Abstract
Amidst the prevalent and notable characteristic of a hypoxic microenvironment present in the majority of solid tumors, a burgeoning number of studies have revealed the significance of noncoding RNAs (ncRNAs) in hypoxic tumor regions. The transcriptome of cancers is highly heterogeneous, with noncoding transcripts playing crucial roles. Long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) are two distinctive classes of ncRNA that are garnering increasing attention. Biologically, they possess intriguing properties and possess significant regulatory functions. Clinically, they present as promising biomarkers and therapeutic targets. Additionally, recent research has evaluated the clinical applications of these ncRNAs in RNA-based treatments and noninvasive liquid biopsies. This review provides a comprehensive summary of recent studies on lncRNAs and circRNAs within the hypoxic tumor microenvironment. Furthermore, the clinical significance of lncRNAs and circRNAs in cancer diagnosis and treatment is emphasized, which could pave the way for the development of effective targeted therapies.
1. Introduction
The tumor microenvironment (TME) is composed of various components, including the blood vessels, extracellular matrix, and stromal cells (e.g., endothelial cells, fibroblasts, immune cells, and mesenchymal stem cells), as well as secreted factors, like growth factors and cytokines [1]. Hypoxia constitutes a notable feature within the microenvironment of solid tumors [2]. Notably, hundreds or even thousands of coding and noncoding genes participate in complex adaptive mechanisms through which rapidly growing tumor cells adapt to hypoxia [3,4].
Noncoding RNAs (ncRNAs) mainly include microRNAs (miRNAs), long ncRNAs (lncRNAs), circular RNAs (circRNAs), and small nucleolar RNAs (snoRNAs) [5]. To date, the human database project has estimated the presence of 2654 miRNAs, 96,411 lncRNAs, 768,986 circRNAs, and 2064 snoRNAs [6,7,8,9]. Their presence can be readily detected in bodily fluids, such as tissues, serum, plasma, urine, and saliva [10]. Advancements in high-throughput technology and extensive research have shown that dysregulated ncRNAs participate in tumor initiation and metastasis through diverse molecular pathways [11,12,13]. In recent years, the clinical significance of ncRNAs in cancer has attracted considerable attention.
Although numerous protein-coding genes regulated by HIFs (hypoxia-inducible factors) contribute to the adaptation of tumor cells to hypoxia, the response of ncRNAs to hypoxia and their role in hypoxia-related cancer remain largely elusive. Recent developments in miRNAs in the context of hypoxia have been summarized in a previous review [14,15,16]. However, the role and mechanisms of hypoxia-related lncRNAs and circRNAs in cancer development have not been systematically investigated. In this review, we summarize the regulatory mechanisms exhibited by lncRNAs and circRNAs in their interaction with hypoxia and highlight their clinical significance in the diagnosis and treatment of cancer.
2. LncRNAs and circRNAs Act as Novel Players in the Hypoxic Tumor Microenvironment
Among the ncRNA subcategories, lncRNAs and circRNAs are recognized as essential regulators by diverse mechanisms of action in various pathophysiological processes [17,18,19]. LncRNAs, a diverse and extensive class of regulatory transcripts of at least 200 nucleotides in length, are generated from various genomic regions, encompassing promoter sequences, exons, antisense strands, enhancer elements, untranslated regions (UTRs) at both the 3′ and 5′ ends, and introns, as well as intergenic and intragenic segments [19,20,21]. However, certain scholars contend that a size cutoff of 200 nucleotides serves as a practical threshold in biochemical and biophysical RNA purification procedures, depleting the majority of infrastructural RNAs. Consequently, it is recommended to delineate ncRNAs exceeding 500 nucleotides as lncRNAs [22]. CircRNAs, a subtype of endogenous ncRNAs, are generated via an alternative splicing process known as backsplicing, exhibiting reduced susceptibility to exonuclease degradation due to their resultant circular molecules being devoid of terminal 5′ caps and 3′ polyadenylated tails, thereby conferring upon them enhanced stability relative to their linear RNA counterparts [23,24,25].
LncRNAs and circRNAs regulate gene expression through multiple mechanisms, including epigenetic recruitment, transcription regulation, post-transcriptional modulation, RNA/protein modifications, and encoding functional peptides [26,27,28,29,30,31]. The most prevalent mechanism among them involves lncRNAs and circRNAs functioning as competing endogenous RNAs (ceRNAs) or natural miRNA sponges, which engage in communication and co-regulation through competitive binding to shared miRNAs. A comprehensive understanding of this novel RNA crosstalk will provide profound insights into gene regulatory networks and have profound implications for cancers [32,33]. Additionally, they are attracting considerable attention owing to their substantial abundance, specific expression patterns, crucial functional roles in various diseases, and promising avenues for clinical application [28,34,35,36,37].
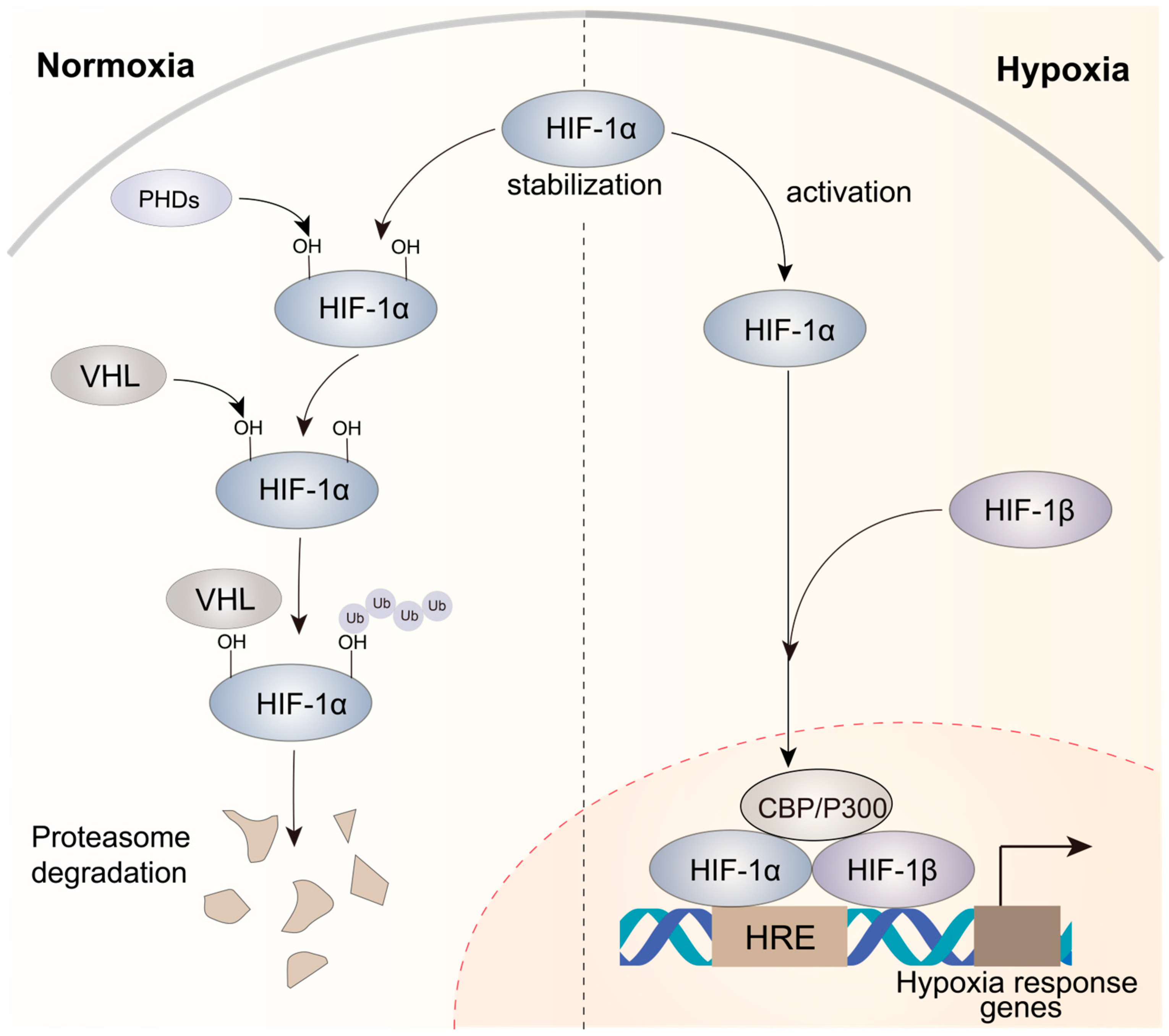
The microenvironment of solid tumors often exhibits hypoxia, which results from unrestrained and rapid tumor cell growth that limits the availability of oxygen [38]. HIFs are crucial for hypoxic responses [39], and increased HIF levels are frequently linked to a poor prognosis in various tumor types [40,41]. HIFs are heterodimeric proteins comprising a constantly expressed subunit (HIF-1β) and an oxygen-sensitive subunit (HIF-1α, HIF-2α, or HIF-3α). Under normal oxygen conditions, the HIF-1α subunit undergoes hydroxylation by dioxygenases called PHD1, PHD2, and PHD3. Subsequently, the hydroxylated prolines (PHDs) are recognized by the von Hippel–Lindau (VHL) E3 ubiquitin ligase, resulting in the ubiquitination of HIF-1α (Figure 1). However, under hypoxic conditions, PHDs cannot use oxygen as their co-substrate, resulting in the inhibition of hydroxylation. This inhibition leads to the accumulation of non-hydroxylated HIF-α subunits, which eventually translocate to the nucleus (Figure 1). In the nucleus, a heterodimeric complex consisting of HIF-1α and HIF-1β binds to hypoxia-responsive elements (HREs) on the promoters of hypoxia-responsive genes and drives the transcriptional activation of target genes after the recruitment of CBP/P300, a co-activator [42,43]. This process initiates a sequential cascade of cellular adaptations to hypoxia, encompassing amplified cell proliferation and angiogenesis, diminished apoptosis, and increased invasion and metastasis.
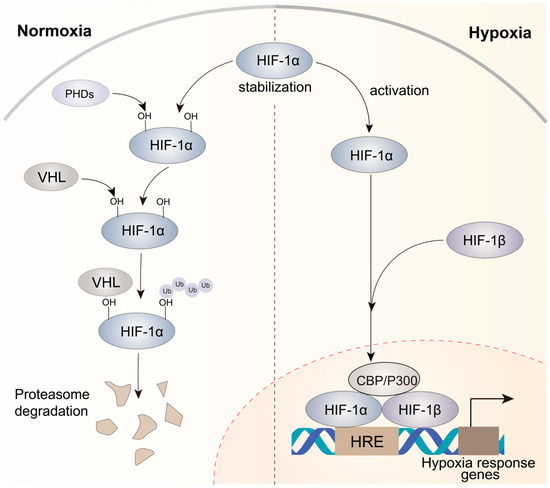
Figure 1.
Mechanisms of HIF-1α activation under normoxic and hypoxic conditions in cancer. Under normoxic circumstances, HIF-1α displays instability, while under hypoxic conditions, it attains stability. The HIF-1α–HIF-1β heterodimer is recruited to HREs, activating the expression of hypoxia response genes that contribute to complex processes involved in tumor progression.
LncRNAs and circRNAs modulated by hypoxia have been shown to exert substantial effects on the hypoxic TME, including tumor metabolism, immune escape, angiogenesis, migration/invasion, apoptosis, and growth/proliferation [42,44]. Furthermore, hypoxia signaling can be regulated by lncRNAs and circRNAs through distinct mechanisms, particularly through the stabilization of HIF-1α [45,46]. However, there are still a large number of lncRNAs and circRNAs that have not been identified as regulatory factors for hypoxia signaling. Consequently, the identification and analysis of novel hypoxia-related lncRNAs and circRNAs, specifically those undergoing expression alterations in response to hypoxic conditions or interacting with HIFs, may offer valuable perspectives for the diagnosis and treatment of cancer and promote the development of novel treatments based on ncRNA-targeted strategies.
