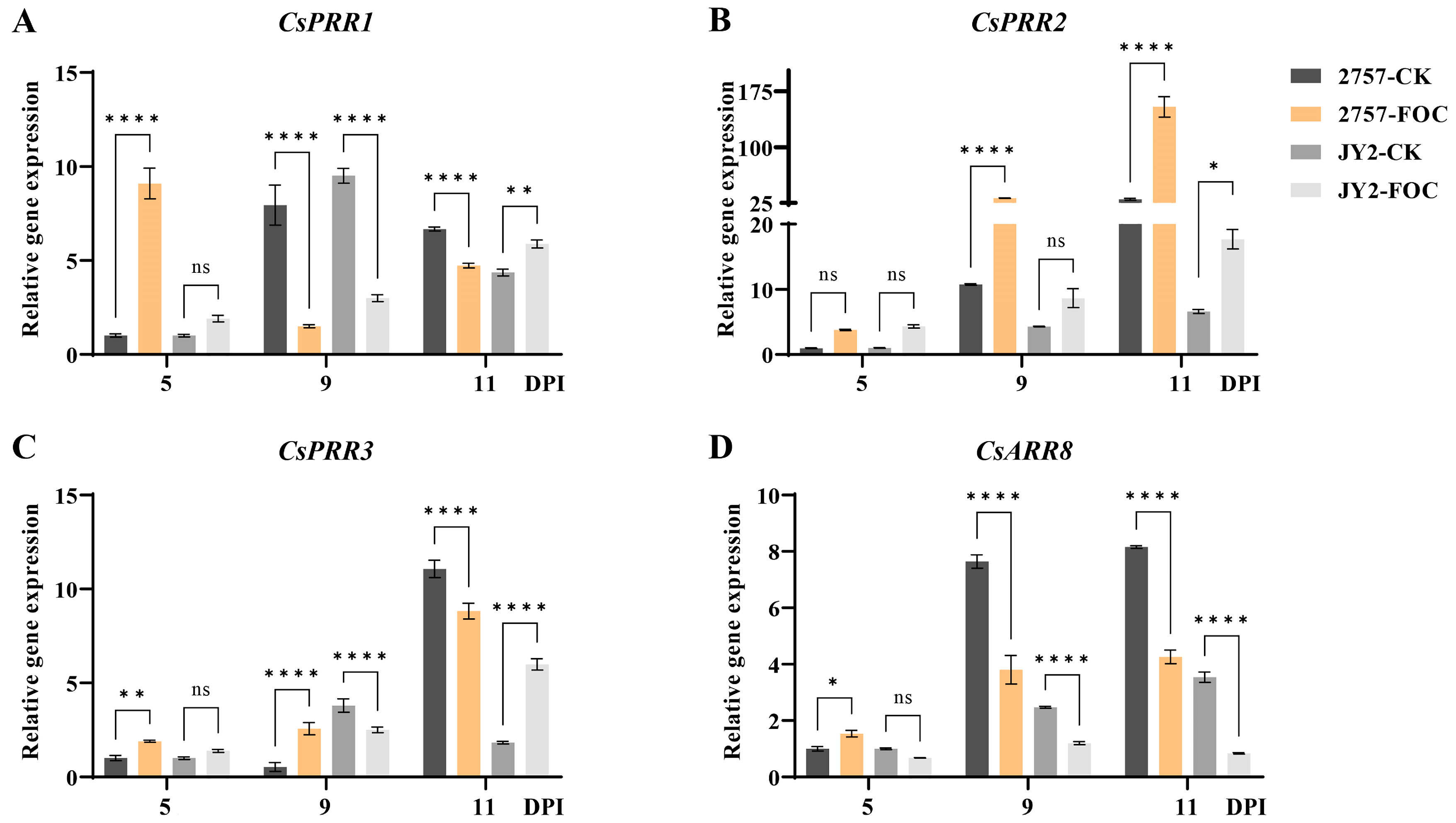
3.5. CsRR Gene Expression in Response to Foc
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yundaeng, C.; Somta, P.; Tangphatsornruang, S.; Chankaew, S.; Srinives, P. A single base substitution in BADH/AMADH is responsible for fragrance in cucumber (Cucumis sativus L.), and development of SNAP markers for the fragrance. Theor. Appl. Genet. 2015, 128, 1881–1892. [Google Scholar] [CrossRef]
- Pan, Y.; Qu, S.; Bo, K.; Gao, M.; Haider, K.R.; Weng, Y. QTL mapping of domestication and diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor. Appl. Genet. 2017, 130, 1531–1548. [Google Scholar] [CrossRef]
- Che, G.; Zhang, X. Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 2019, 47, 38–46. [Google Scholar] [CrossRef]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef]
- Hui, W.; Wu, H.; Zheng, H.; Wang, K.; Yang, T.; Fan, J.; Wu, J.; Wang, J.; Al Mutairi, A.A.; Yang, H.; et al. Genome-wide characterization of RR gene family members in Zanthoxylum armatum and the subsequent functional characterization of the C-type RR. Plant Physiol. Biochem. 2024, 214, 108943. [Google Scholar] [CrossRef]
- Cortleven, A.; Marg, I.; Yamburenko, M.V.; Schlicke, H.; Hill, K.; Grimm, B.; Schaller, G.E.; Schmülling, T. Cytokinin Regulates the Etioplast-Chloroplast Transition through the Two-Component Signaling System and Activation of Chloroplast-Related Genes. Plant Physiol. 2016, 172, 464–478. [Google Scholar] [CrossRef]
- D’Agostino, I.B.; Deruère, J.; Kieber, J.J. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000, 124, 1706–1717. [Google Scholar] [CrossRef]
- D’Agostino, I.B.; Kieber, J.J. Phosphorelay signal transduction: The emerging family of plant response regulators. Trends Biochem. Sci. 1999, 24, 452–456. [Google Scholar] [CrossRef]
- Ren, B.; Liang, Y.; Deng, Y.; Chen, Q.; Zhang, J.; Yang, X.; Zuo, J. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 2009, 19, 1178–1190. [Google Scholar] [CrossRef]
- Sakai, H.; Aoyama, T.; Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000, 24, 703–711. [Google Scholar] [CrossRef]
- Farré, E.M.; Liu, T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 2013, 16, 621–629. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian Waves of Expression of the APRR1/TOC1 Family of Pseudo-Response Regulators in Arabidopsis thaliana: Insight into the Plant Circadian Clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Makino, S.; Matsushika, A.; Kojima, M.; Yamashino, T.; Mizuno, T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002, 43, 58–69. [Google Scholar] [CrossRef]
- Wang, F.; Han, T.; Jeffrey Chen, Z. Circadian and photoperiodic regulation of the vegetative to reproductive transition in plants. Commun. Biol. 2024, 7, 579. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Liu, Q.; Zhang, Y.; Dong, K. Genome-Wide Identification and Characterization of PRR Gene Family and their Diurnal Rhythmic Expression Profile in Maize. Int. J. Genom. 2022, 2022, 6941607. [Google Scholar] [CrossRef]