3. Hypoxia-Related lncRNAs in Cancers
In the context of oxygen deprivation, dysregulation of hypoxia-associated lncRNAs facilitates the adaptation of tumor cells to microenvironmental stress, leading to enhanced aggressiveness, immune evasion, invasive angiogenesis, and glycolysis. These effects may occur in a HIF-dependent or HIF-independent manner. In the following sections, we provide a comprehensive summary of the functions and mechanisms of lncRNAs that interact with hypoxia in cancer (Figure 2 and Table 1).
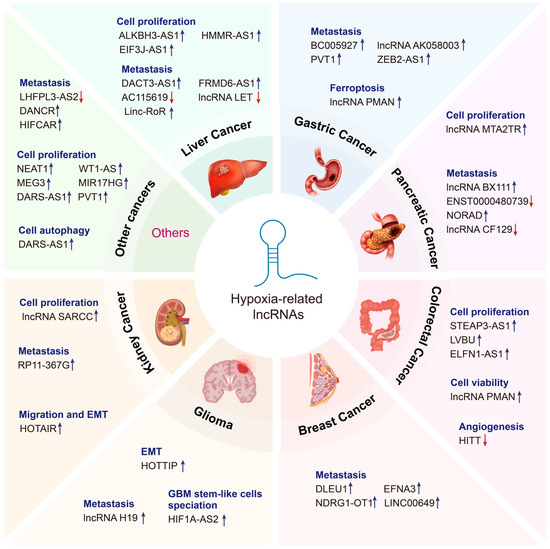
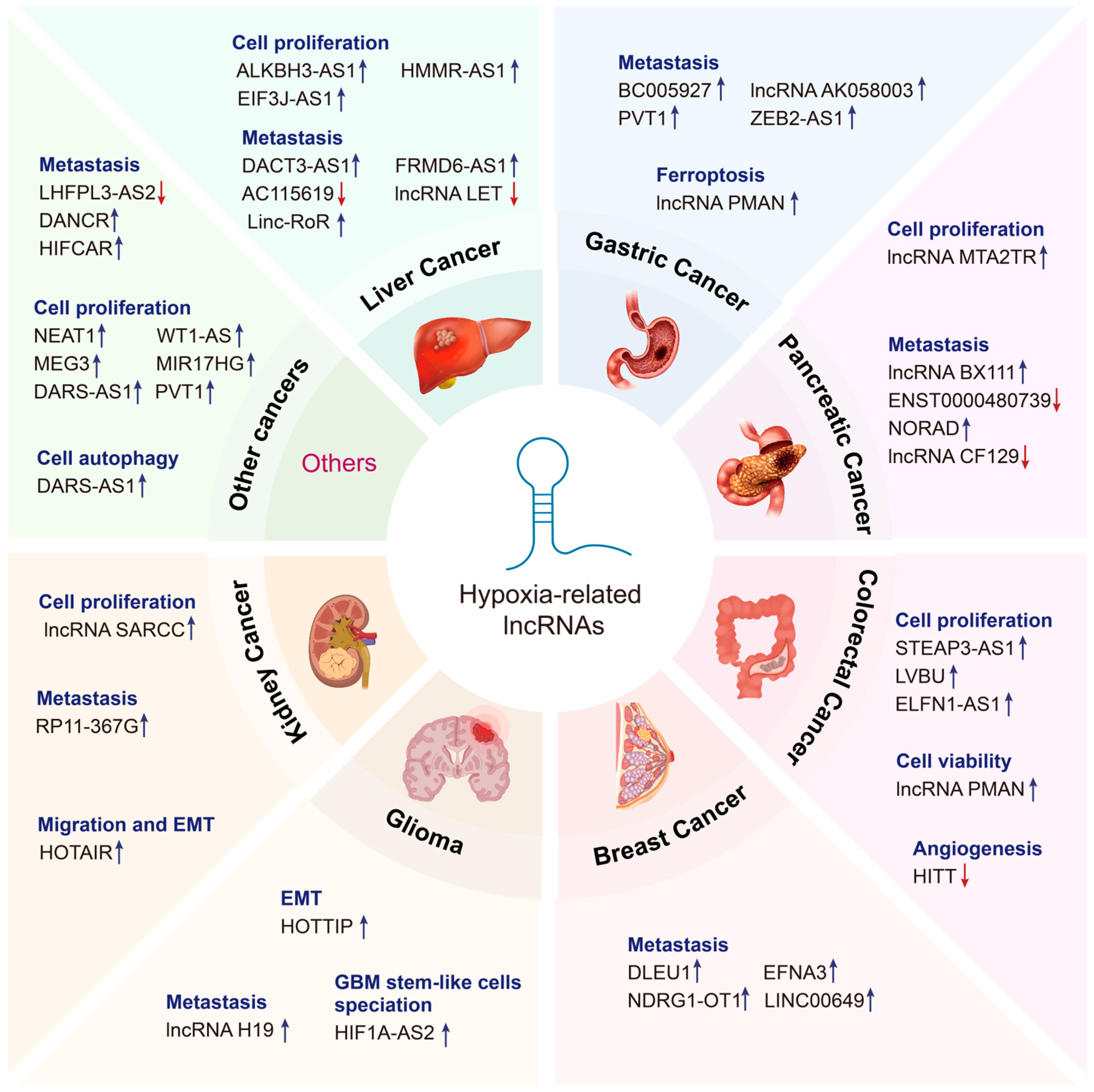
Figure 2.
Emerging roles of hypoxia-related lncRNAs in cancer. Hypoxia-related lncRNAs perform diverse functions in various cancer types, including liver cancer, gastric cancer, pancreatic cancer, colorectal cancer, breast cancer, glioma, kidney cancer, and so on. Upregulated lncRNAs are indicated by the blue arrow, whereas downregulated lncRNAs are indicated by the red arrow.

Table 1.
Summary of hypoxia-related lncRNAs in cancers.
3.1. Gastrointestinal Cancers
3.1.1. Liver Cancer
Hepatocellular carcinoma (HCC) is the most prevalent type of primary liver cancer and is the most common cause of cancer-related death [91]. Multiple lines of studies have revealed that targeting hypoxia-related lncRNAs may represent a novel strategy for HCC. These lncRNAs can influence the stability or functionality of their partners through their interactions with specific proteins, thereby modulating the biological behavior of HCC cells. For instance, the lncRNA DACT3-AS1 has been shown to promote metastasis in HCC. Mechanistically, DACT3-AS1 increases the deacetylation of FOXA3 by facilitating its interaction with HDAC2, leading to a decrease in FOXA3 protein levels. Furthermore, by binding to the pyruvate kinase M2 (PKM2) promoter area, the transcription factor FOXA3 might prevent PKM2, an enzyme that plays an important role in promoting HCC invasiveness, from being expressed [47]. Beyond their interactions with specific proteins, hypoxia-related lncRNAs also participate in tumorigenesis and progression by modulating the degradation or accumulation of mRNAs. The lncRNA-LET facilitates hypoxia-induced metastasis in HCC by modulating the accumulation and stability of HIF-1α mRNA. This modulation occurs through the downregulation of nuclear factor 90 (NF90) protein via the ubiquitin–proteasome pathway [50]. Similarly, FRMD6-AS1 was found to increase SENP1 protease activity, thereby increasing the stability of HIF-1α under hypoxia [51]. HIF-1α-induced ALKBH3-AS1 played a tumorigenic role by enhancing the stability of ALKBH3 mRNA. ALKBH3 is an oncogene that promotes the invasion and proliferation of HCC cells [48]. A few studies have shown that some hypoxia-related lncRNAs contain short ORFs (sORFs, short open reading frames, length < 300 nt) and are capable of encoding a small peptide with crucial biological functions in hypoxia. For instance, the hypoxia-responsive lncRNA AC115619 inhibits the growth of HCC cells through its peptide-coding functions. Mechanistically, through its interaction with WTAP, the encoded micropeptide AC115619–22aa prevents the m6A methyltransferase complex from forming. This interaction effectively reduces the increase in m6A modification, which is associated with high malignancy in tumors [49]. Hypoxia-related lncRNAs also function as sponges for miRNAs, thereby chestrating the intricate regulation of downstream gene expression and influencing the progression of HCC. Some lncRNAs (e.g., linc-RoR, EIF3J-AS1, and HMMR-AS1) can be induced by hypoxia and exert a vital role in hypoxia-induced cancer development by sponging miRNAs and influencing the expression of their targets [52,53,54]. These findings have illuminated the pivotal role of hypoxia-related lncRNAs in HCC through a myriad of mechanisms, primarily influencing gene expression and signaling pathway activity to promote HCC progression and metastasis.
3.1.2. Gastric Cancer
Gastric cancer (GC) ranks among the top five most widespread types of cancer globally and constitutes the third principal cause of cancer-related mortality worldwide [92]. Studies have demonstrated that hypoxia-related lncRNAs can modulate GC progression by influencing the expression of downstream genes. For instance, HIF-1α-induced PMAN enhances ferroptosis resistance by facilitating the cytoplasmic translocation of ELAVL1 and enhancing SLC7A11 expression under hypoxic conditions, thereby favoring the proliferation and progression of GC cells [55]. Furthermore, DNA methylation stands as a crucial epigenetic modification, wielding a profound influence over the regulation of a myriad of oncogenes and tumor suppressor genes. Notably, hypoxia-related lncRNAs are deeply entwined in the regulation of this methylation process, underscoring their significance in the complex landscape of cancer biology. In hypoxic GC cells, the lncRNA BC005927 regulates the ephrin type B receptor 4 (EPHB4), a metastasis-related gene, to influence the metastatic and invasive abilities of GC cells. Knockdown of BC005927 reduces EPHB4 expression by modulating the DNA methylation of EPHB4 in GC cells [56]. Similarly, the lncRNA AK058003 is upregulated under hypoxic conditions and regulates the metastasis-associated gene SNCG via DNA demethylation to modulate the hypoxia-induced metastasis of GC [57]. Numerous investigations have illuminated the intricate role of hypoxia-related lncRNAs in the modulation of HIF-1α expression via miRNA sponges, thereby influencing GC progression. For example, the lncRNAs PVT1 (plasmacytoma variant translocation 1) and ZEB2-AS1 increase HIF-1α expression and contribute to GC cell growth and invasion by influencing miRNA targets [58,59]. It is commonly known that a promising treatment approach for cancer involves strategically targeting HIF-1α and HIF-2α. However, resistance to treatment is an inevitable aspect of cancer therapy [93]. The aforementioned studies indicate that hypoxia-related lncRNAs extensively modulate the progression of GC, offering a novel perspective for guiding GC therapy by targeting these lncRNAs.
3.1.3. Pancreatic Cancer
The incidence and mortality of pancreatic cancer have been increasing steadily, while the rates of other prevalent cancers have been on the decline [94]. Numerous studies have found the potential clinical applications of hypoxia-related lncRNAs in pancreatic cancer. These lncRNAs are capable of regulating the expression level of oncogenes by recruiting transcription factors to their promoter regions. For example, lncRNA MTA2TR upregulated the transcription of metastasis-associated protein 2 (MTA2) by recruiting activating transcription factor 3 to the MTA2 promoter region in pancreatic cancer. MTA2 facilitates the stabilization of the HIF-1α protein through deacetylation [62]. Furthermore, the hypoxia-related lncRNAs emerge as pivotal factors, orchestrating the ubiquitin-dependent degradation of transcription factors, thus intricately modulating the expression and activity of target genes. The lncRNA CF129 can effectively suppress FOXC2 transcription by inducing the MKRN1-mediated ubiquitin-dependent degradation of p53, thereby inhibiting cancer cell proliferation and invasion [60]. Hypoxia-related lncRNAs can also indirectly affect the expression level of HIF-1α. Downregulated ENST00000480739 suppresses HIF-1α expression by promoting the transcription of OS-9, thereby contributing to tumor progression in pancreatic cancer [64]. Additionally, a hypoxia-related lncRNA can function as a ceRNA that binds to a specific miRNA, thereby alleviating the inhibitory effect of that miRNA on its targets. Overexpression of NORAD increases cancer cell migration and invasion, whereas its knockdown prevents epithelial-to-mesenchymal transition (EMT) and metastasis. Mechanistically, NORAD modulates the expression of RhoA by acting as a ceRNA of miR-125a-3p [61].