- Liu, H.; Jiao, J.; Liang, X.; Liu, J.; Meng, H.; Chen, S.; Li, Y.; Cheng, Z. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Holdsworth, W.L.; Klein, C.L.; Barry, C.S. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 2014, 78, 1022–1033. [Google Scholar] [CrossRef]
- Huang, J.; Liu, F.; Ren, R.; Deng, J.; Zhu, L.; Li, H.; Cai, F.; Meng, Z.; Chen, Q.; Shi, T. QTL Mapping and Candidate Gene Analysis for Starch-Related Traits in Tartary Buckwheat (Fagopyrum tataricum (L.) Gaertn). Int. J. Mol. Sci. 2024, 25. [Google Scholar] [CrossRef]
- Thakur, O.; Randhawa, G.S. Identification and characterization of SSR, SNP and InDel molecular markers from RNA-Seq data of guar (Cyamopsis tetragonoloba, L. Taub.) roots. BMC Genom. 2018, 19, 951. [Google Scholar] [CrossRef]
- Bhavani, P.; Nandini, C.; Maharajan, T.; Ningaraju, T.M.; Nandini, B.; Parveen, S.G.; Pushpa, K.; Ravikumar, R.L.; Nagaraja, T.E.; Ceasar, S.A. Brown-top millet: An overview of breeding, genetic, and genomic resources development for crop improvement. Planta 2024, 260, 10. [Google Scholar] [CrossRef]
- Liu, J.; Qu, J.; Yang, C.; Tang, D.; Li, J.; Lan, H.; Rong, T. Development of genome-wide insertion and deletion markers for maize, based on next-generation sequencing data. BMC Genom. 2015, 16, 601. [Google Scholar] [CrossRef]
- Wu, D.-H.; Wu, H.-P.; Wang, C.-S.; Tseng, H.-Y.; Hwu, K.-K. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Liao, P.-Y.; Lee, K.H. From SNPs to functional polymorphism: The insight into biotechnology applications. Biochem. Eng. J. 2010, 49, 149–158. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Welsh, J.; McClelland, M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990, 18, 7213–7218. [Google Scholar] [CrossRef]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Moore, S.S.; Sargeant, L.L.; King, T.J.; Mattick, J.S.; Georges, M.; Hetzel, D.J.S. The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics 1991, 10, 654–660. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Weber, J.L.; David, D.; Heil, J.; Fan, Y.; Zhao, C.; Marth, G. Human diallelic insertion/deletion polymorphisms. Am. J. Hum. Genet. 2002, 71, 854–862. [Google Scholar] [CrossRef]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef]
- Lander, E.S. The new genomics: Global views of biology. Science 1996, 274, 536–539. [Google Scholar] [CrossRef]
- Yang, J.; He, J.; Wang, D.-B.; Shi, E.; Yang, W.; Geng, Q.; Wang, Z. Progress in research and application of InDel markers. Biodivers. Sci. 2016, 24, 237–243. [Google Scholar] [CrossRef]
- Wen, C.; Zhao, W.; Liu, W.; Yang, L.; Wang, Y.; Liu, X.; Xu, Y.; Ren, H.; Guo, Y.; Li, C.; et al. CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 2019, 146, dev180166. [Google Scholar] [CrossRef]
- Song, W.; Xie, Y.; Liu, B.; Huang, Y.; Cheng, Z.; Zhao, Z.; Tian, D.; Geng, Y.; Guo, J.; Li, C.; et al. Single nucleotide polymorphisms in SEPALLATA 2 underlie fruit length variation in cucurbits. Plant Cell 2024, 36, 4607–4621. [Google Scholar] [CrossRef]
- Xu, X.; Zhu, Y.; Yuan, Y.; Sohail, H.; He, S.; Ye, Y.; Wang, M.; Lv, M.; Qi, X.; Yang, X.; et al. R2R3-MYB transcription factor CsMYB60 controls mature fruit skin color by regulating flavonoid accumulation in cucumber. Plant J. 2024, 119, 796–813. [Google Scholar] [CrossRef]
- Zhang, R.J.; Liu, B.; Song, S.S.; Salah, R.; Song, C.J.; Xia, S.W.; Hao, Q.; Liu, Y.J.; Li, Y.; Lai, Y.S. Lipid-Related Domestication Accounts for the Extreme Cold Sensitivity of Semiwild and Tropic Xishuangbanna Cucumber (Cucumis sativus L. var. xishuangbannanesis). Int. J. Mol. Sci. 2023, 25, 79. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Du, H.; Zhao, H.; Li, H.; Xu, Y.; Mao, A.; Zhang, X.; Fu, Y.; Xia, Y.; et al. The vegetable SNP database: An integrated resource for plant breeders and scientists. Genomics 2022, 114, 110348. [Google Scholar] [CrossRef]
- Xu, X.; Du, Y.; Li, S.; Tan, M.; Sohail, H.; Liu, X.; Qi, X.; Yang, X.; Chen, X. A genome-wide association study reveals molecular mechanism underlying powdery mildew resistance in cucumber. Genome Biol. 2024, 25, 252. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 2006, 6, 1. [Google Scholar] [CrossRef]
- Murakami, M.; Matsushika, A.; Ashikari, M.; Yamashino, T.; Mizuno, T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci. Biotechnol. Biochem. 2005, 69, 410–414. [Google Scholar] [CrossRef]
- Jeong, H.-B.; Jang, S.-J.; Kang, M.-Y.; Kim, S.; Kwon, J.-K.; Kang, B.-C. Candidate Gene Analysis Reveals That the Fruit Color Locus C1 Corresponds to PRR2 in Pepper (Capsicum frutescens). Front. Plant Sci. 2020, 11, 399. [Google Scholar] [CrossRef]
- Para, A.; Farré, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 Is a Vascular Regulator of TOC1 Stability in the Arabidopsis Circadian Clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef]
- Osakabe, Y.; Miyata, S.; Urao, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 2002, 293, 806–815. [Google Scholar] [CrossRef]
- Zhong, L.; Hao, S.; Zhai, T.; Yang, Y.; Lin, H.; Lin, B.; Shen, B.; Liu, S.; Hu, Y.; Chen, X. The role of LED supplementary lighting in promoting graft necrotic layer formation in pumpkin-cucumber grafts. Sci. Hortic. 2024, 330, 112953. [Google Scholar] [CrossRef]
- Xue, W.; Liu, N.; Zhang, T.; Li, J.; Chen, P.; Yang, Y.; Chen, S. Substance metabolism, IAA and CTK signaling pathways regulating the origin of embryogenic callus during dedifferentiation and redifferentiation of cucumber cotyledon nodes. Sci. Hortic. 2022, 293, 110680. [Google Scholar] [CrossRef]
- Tang, H.-Y.; Dong, X.; Wang, J.-K.; Xia, J.-H.; Xie, F.; Zhang, Y.; Yao, X.; Xu, Y.-J.; Wan, Z.-J. Fine Mapping and Candidate Gene Prediction for White Immature Fruit Skin in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2018, 19, 1493. [Google Scholar] [CrossRef]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arab. Book. 2008, 6, e0112. [Google Scholar] [CrossRef]
- He, Y.; Liu, X.; Ye, L.; Pan, C.; Chen, L.; Zou, T.; Lu, G. Genome-Wide Identification and Expression Analysis of Two-Component System Genes in Tomato. Int. J. Mol. Sci. 2016, 17, 1204. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Dai, C.B.; Wang, W.F.; Sun, Y.H. Genome-wide identification of the ARRs gene family in tobacco (Nicotiana tabacum). Genes. Genom. 2021, 43, 601–612. [Google Scholar] [CrossRef]