3.1.4. Colorectal Cancer
Accumulating research has found that hypoxia-related lncRNAs can modulate the advancement of colorectal cancer (CRC). Hypoxia-related lncRNAs also affect CRC development by influencing post-transcriptional processes such as alternative splicing and mRNA stabilization. A study showed that higher LUCAT1 expression indicated a worse prognosis in CRC. Mechanistically, the interaction between LUCAT1 and polypyrimidine tract-binding protein 1 (PTBP1) facilitates the binding of some DNA damage-related genes with PTBP1, consequently leading to changes in the alternative splicing of these genes [66]. Hypoxia-induced lncRNA STEAP3-AS1 activates the Wnt/β-catenin signaling pathway through the inhibition of m6A-mediated STEAP3 mRNA degradation, ultimately facilitating the progression of CRC [67]. In addition, hypoxia-related lncRNAs can also affect crucial signaling cascades to influence CRC progression by functioning as ceRNAs. For instance, HIF-1α-induced lncRNA LVBU, acting as a sponge of miR-10a/miR-34c, leads to the reprogramming of the urea cycle and the phenotype of polyamine synthesis by modulating BCL6 and p53, thereby facilitating CRC progression [68]. Similarly, ELFN1-AS1, as a downstream effector of hypoxia, promotes CRC cell proliferation and invasion by upregulating TRIM14 through miR-191-5p sponging [69]. Notably, hypoxia-related lncRNAs possess the capacity to regulate HIF-1α expression. For instance, the lncRNA HITT is significantly downregulated and directly binds to YB-1, hindering the interaction of the 5′-UTR of HIF-1α with YB-1 and inhibiting HIF-1α translation [65]. These investigations underscore the essential roles that hypoxia-related lncRNAs play in the modulation of oncogene expression and vital signaling pathways within CRC, ultimately contributing to the trajectory of cancer progression and prognostic outcomes. Thus, the strategic targeting of these hypoxia-related lncRNAs emerges as a promising therapeutic avenue.
3.2. Breast Cancer
Breast cancer (BC) ranks among the three most prevalent cancers globally. Early-stage BC is regarded as potentially treatable. Studies have revealed the potential of hypoxia-related lncRNAs as targets for the diagnosis and therapy of BC disease. Hypoxia-related lncRNAs emerge as pivotal players in the malignant behavior and progression of BC, intricately modulating HIF-1α and its downstream signaling pathways. For example, lncRNA DLEU1 is upregulated and promotes the malignant behavior of BC cells. Mechanistically, DLEU1 may serve as a cofactor and activate HIF-1α-mediated transcription of CKAP2, thereby activating the ERK and STAT3 signaling pathways to promote tumor growth [70]. Studies have confirmed that LINC00649 can influence cancer progression by affecting certain key signaling pathways [95,96]. Notably, LINC00649 is also upregulated and promotes BC progression and metastasis by increasing HIF-1α mRNA stability through interacting with the NF90/NF45 complex [71]. Other lncRNAs can be induced by hypoxic conditions, exerting a crucial influence on BC development through their competitive binding to miRNAs. A study revealed that the high expression of EFNA3 lncRNAs promoted the metastatic dissemination of BC cells. Functional assays revealed that under hypoxic conditions, EFNA3 suppressed the inhibitory effects of miR-210 on EFNA3 mRNA and facilitated efficient transcription, leading to the accumulation of Ephrin-A3 [72]. Similar to EFNA3, the transactivation of NDRG1-OT1 by HIF-1α can enhance cancer cell growth and migration by sponging miR-875-3p. Moreover, NDRG1-OT1 encodes a 66-amino acid peptide in the 29–226 bp region [73]. Thus, the intricate interplay between lncRNAs and hypoxia intricately contributes to the malignant behavior of BC cells at both transcriptional and post-transcriptional levels. These findings uncover the complex regulatory network governing hypoxia-related lncRNAs in BC progression and offer new potential targets and research avenues for BC treatment.
3.3. Glioma
Glioma is the most prevalent and fatal brain tumor in adults [97]. Consequently, novel and curative treatment approaches are urgently required. Hypoxia-related lncRNAs engage in intricate interactions with RNA-binding protein complexes, playing a pivotal role in the adaptive and malignant behaviors of glioma cells by stimulating the expression of oncogenes. A study showed that HIF1A-AS2 facilitates the specialization and hypoxic adaptation of glioma stem cells in the TME. Mechanistically, the direct interaction between HIF1A-AS2 and the mRNA-binding complexes of DHX9 and IGF2BP2 stimulates the expression of HMGA1, which possesses oncogenic properties [74]. Additionally, hypoxia-related lncRNAs also function as ceRNAs to promote glioma progression. Overexpression of the lncRNA H19 can increase the expression of β-catenin by acting as a ceRNA of miR-181d [75]. Similarly, hypoxia-induced upregulation of HOTTIP can promote metastasis in gliomas. Mechanistically, HOTTIP functions as a sponge of miR-101, inhibiting its endogenous activity and subsequently enhancing Zinc Finger E-Box Binding Homeobox 1 (ZEB1) expression to promote EMT [76]. These results support the notion that hypoxia-related lncRNAs facilitate the specialization, hypoxic adaptation, and EMT of glioma cells, ultimately contributing to tumor progression. Consequently, hypoxia-related lncRNAs emerge as promising novel therapeutic targets for patients grappling with glioma.
3.4. Renal Cell Carcinoma
Accumulating studies have found that hypoxia-related lncRNAs can regulate renal cell carcinoma (RCC) progression. These lncRNAs engage in intricate interactions with nuclear proteins, thereby influencing their stability and functional dynamics. Such interactions, in turn, intricately modulate downstream signaling pathways, ultimately impacting the biological behavior of tumor cells. For example, LncRNA-SARCC inhibits hypoxic cell cycle progression in RCC cells harboring VHL mutations, yet it reverses this inhibition in RCC cells with restored VHL function. Mechanistic analysis demonstrates that LncRNA-SARCC post-transcriptionally modulates the androgen receptor (AR) by physically associating with and destabilizing the AR protein, subsequently suppressing the AR/HIF-2α/C-MYC signaling pathway. Conversely, HIF-2α transcriptionally regulates the expression of LncRNA-SARCC by binding to hypoxia-responsive elements located on the LncRNA-SARCC promoter [77]. A study showed that the RP11-367G18.1 variant 2 played an indispensable role in promoting the hypoxia-induced development and metastasis of RCC. Mechanistically, the RP11-367G18.1 variant 2 regulates the expression of hypoxia-related genes by interacting with the histone acetyltransferase p300 to promote acetylation of histone 4 at lysine 16 (H4K16Ac) [78]. Besides interacting with proteins, hypoxia-related lncRNAs can also function as sponges for miRNAs, indirectly regulating downstream signaling pathways and further impacting the trajectory of RCC progression. A study demonstrated a positive correlation between increased HOTAIR levels and tumor progression. Functional experiments indicated that HOTAIR was sponging miR-217, promoting RCC progression partially through the HIF-1α/AXL signaling pathway [79]. Notably, a recent study has also shown HOTAIR modulates several target genes through epigenetic mechanisms and regulates diverse oncogenic cellular and signaling pathways [98].
3.5. Other Cancers
Increasing numbers of research studies have also demonstrated that hypoxia-related lncRNAs can modulate the progression of other cancers through the mechanisms described above, primarily through their modulation of the HIF-α signaling pathway. In cervical cancer, lincRNA-p21, a lncRNA that responds to hypoxia, plays a crucial role in enhancing glycolysis under hypoxic conditions. Hypoxia/HIF-1α-induced lincRNA-p21 disrupts the protein–protein interaction between VHL and HIF-1α by binding to both factors, thereby inhibiting the VHL-mediated ubiquitination of HIF-1α and leading to the accumulation of HIF-1α [86]. Additionally, studies have also reported that hypoxia-related lncRNAs such as PVT1, DANCR, and HIFCAR facilitate the stabilization and transcriptional network of HIF-1α to facilitate cancer progression [80,81,85]. Several other hypoxia-related lncRNAs have emerged as promising candidates for diagnostic and prognostic biomarkers across a spectrum of cancers, functioning as miRNA sponges or chromatin scaffolds that facilitate protein interactions [82,83,84,99]. Furthermore, WT1-AS and MEG3 have been shown to modulate tumor growth through their association with histone methylation. Beyond histone methylation, hypoxia-related DARS-AS1 also regulates protein stability through m6A modification and ubiquitination [87,88]. In summary, these mechanisms mediated by hypoxia-related lncRNAs ultimately influence oncogenic signaling pathways and tumor behavior, thereby unveiling promising avenues for potential therapeutic interventions.
4. Hypoxia-Related circRNAs in Cancers
CircRNAs are crucial for the regulation of genes, and their dysregulation may promote tumorigenesis [100]. Recent studies on RNA molecules have revealed the complex mechanisms underlying the expression of cancer-related genes [101]. Tumor stem cell maintenance, vascularization, invasion, metastasis, energy metabolism, cell immortalization, genetic instability, and chemotherapy resistance are influenced by hypoxic TME and circRNA functions [102]. To date, the relationship among circRNAs, hypoxia/HIFs, and cancer has been examined only in preliminary studies. Therefore, in-depth investigation is necessary to discover new biomarkers and targets for cancer diagnosis and treatment. The following sections discuss the latest research results about the function of circRNAs that interact with hypoxia in various malignancies (Figure 3 and Table 2).
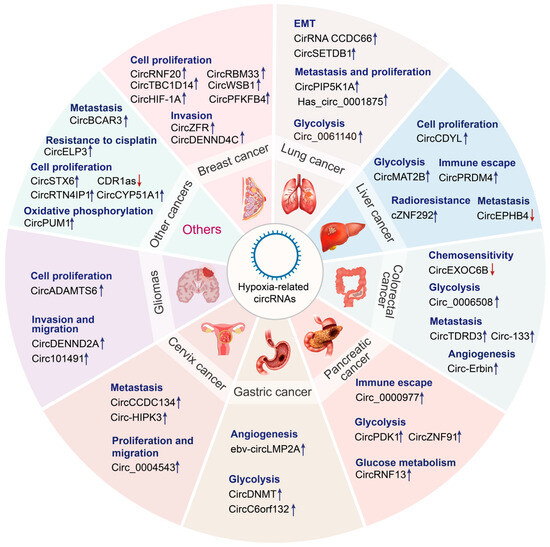
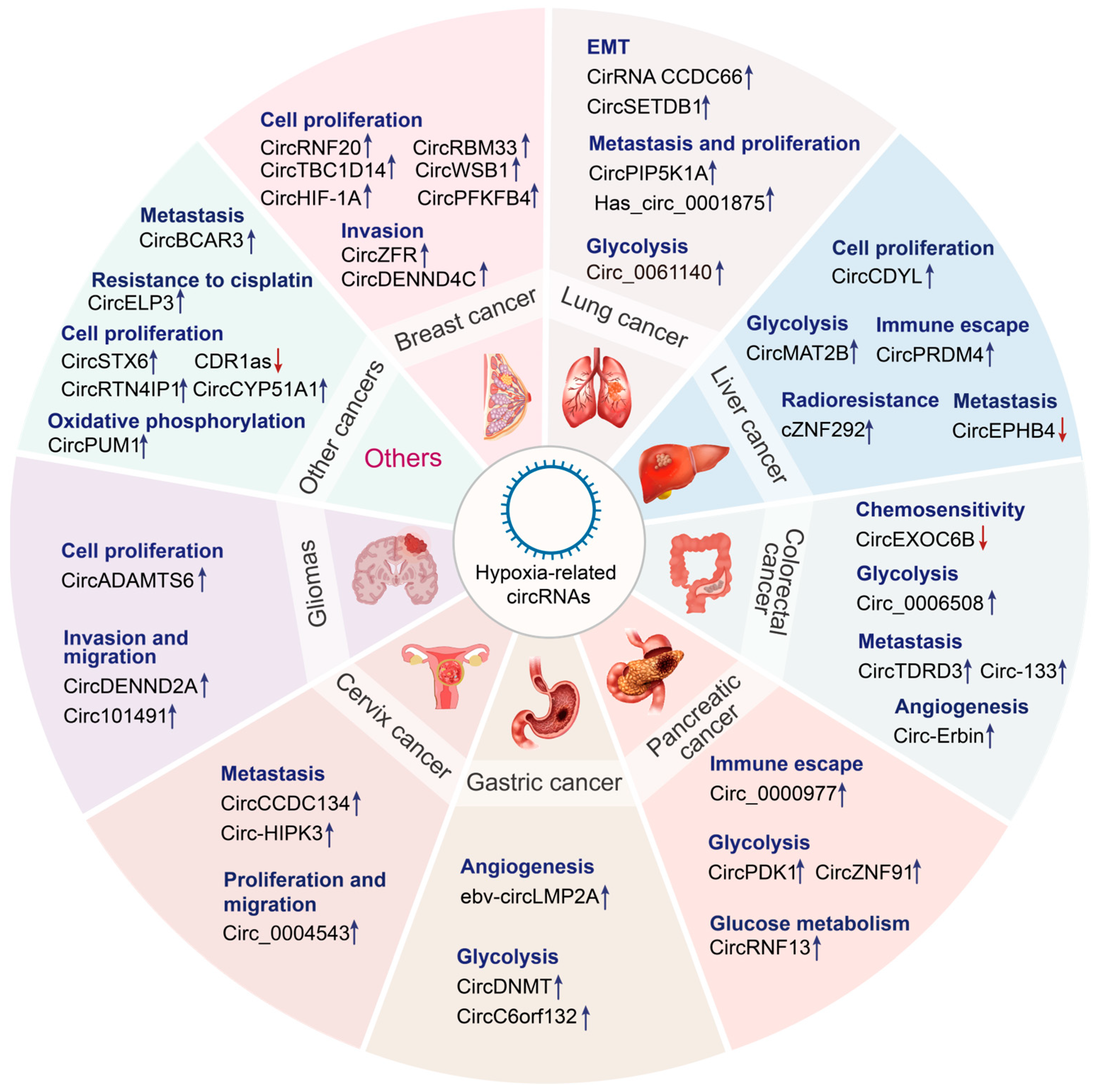
Figure 3.
Emerging roles of hypoxia-related circRNAs in cancers. Hypoxia-related circRNAs perform diverse functions in various cancer types, including lung cancer, liver cancer, colorectal cancer, pancreatic cancer, gastric cancer, cervical cancer, glioma, and so on. Upregulated circRNAs are indicated by the blue arrow, whereas downregulated circRNAs are indicated by the red arrow.

Table 2.
Summary of hypoxia-related circRNAs in cancers.
4.1. Gastrointestinal Cancers
4.1.1. Liver Cancer
HCC is a prevalent cancer, posing a significant challenge to global health care [146]. Research has confirmed that hypoxia-related circRNAs may have a role in the development and advancement of liver cancer. Hypoxia-related circRNAs often function as miRNA sponges to derepress their target genes. For example, upregulated circMAT2B increases the expression of its downstream gene PKM2 by sequestering miR-338-3p in HCC. PKM2 is a crucial enzyme contributing to glycolysis and HCC progression [116]. Certain hypoxia-related circRNAs regulate the immune microenvironment of HCC, influencing the infiltration and activity of immune cells. A study showed that circPRDM4 facilitated the immune evasion of HCC cells by promoting the recruitment of HIF-1α to the promoter region of the CD274 gene, which encodes programmed death ligand 1, consequently suppressing the infiltration of CD8+ T cells in the hypoxic TME [117].
Hypoxia-related circRNAs have been reported to modulate the onset and progression of HCC by regulating the Wnt/β-catenin and phosphatidylinositol 3-kinase (PI3K) signaling pathways. For instance, hypoxia-induced cZNF292 promotes HCC development in a time-dependent manner. Downregulation of cZNF292 promotes the nuclear translocation of SOX9 and concomitantly suppresses the Wnt/β-catenin pathway, consequently attenuating the proliferation and radiosensitivity of hypoxic hepatocytes [118]. Circ-CDYL is overexpressed in HCC and functions as a ceRNA by interacting with mRNAs encoding HIF-1AN and HDGF, thereby sequestering miR-328-3p and miR-892a, respectively. This phenomenon results in the PI3K-threonine kinase (AKT)—mechanistic target of rapamycin complex 1 (mTORC1)/β-catenin and NOTCH2 signaling pathways—being activated, triggering the expression of important effector molecules, such as BIRC5 and MYC proto-oncogene, which consequently promote the self-renewal of HCC cells [119]. Conversely, circ-EPHB4 downregulation inhibits the growth of HCC by regulating the PI3K-AKT pathway and HIF-1α [120]. Therefore, an increasing amount of research demonstrates that hypoxia-related circRNAs are crucial players in HCC progression. The strategic targeting of these hypoxia-related circRNAs presents a promising avenue for therapeutic intervention in HCC.
4.1.2. Colorectal Cancer
The CRC ranks as the second primary contributor to cancer-related mortality globally and the third most prevalent malignancy [147]. Increasing numbers of research studies have also revealed that hypoxia-related lncRNAs can modulate the progression of colorectal cancers. A fundamental mechanism through which hypoxia-related circRNAs exert their influence on CRC progression lies in the modulation of HIF-1α expression. A study demonstrated that circTDRD3 enhanced the malignant progression of CRC by absorbing miR-1231 to increase HIF-1α expression [121]. Li et al. found that clinical samples from patients with CRC exhibited increased expression of circ-ERBIN, which showed a positive correlation with aggressive phenotypes. Functional assays revealed that circ-Erbin facilitated cap-independent translation of HIF-1α by sequestering miR-138-5p and miR-125a-5p, which collectively regulated the eukaryotic translation initiation factor 4EBP-1 [123]. Another study showed that circ_0006508 is remarkably upregulated in CRC and is related to the TNM stage and overall survival. Mechanistically, HIF-1α-induced circ_0006508 directly sequesters miR-1272, suppressing glycolysis [122]. Hypoxia-related circRNAs also modulate key signaling pathways involved in CRC cell growth, survival, and metastasis. CRC tissues exhibit decreased expression of circEXOC6B, which is correlated with an unfavorable prognosis. Mechanistically, circEXOC6B binds to RRAGB and suppresses its heterodimer formation, thereby inhibiting the mTORC1 pathway and decreasing HIF-1α expression. Additionally, HIF-1α interacts with a specific region in the RRAGB promoter to increase its transcription [124]. Circ-133 is upregulated in plasma exosomes from patients with CRC, and its expression increases with disease progression. Hypoxia-induced exosomal circ-133 is transferred to normoxic cells, leading to the promotion of cancer metastasis through modulation of the miR-133a/GEF-H1/RhoA axis. However, knockdown of circ-133 has been shown to prevent CRC metastasis in animal models [125].
4.1.3. Pancreatic Cancer
Hypoxia-related circRNAs have also been shown in a growing body of research to modulate the progression of pancreatic cancer. Hypoxia-related circRNAs often function as miRNA sponges to regulate the expression of HIF-1α. Research revealed that circ_0000977 promoted immune responses in pancreatic cancer under hypoxic settings. Upregulated circ_0000977 increased the expression of ADAM10 and HIF-1α by competitively binding to miR-153, leading to the suppression of natural killer cells in the hypoxic TME [126]. Meanwhile, HIF-1α plays a pivotal role in modulating the biogenesis of these circRNAs, thereby impacting tumor growth and metastasis through their intricate interplay with miRNAs. A study showed that the biogenesis of circRNF13 was mediated by HIF-1α and EIF4A3. Functional analysis revealed that circRNF13 promoted glucose metabolism and accelerated the development of pancreatic cancer. Mechanistically, circRNF13 regulates the miR-654-3p/PDK3 pathway to facilitate tumor formation and metastasis in pancreatic cancer [127]. Similarly, circPDK1 is upregulated in both tumor tissues and serum exosomes in pancreatic cancer and is correlated with an unfavorable prognosis. CircPDK1 is activated by HIF-1α through transcriptional regulation and serves as a ceRNA of miR-628-3p, leading to the activation of the BPTF/c-myc axis. Furthermore, circPDK1 functions as a scaffold to enhance the interaction between 65 kDa Myc box-dependent interacting protein 1 (BIN1), a tumor suppressor that interacts with c-myc to limit its transcriptional activity, and UBE2O, thereby facilitating BIN1 degradation by UBE2O [128]. CircZNF91 has been demonstrated to promote gemcitabine (GEM) resistance. Mechanistically, HIF-1α-induced exosomal circZNF91 is transferred to normoxic pancreatic cancer cells and serves as a ceRNA of miR-23b-3p, thereby relieving the inhibitory effects of miR-23b-3p on Sirtuin1 (SIRT1) expression. Consequently, upregulated SIRT1 enhances HIF-1α stability through deacetylation, resulting in glycolysis and GEM resistance in recipient pancreatic cancer cells [129]. Collectively, these studies revealed the intricate interplay between hypoxia-related circRNAs and HIF-1α, accelerating the deterioration process and influencing the therapeutic response in pancreatic cancer through a myriad of mechanisms.
4.1.4. Gastric Cancer
In developing nations, GC is a major cause of cancer-related mortality. Hypoxia-related circRNAs are considered novel and promising therapeutic targets for GC [147]. Ebv-circLMP2A expression is positively related to microvessel density and the expression of HIF-1α and VEGFA in EBV-associated GC. HIF-1α-induced ebv-circLMP2A facilitates angiogenesis through the KHSRP/VHL/HIF-1α/VEGFA axis under hypoxic conditions [130]. Additionally, hypoxia-related circRNAs regulate the expression of key oncogenes by acting as a ceRNA, which in turn influences the malignant progression of tumors. A study showed that circDNMT1 knockdown attenuated the malignant progression of GC cells. The oncogenic role of circDNMT1 is attributed to its ability to sequester miR-576-3p, which binds to the 3′-untranslated region of HIF-1α mRNA and adversely regulates it. CircDNMT1 modulates the miR-576-3p/HIF-1α axis, resulting in malignant progression and metabolic reprogramming in GC both in vivo and in vitro [131]. Under hypoxic conditions, circC6orf132 facilitates the progression and glycolytic activity of GC cells by sequestering miR-873-5p, resulting in increased expression of PRKAA1. PRKAA1 constitutes a critical subunit of the 5′-AMP-activated protein kinase (AMPK), which plays a pivotal role in promoting energy metabolism and tumor progression in GC [132].
4.1.5. Other Gastrointestinal Cancers
In addition to the tumor types described above, hypoxia-related circRNAs also have a significant impact on other gastrointestinal cancers. Hypoxia-related circRNAs emerge as pivotal regulators of cancer progression, functioning as miRNA sponges that intricately modulate the expression of critical downstream oncogenes. In esophageal cancer, the splicing factor QKI promoted the generation of circBCAR3, which in turn facilitated tumorigenesis. This effect was achieved upon the binding of circBCAR3 to miR-27a-3p, which resulted in the upregulation of transportin-1 (TNPO1). The upregulation of TNPO1, in turn, enhances the proliferation, migration, and invasion of esophageal cancer cells [139]. CircSTX6 regulates the expression of non-muscle myosin heavy chain 9 (MYH9) via the circSTX6/miR-449b-5p and circSTX6/CUL2/HIF-1α pathways, promoting the proliferation and migration of pancreatic ductal adenocarcinoma (PDAC) cells [143]. Similarly, Huang et al. showed that circRTN4IP1 knockdown in intrahepatic cholangiocarcinoma (ICC) cells suppressed cell proliferation and glucose metabolism, whereas cell apoptosis was promoted by the miR-541-5p/HIF-1α axis [145]. Certain circRNAs promote energy metabolism and viability of cancer cells by maintaining mitochondrial stability and function. In esophageal squamous cell carcinoma (ESCC), circPUM1 promoted the progression of ESCC by inhibiting cancer cell pyroptosis. Mechanistically, circPUM1 maintains the stability of mitochondrial complex III, thereby enhancing oxidative phosphorylation and subsequent ATP production in ESCC cells. Additionally, ESCC cells use circPUM1 for cell adaptation [140]. In summary, hypoxia-related circRNAs, which regulate biological processes such as proliferation, migration, metabolism, and apoptosis of cancer cells, may serve as potential prognostic markers and therapeutic targets for patients with gastrointestinal cancers.