- Rehman, O.U.; Uzair, M.; Chao, H.; Fiaz, S.; Khan, M.R.; Chen, M. Role of the type-B authentic response regulator gene family in fragrant rice under alkaline salt stress. Physiol. Plant 2022, 174, e13696. [Google Scholar] [CrossRef]
- Zeng, R.; Li, Z.; Shi, Y.; Fu, D.; Yin, P.; Cheng, J.; Jiang, C.; Yang, S. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, F.; Wang, X.; Feng, J.; Yi, Q.; Zhu, S.; Zhao, X. Mining key genes related to root morphogenesis through genome-wide identification and expression analysis of RR gene family in citrus. Front. Plant Sci. 2022, 13, 1068961. [Google Scholar] [CrossRef]
- Li, Y.; Luo, X.; Peng, X.; Jin, Y.; Tan, H.; Wu, L.; Li, J.; Pei, Y.; Xu, X.; Zhang, W. Development of SNP and InDel markers by genome resequencing and transcriptome sequencing in radish (Raphanus sativus L.). BMC Genom. 2023, 24, 445. [Google Scholar] [CrossRef]
- Salomé, P.A.; To, J.P.C.; Kieber, J.J.; McClung, C.R. Arabidopsis Response Regulators ARR3 and ARR4 Play Cytokinin-Independent Roles in the Control of Circadian Period. Plant Cell 2005, 18, 55–69. [Google Scholar] [CrossRef]
- Matsushika, A.; Kawamura, M.; Nakamura, Y.; Kato, T.; Murakami, M.; Yamashino, T.; Mizuno, T. Characterization of Circadian-Associated Pseudo-Response Regulators: II. The Function of PRR5 and Its Molecular Dissection in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 535–544. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kudo, T.; Makita, N.; Kiba, T.; Kinoshita, T.; Sakakibara, H. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 2020, 84, 970–979. [Google Scholar] [CrossRef]
- Wang, T.; Hu, J.; Ma, X.; Li, C.; Yang, Q.; Feng, S.; Li, M.; Li, N.; Song, X. Identification, evolution and expression analyses of whole genome-wide TLP gene family in Brassica napus. BMC Genom. 2020, 21, 264. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, K.; Ao, W.; Sun, Z.; Li, J.; Gao, Y.; Gan, D.; Yang, J. Resequencing and Transcriptome Analyses Reveal Variations and Expression Patterns of the RR Gene Family in Cucumber. Genes 2025, 16, 409. https://doi.org/10.3390/genes16040409
Su K, Ao W, Sun Z, Li J, Gao Y, Gan D, Yang J. Resequencing and Transcriptome Analyses Reveal Variations and Expression Patterns of the RR Gene Family in Cucumber. Genes. 2025; 16(4):409. https://doi.org/10.3390/genes16040409
Chicago/Turabian StyleSu, Ke, Wenhong Ao, Zhaolong Sun, Jing Li, Yu Gao, Defang Gan, and Jingjing Yang. 2025. "Resequencing and Transcriptome Analyses Reveal Variations and Expression Patterns of the RR Gene Family in Cucumber" Genes 16, no. 4: 409. https://doi.org/10.3390/genes16040409
APA StyleSu, K., Ao, W., Sun, Z., Li, J., Gao, Y., Gan, D., & Yang, J. (2025). Resequencing and Transcriptome Analyses Reveal Variations and Expression Patterns of the RR Gene Family in Cucumber. Genes, 16(4), 409. https://doi.org/10.3390/genes16040409