4.2. Breast Cancer
BC has the highest global incidence rate for women [147]. Despite recent advances in the early diagnosis and treatment of BC, a large number of patients with BC develop metastasis. Therefore, identifying novel biomarkers is necessary to provide more effective diagnostic and therapeutic methods for BC. CircRNAs intricately enhance the expression of HIF-1α through the competitive binding of specific miRNAs, consequently facilitating BC cell growth and metastasis. For example, circRNF20 is upregulated in BC and facilitates cancer cell proliferation and the Warburg effect. Mechanistically, the competitive binding of circRNF20 to miR-487a upregulates the expression of HIF-1α and facilitates the transcription of hexokinase II (HK2), a key enzyme that enhances glycolysis [103]. The circZFR has been shown to promote the progression of BC. Silencing of circZFR suppresses the viability, invasiveness, migration, colony-forming ability, and glycolytic activity of BC cells through the miR-578/HIF-1α axis [104]. CircRBM33 is upregulated in BC, and its silencing suppresses miR-542-3p-mediated expression of HIF-1α, subsequently inhibiting glycolysis and proliferation and enhancing apoptosis in BC cells [105]. In addition, circDENND4C and circTBC1D14 can also facilitate the development of BC under hypoxic conditions [106,107,148].
Recent studies have reported that hypoxia-related circRNAs can promote BC progression by decreasing the expression of the cell cycle inhibitors p53, p21, and p27. The hypoxia-inducible circWSB1 is upregulated in BC and promotes cancer cell proliferation. Mechanistically, the interaction between circWSB1 and ubiquitin-specific peptidase 10 (USP10) attenuates the USP10-mediated stabilization of p53 [108]. CircHIF-1α (hsa_circ_0004623) has been shown to facilitate cancer cell proliferation and metastasis in BC. Mechanistically, circHIF1A modulates nuclear factor IB (NFIB) expression and translocation via post-transcriptional and post-translational modifications, thereby leading to the AKT/STAT3 signaling pathway being activated and p21 being suppressed [109]. Additionally, a poor prognosis is linked to elevated circPFKFB4 expression. Mechanistically, hypoxia-induced circPFKFB4 directly interacts with both DDB1 and DDB2, facilitating the assembly of the CRL4DDB2 E3 ubiquitin ligase, which in turn mediates the ubiquitination of p27 [110].
4.3. Lung Cancer
Lung cancer is the primary cause of cancer-related mortality worldwide, and its most common subtype is non-small cell lung cancer (NSCLC) [149]. Hypoxia-related circRNAs exert intricate regulatory influence on lung cancer cell proliferation, metastasis, and cellular morphology via miRNA sponges. Circ-0001875 is markedly increased in NSCLC cell lines and tissues. It not only facilitates NSCLC cell proliferation and metastasis but also triggers filopodia extension in NSCLC cells. Mechanistically, circ-0001875 modulates SP1 expression by sponging miR-31-5p, thereby influencing EMT through the TGFβ/Smad2 signaling pathway. The negative regulation of circ-0001875 is mediated by SP1 through an AluSq-dependent feedback loop. However, this regulation is disrupted under hypoxic conditions owing to the competitive binding of HIF-1α to SP1 [111]. Similarly, circPIP5K1A is upregulated in NSCLC cells and modulates cancer cell proliferation and metastasis by binding to miR-600 and activating HIF-1α [112].
Lung adenocarcinoma (LUAD) is the most prevalent form of non-small cell lung cancer, which comprises around 40% of all lung cancer [150]. LUAD cells exhibit increased expression of circ-0000211, which promotes cancer cell migration through the miR-622/HIF-1α axis [113]. Similarly, upregulated circ-0061140 has been associated with a decreased survival rate in patients with LUAD. Circ-0061140 acts as a sponge of miR-653 to regulate the expression of HK2, thereby promoting the hypoxia-induced glycolytic activity, migration, and invasiveness of LUAD cells [114]. The expression of hypoxia-related circRNAs not only affects the migration of LUAD cells but also correlates closely with LUAD cell adaptation and drug resistance. The increased expression of CCDC66 in LUAD is attributed to the negative modulation of nAchR7α and the positive modulation of c-Met and FAK. In response to hypoxia, cells quickly upregulate phosphorylated c-Met, EMT, and SAE2, which consequently enhance drug resistance and EMT in LUAD [115]. Therefore, identifying hypoxia-related circRNAs that are potentially targetable in lung cancer progression is necessary to improve treatment efficacy.
4.4. Cervical Cancer
The second most frequent cause of cancer death for women between the ages of 20 and 39 is cervical cancer. Hypoxia-related circRNAs have been demonstrated to be crucial in regulating the progress of cervical cancer by modulating HIF-1α expression under hypoxic conditions [151]. For example, circCCDC134 is significantly upregulated in cervical cancer and facilitates tumor growth and metastasis. Mechanistically, circCCDC134 mediates nuclear recruitment of p65 and functions as a sponge of miR-503-5p to modulate myeloblastosis (MYB) expression, which is a transcription factor, in the cytoplasm, thereby facilitating HIF-1α transcription [133]. Upregulated circ_0004543 facilitated the viability, proliferation, migration, and invasiveness of cervical cancer cells while suppressing their apoptosis. These effects are mediated through the sequestration of hsa-miR-217 by circ_0004543, which increases HIF-1α levels in CC cells [134]. Similarly, upregulated circ-HIPK3 in cervical cancer cells sequesters miR-338-3p to increase the expression of HIF-1α, consequently enhancing the proliferation, colony formation, migration, and invasiveness of cervical cancer cells while suppressing their apoptosis [135]. In cervical cancer, circCCDC134, circ_0004543, and circ-HIPK3 all upregulate the expression of HIF-1α by acting as miRNA sponges, thus promoting the proliferation, migration, and invasion of cervical cancer cells and thereby accelerating tumor growth and metastasis.
4.5. Glioma
Gliomas, which originate from progenitor or neuroglial stem cells, are predominant tumors in the central nervous system. Therefore, identifying novel and effective biomarkers for the diagnosis and treatment of glioma is necessary [152]. Hypoxia-related circRNAs have the ability to function as miRNA sponges to regulate the expression of downstream genes. Circ101491 is found to be upregulated in both tumor tissues and peripheral plasma of glioma patients. Upregulated circ101491 is strongly linked to a poor prognosis in glioma. Hypoxia-induced upregulation of circ101491 can promote the invasiveness, metastasis, and viability of neighboring or residual glioma cells through exosomes. Mechanistically, circ101491 mediates the upregulation of the downstream target gene endothelin-1 (EDN1) by acting as a ceRNA of miR-125b-5p. Notably, high expression of EDN1 promotes the viability and invasion of glioma cells [138]. Similarly, another study showed that hypoxia-induced upregulation of circDENND2A promoted the invasiveness and migration of gliomas by sequestering miR-625-5p [136]. In addition to acting as miRNA sponges, circRNAs promote glioma cell proliferation and inhibit apoptosis through other mechanisms. CircADAMTS6 can drive the malignant progression of glioblastoma by suppressing apoptosis and facilitating cancer cell proliferation. Mechanistically, hypoxia-induced upregulation of circADAMTS6 is mediated by the transcription factor AP-1 and the RNA-binding protein TDP43 in glioblastoma. CircADAMTS6 promotes the progression of glioblastoma by facilitating the recruitment and stabilization of annexin A2 (ANXA2) through a proteasome-dependent mechanism. ANXA2 exerts its effects by inhibiting apoptosis and promoting cell proliferation and self-renewal of stem cells [137]. Consequently, hypoxia-related circRNAs may serve as potential prognostic markers and therapeutic targets for patients with glioma.
4.6. Other Cancers
Recent research has revealed that hypoxia-related circRNAs are dysregulated in most cancer types, thus playing an indispensable role in the development of cancer, either as tumor suppressors or as oncogenes [101]. For example, in bladder cancer, circELP3 is upregulated in the hypoxic microenvironment and is positively correlated with tumor development. Bladder cancer cells with reduced circELP3 expression have impaired self-renewal capacity. In addition, silencing of circELP3 enhances apoptosis and reduces cisplatin resistance in bladder cancer cells [141]. CircRNA CDR1as is downregulated in ovarian cancer and serves as a tumor suppressor. It sequesters miR-135b-5p, increasing the expression of HIF-1AN and suppressing the proliferation of ovarian cancer cells [144]. In hypoxic osteosarcoma cells, circCYP51A1 can regulate glycolysis, proliferation, migration, and invasion by modulating the miR-490-3p/KLF12 axis [142]. These studies revealed that hypoxia-related circRNAs exert pivotal roles in cancer by regulating tumor progression and even chemotherapy drug sensitivity. Their influence on cancer cell behavior underscores their importance as potential therapeutic targets in cancer treatment.
5. Clinical Relevance of Hypoxia-Related lncRNAs and circRNAs in Cancers
Numerous hypoxia-related lncRNAs and circRNAs have been reported to be dysregulated in various cancer types. Additionally, they are highly specific and can be easily detected in tissues, serum, plasma, urine, and saliva. Therefore, hypoxia-related lncRNAs and circRNAs hold great promise as diagnostic and prognostic biomarkers and therapeutic targets in various cancers (Figure 4).
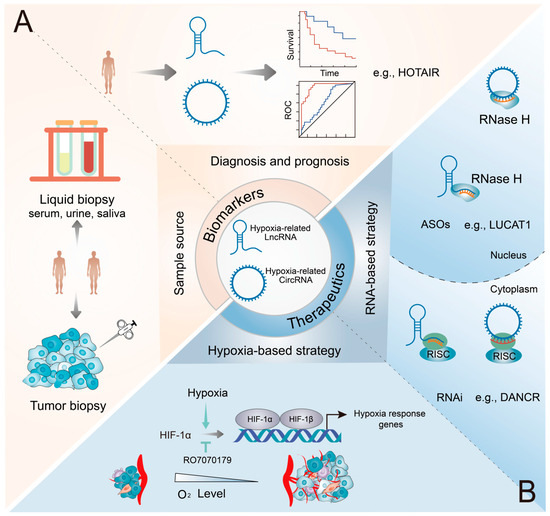
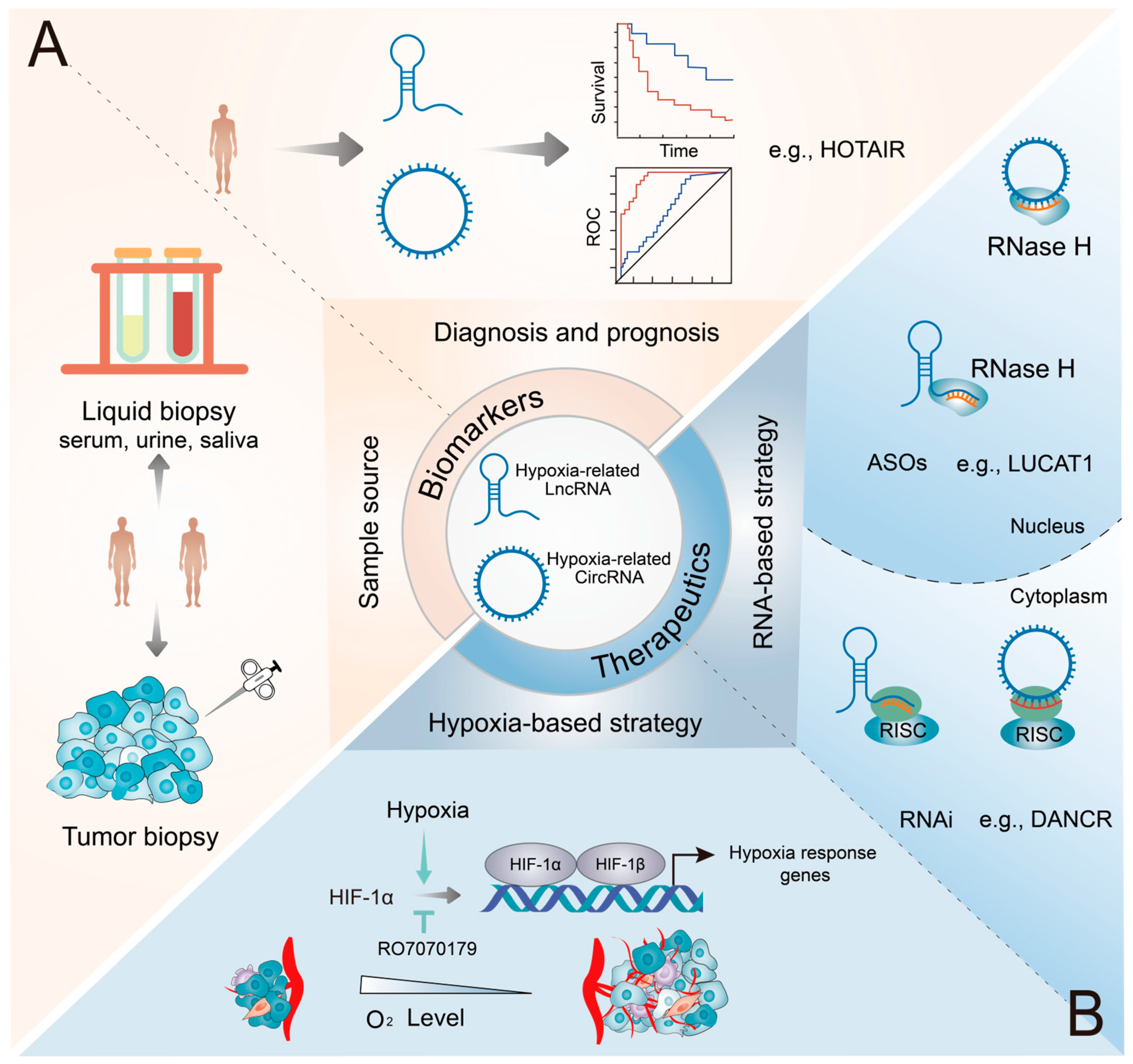
Figure 4.
Potential applications of hypoxia-related lncRNAs and circRNAs as biomarkers and therapeutic targets in cancers. (A) Diagnostic potential of hypoxic ncRNAs. These ncRNAs can be reliably detected in a diverse range of biospecimens, encompassing both conventional tumor biopsy samples and liquid biopsy samples (such as blood and urine). (B) Therapeutic potential of hypoxic ncRNAs. Inhibitors effectively modulate the expression and function of HIF-1α, while RNAi and ASO therapies underscore the precise targeting prowess of ncRNAs, specifically in the cytoplasm and nucleus, respectively.
5.1. Biomarker Potentials
Ongoing clinical trials are actively recruiting patients to ascertain the potential of ncRNAs as diagnostic and prognostic biomarkers across various cancer types. Clinical research focusing on hypoxia-related lncRNAs and circRNAs offers a promising avenue for future exploration (Table 3). Thus, hypoxia-related lncRNAs and circRNAs have emerged as potential biomarkers for the diagnosis and prognosis of cancer (Figure 4A). A clinical study (NCT03469544) is currently investigating the potential of HOTAIR as a biomarker for thyroid cancer. Notably, HOTAIR, one of the most extensively investigated lncRNA, exhibits high specificity for RCC, and there is a positive correlation between its expression and the TNM stage [153]. Additionally, a notable elevation of HOTAIR levels was observed in CRC tissues relative to adjacent normal tissues (p = 0.00099), and survival analysis indicated that patients exhibiting a high level of HOTAIR had a poorer prognosis in CRC (p = 0.032) [154]. Another clinical trial (NCT04729855) is actively recruiting participants to examine the relationship between HOTTIP and CRC. In glioma patients, high HOTTIP expression was linked to poor overall survival, according to Kaplan–Meier survival analysis (p = 0.032) [76]. Additionally, ROC (receiver operating characteristics) analysis demonstrated that the diagnostic performance of the combination of circ-CDYL, HDGF, and HIF1AN for early-stage HCC (area under the curve [AUC] = 0.73) surpassed that of the traditional biomarker α-fetoprotein (AFP) (AUC = 0.59) [119]. In addition to these ncRNAs described above, several other hypoxia-related lncRNAs and circRNAs, including PVT1, HITT, BX111, circTDRD3, and circDENND4C, have shown promise as diagnostic and prognostic biomarkers in various cancers [63,65,80,106,121]. Notably, circDENND4C has been extensively evaluated in numerous independent studies as a prognostic biomarker for BC [148].

Table 3.
Examples of preclinical and clinical trials exploring the application of hypoxia-related ncRNAs in cancers.
Clinically, liquid biopsy is receiving substantial attention owing to its less invasive nature relative to conventional tissue biopsy and its feasibility for real-time disease monitoring. Hypoxia-related circ101491, circ-133, and circHIF1A are overexpressed in peripheral blood plasma in a variety of malignancies, and this dysregulation correlates with a poor prognosis [109,125,138]. A clinical trial (NCT04584996) is investigating the expression of candidate circRNAs in tissue, blood, bile, and biopsy samples from patients with pancreaticobiliary cancer. Altogether, hypoxia-related lncRNAs and circRNAs can be used as biomarkers to improve the accuracy of cancer diagnosis and prognosis.
5.2. Therapeutic Potential
Low oxygen concentration, or hypoxia, is an important factor driving tumor aggressiveness. Clinical trials are underway to address hypoxia in cancer with investigational drugs (Figure 4B). In particular, a clinical study (NCT02564614) is examining the role of the HIF-1α mRNA antagonist RO7070179 in adult patients with liver cancer. Additionally, a phase 3 clinical trial (NCT04195750) is investigating the efficacy of the HIF-2α inhibitor MK-6482 in advanced renal clear cell carcinoma. Targeting HIF-1α and HIF-2α represents a promising therapeutic strategy for cancer. However, the emergence of resistance is inevitable. For instance, prolonged exposure to PT2399, a selective inhibitor of HIF-2, leads to the emergence of resistance associated with elevated tumor vascularity and increased levels of VEGF [93]. Therefore, developing novel and more effective therapeutic approaches is necessary to enhance treatment outcomes. Targeting the downstream effectors of the HIF signaling pathway is another promising approach to counteracting the hypoxia-induced invasiveness of cancer. The association of hypoxia-related lncRNAs and circRNAs with the onset and progression of cancer highlights their potential as therapeutic targets for cancer. Therefore, specific targeting of lncRNAs and circRNAs in the hypoxic TME represents a promising therapeutic strategy for complex and diverse cancer types.
Effective gene overexpression or knockdown techniques may facilitate the targeting of hypoxia-related lncRNAs and circRNAs for therapeutic purposes (Figure 4B). At present, RNA-based therapies primarily involve the use of RNA interference (RNAi) and antisense oligonucleotides (ASOs) to target specific regions and RNA molecules. Studies using mouse models have shown that targeting oncogenic hypoxia-related lncRNAs and circRNAs with ASOs and RNAi can effectively inhibit tumor growth. For instance, shRNAs can effectively target DANCR, a recently identified oncogenic lncRNA in NPC, to suppress tumor metastasis in nude mouse models [81]. Similarly, intravenous administration of shRNAs targeting circ-0001875 and circWSB1 can inhibit tumor proliferation in nude mouse models [108,111]. Various hypoxia-related circRNAs have been shown to regulate radiosensitivity and chemotherapy resistance in cancer. For instance, targeting cZNF292 with shRNAs enhances the radiosensitivity of HCC cells under hypoxic conditions in mice. Notably, the combination of radiotherapy and cZNF292 knockdown can reduce tumor volume in mice more effectively than radiotherapy or cZNF292 knockdown alone [118]. Targeting hypoxia-related circZNF91 with siRNAs can enhance GEM sensitivity in mice, whereas exosomal circZNF91 can transmit GEM resistance from hypoxic pancreatic cancer cells to normoxic pancreatic cancer cells [129]. Recently, a study in mice with Angelman syndrome showed that ASOs can be used to target lncRNAs [155]. The combination of LUCAT1 knockdown via ASO and oxaliplatin is reported to be more effective than oxaliplatin monotherapy in treating CRC in mice [66]. Conversely, the overexpression of suppressive hypoxia-related ncRNAs (e.g., lncRNA-LET and circEPHB4) has been shown to diminish tumor size and reduce metastases in nude mouse models [50,120]. Given their pivotal functional roles, hypoxia-related lncRNAs and circRNAs serve as promising therapeutic targets for cancer.
To date, several anti-hypoxia drugs based on HIF-1α, such as topotecan, and HIF-2α inhibitors, such as PT2399, have been clinically used for the treatment of cancer patients. Although some progress has been made in research on the role of hypoxia-related lncRNAs and circRNAs in cancer, clinical strategies for targeting these molecules are lacking at present. Therefore, large-scale, multi-center prospective cohort studies are warranted to validate the clinical utility of hypoxia-related lncRNAs and circRNAs in the development of new anti-cancer therapies for improving patient outcomes.
6. Concluding Remarks and Perspectives
The high death rate of cancer is due to its quick progression and lack of early diagnostic indicators. Consequently, there is an urgent imperative to identify novel tumor markers and therapeutic targets that can facilitate early diagnosis, accurate prognosis, and effective treatment. The hypoxia-related lncRNAs and circRNAs altered expression in the tumor specimens, and the easy detection of them in bodily fluids makes them attractive candidates for non-invasive liquid biopsy approaches. Moreover, these hypoxia-related ncRNAs have been shown to exert substantial effects in cancer hallmarks, including tumor metabolism, immune escape, tumor angiogenesis, migration/invasion, apoptosis, and growth/proliferation. Therefore, hypoxia-related lncRNAs and circRNAs have the potential to be used as biomarkers or therapeutic targets in solid tumors.
Although substantial progress has been made in examining the functions of lncRNAs and circRNAs in the hypoxic TME, certain unaddressed challenges and limitations warrant further investigation. In addition, the following questions remain unresolved: (i) When targeting hypoxia-related lncRNAs and circRNAs in cancer, how can we prevent their impact on essential gene expression in normal tissues? The identification of target ligands with high affinity and stability may help mitigate potential adverse effects and toxicity. Moreover, the refinement of delivery systems specifically engineered for the precise targeting of circRNA is paramount. (ii) Which factors contribute to the significant differences in hypoxic responses observed between cell lines and animal models? To date, most studies have predominantly focused on two-dimensional cell culture models; therefore, future studies should involve the use of three-dimensional tumor organ models that incorporate the TME [156]. (iii) Does the combination of hypoxia-related lncRNA- or circRNA-targeting therapies with other anti-tumor strategies yield superior therapeutic effects? In a recent study, the combination of ASO-mediated lncRNA downregulation with conventional chemotherapeutic agents demonstrated higher efficacy than chemotherapeutic agents alone in the treatment of CRC [66,118]. Therefore, the combination of hypoxia-related lncRNA- or circRNA-targeting therapies with other anti-cancer treatments holds substantial promise for enhancing treatment efficacy. (iv) Utilizing hypoxia-associated lncRNAs and circRNAs could be converted into RNA vaccines for tumor management in clinical settings. The advent of mRNA as a secure and efficacious platform in the race to develop a COVID-19 vaccine has imparted a swift understanding of the merits and potential risks associated with mRNA–LNP (a representative lipid nanoparticle) technology across the globe [157]. Notably, circRNAs, characterized by their substantial stability and attainable high translation efficiencies, have garnered considerable attention [158].
Notably, a gap exists between basic research findings and the clinical application of hypoxia-related lncRNAs and circRNAs as diagnostic, prognostic, and therapeutic biomarkers. Therefore, the therapeutic value and safety of hypoxia-related lncRNAs or circRNAs should be extensively investigated in future clinical studies with a large sample size.
Author Contributions
Z.M. conceived the review article; Z.M. and G.Q. designed the overall research framework, provided overall supervision, and critically revised the manuscript; L.G. contributed to collecting relevant papers and the overall editing of the manuscript; C.Z. and H.Z. created the table and corresponding visualizations; F.Y. helped to modify the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the following research grants from the National Foreign Experts Program of China (DL2023027001L to Z.M.); and Yangtze University Science and Technology Aid to Tibet Medical Talent Training Program Project (2023YZ18 to Z.M.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Casey, S.C.; Amedei, A.; Aquilano, K.; Azmi, A.S.; Benencia, F.; Bhakta, D.; Bilsland, A.E.; Boosani, C.S.; Chen, S.; Ciriolo, M.R.; et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin. Cancer Biol. 2015, 35, S199–S223. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Harris, A.L.; McIntyre, A. The tumour hypoxia induced non-coding transcriptome. Mol. Asp. Med. 2016, 47–48, 35–53. [Google Scholar] [CrossRef]
- Shih, J.W.; Kung, H.J. Long non-coding RNA and tumor hypoxia: New players ushered toward an old arena. J. Biomed. Sci. 2017, 24, 53. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhao, F.; Zhang, J. circAtlas 3.0: A gateway to 3 million curated vertebrate circular RNAs based on a standardized nomenclature scheme. Nucleic Acids Res. 2024, 52, D52–D60. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Bourelle, P.; Desjardins-Henri, C.; Mathurin-St-Pierre, D.; Deschamps-Francoeur, G.; Fafard-Couture, É.; Garant, J.M.; Elela, S.A.; Scott, M.S. snoDB: An interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res. 2020, 48, D220–D225. [Google Scholar] [CrossRef]
- Chandra Gupta, S.; Nandan Tripathi, Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int. J. Cancer 2017, 140, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Rajappa, A.; Banerjee, S.; Sharma, V.; Khandelia, P. Circular RNAs: Emerging Role in Cancer Diagnostics and Therapeutics. Front. Mol. Biosci. 2020, 7, 577938. [Google Scholar] [CrossRef]
- Shen, G.; Li, X.; Jia, Y.F.; Piazza, G.A.; Xi, Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013, 34, 336–341. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, Y.; Li, J.; Zhu, Q. Small Non-Coding RNAs in Human Cancer. Genes 2022, 13, 2072. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef] [PubMed]
- Coan, M.; Haefliger, S.; Ounzain, S.; Johnson, R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat. Rev. Genet. 2024, 25, 578–595. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going circular: History, present, and future of circRNAs in cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Li Santi, A.; Gorrasi, A.; Alfieri, M.; Montuori, N.; Ragno, P. A novel oncogenic role for urokinase receptor in leukemia cells: Molecular sponge for oncosuppressor microRNAs. Oncotarget 2018, 9, 27823–27834. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Kanojia, D.; Kirtonia, A.; Srujana, N.S.V.; Jeevanandan, S.P.; Shyamsunder, P.; Sampath, S.S.; Dakle, P.; Mayakonda, A.; Kaur, H.; Yanyi, J.; et al. Transcriptome analysis identifies TODL as a novel lncRNA associated with proliferation, differentiation, and tumorigenesis in liposarcoma through FOXM1. Pharmacol. Res. 2022, 185, 106462. [Google Scholar] [CrossRef]
- Dinescu, S.; Ignat, S.; Lazar, A.D.; Constantin, C.; Neagu, M.; Costache, M. Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes 2019, 10, 52. [Google Scholar] [CrossRef]
- Shao, C.; Yang, F.; Miao, S.; Liu, W.; Wang, C.; Shu, Y.; Shen, H. Role of hypoxia-induced exosomes in tumor biology. Mol. Cancer 2018, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Cowman, S.J.; Koh, M.Y. Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer 2022, 8, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Moreno Roig, E.; Yaromina, A.; Houben, R.; Groot, A.J.; Dubois, L.; Vooijs, M. Prognostic Role of Hypoxia-Inducible Factor-2α Tumor Cell Expression in Cancer Patients: A Meta-Analysis. Front. Oncol. 2018, 8, 224. [Google Scholar] [CrossRef]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, D.; Xie, H.; Hu, Y. Interplay of long non-coding RNAs and HIF-1α: A new dimension to understanding hypoxia-regulated tumor growth and metastasis. Cancer Lett. 2021, 499, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.; Liu, S.; Zhao, H.; Zhuang, Y.; Ma, S.; Lin, C.; Hu, J.; Liu, X. Hypoxia-responsive circRNAs: A novel but important participant in non-coding RNAs ushered toward tumor hypoxia. Cell Death Dis. 2022, 13, 666. [Google Scholar] [CrossRef]
- Wang, L.; Li, B.; Bo, X.; Yi, X.; Xiao, X.; Zheng, Q. Hypoxia-induced LncRNA DACT3-AS1 upregulates PKM2 to promote metastasis in hepatocellular carcinoma through the HDAC2/FOXA3 pathway. Exp. Mol. Med. 2022, 54, 848–860. [Google Scholar] [CrossRef]
- Lu, Q.; Wang, H.; Lei, X.; Ma, Q.; Zhao, J.; Sun, W.; Guo, C.; Huang, D.; Xu, Q. LncRNA ALKBH3-AS1 enhances ALKBH3 mRNA stability to promote hepatocellular carcinoma cell proliferation and invasion. J. Cell Mol. Med. 2022, 26, 5292–5302. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, T.; Yan, L.; Zhu, S.; Jin, W.; Bai, Y.; Zeng, Y.; Zhang, X.; Yin, Z.; Yang, J.; et al. Hypoxia-Responsive lncRNA AC115619 Encodes a Micropeptide That Suppresses m6A Modifications and Hepatocellular Carcinoma Progression. Cancer Res. 2023, 83, 2496–2512. [Google Scholar] [CrossRef]
- Yang, F.; Huo, X.S.; Yuan, S.X.; Zhang, L.; Zhou, W.P.; Wang, F.; Sun, S.H. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lei, X.; Lu, Q.; Wu, Q.; Ma, Q.; Huang, D.; Zhang, Y. LncRNA FRMD6-AS1 promotes hepatocellular carcinoma cell migration and stemness by regulating SENP1/HIF-1alpha axis. Pathol. Res. Pract. 2023, 243, 154377. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.K.; Haga, H.; Patel, T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014, 127, 1585–1594. [Google Scholar] [CrossRef]
- Yang, X.; Yao, B.; Niu, Y.; Chen, T.; Mo, H.; Wang, L.; Guo, C.; Yao, D. Hypoxia-induced lncRNA EIF3J-AS1 accelerates hepatocellular carcinoma progression via targeting miR-122-5p/CTNND2 axis. Biochem. Biophys. Res. Commun. 2019, 518, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Dong, K.; Zhang, H.; Gong, J.; Wang, S. Exosomal lncRNA HMMR-AS1 mediates macrophage polarization through miR-147a/ARID3A axis under hypoxia and affects the progression of hepatocellular carcinoma. Environ. Toxicol. 2022, 37, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Song, J.; Gao, Y.; Huang, S.; Dou, R.; Zhong, P.; Huang, G.; Han, L.; Zheng, J.; Zhang, X.; et al. Hypoxia-induced HIF-1alpha/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol. 2022, 52, 102312. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Sun, L.; Min, J.; Liu, J.; Chen, D.; Zhang, H.; Zhang, H.; Zhang, H.; Zhou, Y.; et al. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018, 109, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Zhang, H.; Sun, L.; Zhou, Y.; Jin, H.; Zhang, H.; Zhang, H.; Liu, J.; Guo, H.; et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia 2014, 16, 1094–1106. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Liu, H.W.; Chen, J.Q.; Wang, S.H.; Hao, L.Q.; Liu, M.; Wang, B. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed. Pharmacother. 2017, 88, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Gao, H.; Liu, K.; Gao, B.; Ren, H.; Li, Z.; Liu, F. The lncRNA ZEB2-AS1 is upregulated in gastric cancer and affects cell proliferation and invasion via miR-143-5p/HIF-1alpha axis. Onco Targets Ther. 2019, 12, 657–667. [Google Scholar] [CrossRef]
- Liu, M.; Zhong, J.; Zeng, Z.; Huang, K.; Ye, Z.; Deng, S.; Chen, H.; Xu, F.; Li, Q.; Zhao, G. Hypoxia-induced feedback of HIF-1alpha and lncRNA-CF129 contributes to pancreatic cancer progression through stabilization of p53 protein. Theranostics 2019, 9, 4795–4810. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Wen, C.; Huo, Z.; Wang, W.; Zhan, Q.; Cheng, D.; Chen, H.; Deng, X.; Peng, C.; et al. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol. Cancer 2017, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xu, F.Y.; Zheng, H.; Cheng, P.; Chen, Q.Y.; Ye, Z.; Zhong, J.X.; Deng, S.J.; Liu, M.L.; Huang, K.; et al. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1alpha. Theranostics 2019, 9, 5298–5314. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.J.; Chen, H.Y.; Ye, Z.; Deng, S.C.; Zhu, S.; Zeng, Z.; He, C.; Liu, M.L.; Huang, K.; Zhong, J.X.; et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene 2018, 37, 5811–5828. [Google Scholar] [CrossRef]
- Sun, Y.W.; Chen, Y.F.; Li, J.; Huo, Y.M.; Liu, D.J.; Hua, R.; Zhang, J.F.; Liu, W.; Yang, J.Y.; Fu, X.L.; et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma. Br. J. Cancer 2014, 111, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Zhao, K.; Lin, Q.; Li, H.; Xue, X.; Ge, W.; He, H.; Liu, D.; Xie, H.; et al. A novel LncRNA HITT forms a regulatory loop with HIF-1alpha to modulate angiogenesis and tumor growth. Cell Death Differ. 2020, 27, 1431–1446. [Google Scholar] [CrossRef] [PubMed]
- Huan, L.; Guo, T.; Wu, Y.; Xu, L.; Huang, S.; Xu, Y.; Liang, L.; He, X. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol. Cancer 2020, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Peng, J.; Xie, X.; Yu, F.; Wang, W.; Pan, Q.; Jin, H.; Huang, X.; Yu, H.; Li, S.; et al. Roles of lncRNA LVBU in regulating urea cycle/polyamine synthesis axis to promote colorectal carcinoma progression. Oncogene 2022, 41, 4231–4243. [Google Scholar] [CrossRef]
- Jing, X.; Du, L.; Shi, S.; Niu, A.; Wu, J.; Wang, Y.; Wang, C. Hypoxia-Induced Upregulation of lncRNA ELFN1-AS1 Promotes Colon Cancer Growth and Metastasis Through Targeting TRIM14 via Sponging miR-191-5p. Front. Pharmacol. 2022, 13, 806682. [Google Scholar] [CrossRef]
- Ma, H.N.; Chen, H.J.; Liu, J.Q.; Li, W.T. Long non-coding RNA DLEU1 promotes malignancy of breast cancer by acting as an indispensable coactivator for HIF-1alpha-induced transcription of CKAP2. Cell Death Dis. 2022, 13, 625. [Google Scholar] [CrossRef]
- Zhang, J.; Du, C.; Zhang, L.; Wang, Y.; Zhang, Y.; Li, J. LncRNA LINC00649 promotes the growth and metastasis of triple-negative breast cancer by maintaining the stability of HIF-1alpha through the NF90/NF45 complex. Cell Cycle 2022, 21, 1034–1047. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Maldonado, L.; Tiana, M.; Roche, O.; Prado-Cabrero, A.; Jensen, L.; Fernandez-Barral, A.; Guijarro-Munoz, I.; Favaro, E.; Moreno-Bueno, G.; Sanz, L.; et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 2015, 34, 2609–2620. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.H.; Luo, J.L.; Hsu, M.H.; Chen, L.H.; Lu, T.P.; Tsai, M.H.; Chuang, E.Y.; Chuang, L.L.; Lai, L.C. Regulatory mechanisms and function of hypoxia-induced long noncoding RNA NDRG1-OT1 in breast cancer cells. Cell Death Dis. 2022, 13, 807. [Google Scholar] [CrossRef]
- Mineo, M.; Ricklefs, F.; Rooj, A.K.; Lyons, S.M.; Ivanov, P.; Ansari, K.I.; Nakano, I.; Chiocca, E.A.; Godlewski, J.; Bronisz, A. The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Rep. 2016, 15, 2500–2509. [Google Scholar] [CrossRef]
- Wu, W.; Hu, Q.; Nie, E.; Yu, T.; Wu, Y.; Zhi, T.; Jiang, K.; Shen, F.; Wang, Y.; Zhang, J.; et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci. Rep. 2017, 7, 45029. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, W.; Liu, G.; Xie, S.; Li, Q.; Li, Y.; Lin, Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed. Pharmacother. 2017, 95, 711–720. [Google Scholar] [CrossRef]
- Zhai, W.; Sun, Y.; Jiang, M.; Wang, M.; Gasiewicz, T.A.; Zheng, J.; Chang, C. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2alpha/C-MYC axis under hypoxia. Oncogene 2016, 35, 4866–4880. [Google Scholar] [CrossRef] [PubMed]
- Shao, I.H.; Peng, P.H.; Wu, H.H.; Chen, J.L.; Lai, J.C.; Chang, J.S.; Wu, H.T.; Wu, K.J.; Pang, S.T.; Hsu, K.W. RP11-367G18.1 V2 enhances clear cell renal cell carcinoma progression via induction of epithelial-mesenchymal transition. Cancer Med. 2023, 12, 9788–9801. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Li, O.; Zheng, W.; Xiao, W.Z.; Zhang, L.; Wu, D.; Cai, G.Y.; He, J.C.; Chen, X.M. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017, 8, e2772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, W.; Lian, J.; Zhang, H.; Yu, B.; Zhang, M.; Wei, F.; Wu, J.; Jiang, J.; Jia, Y.; et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2020, 27, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Liu, X.; Mao, Y.P.; Yang, X.J.; Wang, Y.Q.; Zhang, P.P.; Lei, Y.; Hong, X.H.; He, Q.M.; Ma, J.; et al. Long non-coding RNA DANCR stabilizes HIF-1alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics 2018, 8, 5676–5689. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhao, Y.; Li, X.; Tao, Z.; Hou, M.; Ma, H. Overexpression of HIF-2alpha-Dependent NEAT1 Promotes the Progression of Non-Small Cell Lung Cancer through miR-101-3p/SOX9/Wnt/β-Catenin Signal Pathway. Cell Physiol. Biochem. 2019, 52, 368–381. [Google Scholar] [CrossRef]
- Cheng, Z.; Lu, C.; Wang, H.; Wang, N.; Cui, S.; Yu, C.; Wang, C.; Zuo, Q.; Wang, S.; Lv, Y.; et al. Long noncoding RNA LHFPL3-AS2 suppresses metastasis of non-small cell lung cancer by interacting with SFPQ to regulate TXNIP expression. Cancer Lett. 2022, 531, 1–13. [Google Scholar] [CrossRef]
- Yan, J.; Deng, Y.X.; Cai, Y.L.; Cong, W.D. LncRNA MIR17HG promotes the proliferation, migration, and invasion of retinoblastoma cells by up-regulating HIF-1alpha expression via sponging miR-155-5p. Kaohsiung J. Med. Sci. 2022, 38, 554–564. [Google Scholar] [CrossRef]
- Shih, J.W.; Chiang, W.F.; Wu, A.T.H.; Wu, M.H.; Wang, L.Y.; Yu, Y.L.; Hung, Y.W.; Wang, W.C.; Chu, C.Y.; Hung, C.L.; et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat. Commun. 2017, 8, 15874. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, H.; Mei, Y.; Wu, M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell 2014, 53, 88–100. [Google Scholar] [CrossRef]
- Shen, W.; Zhu, M.; Wang, Q.; Zhou, X.; Wang, J.; Wang, T.; Zhang, J. DARS-AS1 recruits METTL3/METTL14 to bind and enhance DARS mRNA m(6)A modification and translation for cytoprotective autophagy in cervical cancer. RNA Biol. 2022, 19, 751–763. [Google Scholar] [CrossRef]
- Tong, J.; Xu, X.; Zhang, Z.; Ma, C.; Xiang, R.; Liu, J.; Xu, W.; Wu, C.; Li, J.; Zhan, F.; et al. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma malignancy. Haematologica 2020, 105, 1630–1640. [Google Scholar] [CrossRef]
- McCarty, G.; Loeb, D.M. Hypoxia-sensitive epigenetic regulation of an antisense-oriented lncRNA controls WT1 expression in myeloid leukemia cells. PLoS ONE 2015, 10, e0119837. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Y.; Lei, B.X.; Lin, Y.Y.; Sui, M.H.; Zhang, M.L.; Zhuang, Z.Q.; Dong, J.T.; Jin, D.M.; Yan, T.B. Long noncoding RNA MEG3 silencing protects against hypoxia-induced pheochromocytoma-12 cell injury through inhibition of TIMP2 promoter methylation. J. Cell Physiol. 2020, 235, 1649–1662. [Google Scholar] [CrossRef]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016, 539, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S. LINC00649 promotes bladder cancer malignant progression by regulating the miR-15a-5p/HMGA1 axis. Oncol. Rep. 2021, 45, 8. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; Yang, L.; Cai, X. LncRNA LINC00649 aggravates the progression of cervical cancer through sponging miR-216a-3p. J. Obstet. Gynaecol. Res. 2022, 48, 2853–2862. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Hakami, M.A.; Hazazi, A.; Abdulaziz, O.; Almasoudi, H.H.; Alhazmi, A.Y.M.; Alkhalil, S.S.; Alharthi, N.S.; Alhuthali, H.M.; Almalki, W.H.; Gupta, G.; et al. HOTAIR: A key regulator of the Wnt/β-catenin signaling cascade in cancer progression and treatment. Pathol. Res. Pract. 2024, 253, 154957. [Google Scholar] [CrossRef] [PubMed]
- Morelli, E.; Fulciniti, M.; Samur, M.K.; Ribeiro, C.F.; Wert-Lamas, L.; Henninger, J.E.; Gullà, A.; Aktas-Samur, A.; Todoerti, K.; Talluri, S.; et al. A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth. Blood 2023, 141, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Investig. 2013, 123, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, M.; Dong, Y.; Xu, B.; Chen, J.; Ding, Y.; Qiu, S.; Li, L.; Karamfilova Zaharieva, E.; Zhou, X.; et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1alpha/HK2. Cell Death Dis. 2020, 11, 145. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, F.; Xiong, Y.; Wang, N.; Gu, Y.; Qiu, X. CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int. 2020, 20, 400. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, M.; Yu, D.; Hou, G.; Wu, J.; Li, F. CircRBM33 downregulation inhibits hypoxia-induced glycolysis and promotes apoptosis of breast cancer cells via a microRNA-542-3p/HIF-1alpha axis. Cell Death Discov. 2022, 8, 126. [Google Scholar] [CrossRef]
- Liang, G.; Liu, Z.; Tan, L.; Su, A.N.; Jiang, W.G.; Gong, C. HIF1alpha-associated circDENND4C Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer. Res. 2017, 37, 4337–4343. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; He, Y.; Zhang, N.; Zhang, S.; Li, Y.; Wang, X.; Liang, Y.; Chen, X.; Zhao, W.; et al. Hypoxia-Induced FUS-circTBC1D14 Stress Granules Promote Autophagy in TNBC. Adv. Sci. 2023, 10, e2204988. [Google Scholar] [CrossRef]
- Yang, R.; Chen, H.; Xing, L.; Wang, B.; Hu, M.; Ou, X.; Chen, H.; Deng, Y.; Liu, D.; Jiang, R.; et al. Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10. Mol. Cancer 2022, 21, 88. [Google Scholar] [CrossRef]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, R.; Xing, L.; Wang, B.; Liu, D.; Ou, X.; Deng, Y.; Jiang, R.; Chen, J. Hypoxia-inducible CircPFKFB4 Promotes Breast Cancer Progression by Facilitating the CRL4(DDB2) E3 Ubiquitin Ligase-mediated p27 Degradation. Int. J. Biol. Sci. 2022, 18, 3888–3907. [Google Scholar] [CrossRef]
- Wu, D.; Chen, T.; Zhao, X.; Huang, D.; Huang, J.; Huang, Y.; Huang, Q.; Liang, Z.; Chen, C.; Chen, M.; et al. HIF1alpha-SP1 interaction disrupts the circ-0001875/miR-31-5p/SP1 regulatory loop under a hypoxic microenvironment and promotes non-small cell lung cancer progression. J. Exp. Clin. Cancer Res. 2022, 41, 156. [Google Scholar] [CrossRef]
- Chi, Y.; Luo, Q.; Song, Y.; Yang, F.; Wang, Y.; Jin, M.; Zhang, D. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1alpha regulation. J. Cell Biochem. 2019, 120, 19019–19030. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Xu, Y.; Hu, J.; Zhang, S.; Li, M.; Xu, L. A novel circular RNA, hsa-circ-0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-α expression. Biochem. Biophys. Res. Commun. 2020, 521, 395–401. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, H.; Xia, L.; Lan, F. Circular RNA circ_0061140 accelerates hypoxia-induced glycolysis, migration, and invasion in lung adenocarcinoma through the microRNA-653/hexokinase 2 (HK2) axis. Bioengineered 2022, 13, 7156–7166. [Google Scholar] [CrossRef]
- Joseph, N.A.; Chiou, S.H.; Lung, Z.; Yang, C.L.; Lin, T.Y.; Chang, H.W.; Sun, H.S.; Gupta, S.K.; Yen, L.; Wang, S.D.; et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J. Hematol. Oncol. 2018, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pan, X.; Zhu, D.; Deng, Z.; Jiang, R.; Wang, X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology 2019, 70, 1298–1316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Q.; Zuo, X.L.; Cai, J.; Zhang, Y.; Han, G.Y.; Zhang, L.; Ding, W.Z.; Wu, J.D.; Wang, X.H. Hypoxia-associated circPRDM4 promotes immune escape via HIF-1alpha regulation of PD-L1 in hepatocellular carcinoma. Exp. Hematol. Oncol. 2023, 12, 17. [Google Scholar] [CrossRef]
- Yang, W.; Liu, Y.; Gao, R.; Xiu, Z.; Sun, T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell Signal 2019, 60, 122–135. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Du, B.; Zhan, Y.; Wang, K.; Wang, X.; Chen, B.; Wei, X.; Xiao, J. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1alpha. Mol. Carcinog. 2019, 58, 875–886. [Google Scholar] [CrossRef]
- Fu, Z.; Zhang, P.; Zhang, R.; Zhang, B.; Xiang, S.; Zhang, Y.; Luo, Z.; Huang, C. Novel hypoxia-induced HIF1alpha-circTDRD3-positive feedback loop promotes the growth and metastasis of colorectal cancer. Oncogene 2023, 42, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.; Hao, W.; Zhao, W.; Yang, G.; Jing, C. Hypoxia-induced Circular RNA hsa_circ_0006508 Promotes the Warburg Effect in Colorectal Cancer Cells. Balk. Med. J. 2023, 40, 21–27. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Lin, W.; Yuan, Q.; Lu, Y.; Wang, H.; Chen, Y.; Chen, L.; Dai, P.; Long, H.; et al. circEXOC6B interacting with RRAGB, an mTORC1 activator, inhibits the progression of colorectal cancer by antagonizing the HIF1A-RRAGB-mTORC1 positive feedback loop. Mol. Cancer 2022, 21, 135. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, H.; Yang, Y.; Wang, X.; Deng, T.; Liu, R.; Ning, T.; Bai, M.; Li, H.; Zhu, K.; et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics 2020, 10, 8211–8226. [Google Scholar] [CrossRef]
- Ou, Z.L.; Luo, Z.; Wei, W.; Liang, S.; Gao, T.L.; Lu, Y.B. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: Role of circ_0000977/miR-153 axis. RNA Biol. 2019, 16, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhu, Z.; Xiao, W.; Zong, G.; Wang, C.; Jiang, W.; Li, K.; Shen, J.; Guo, X.; Cui, J.; et al. Hypoxia-induced circRNF13 promotes the progression and glycolysis of pancreatic cancer. Exp. Mol. Med. 2022, 54, 1940–1954. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, X.; Zhai, S.; Shi, M.; Peng, C.; Deng, X.; Fu, D.; Wang, J.; Shen, B. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 2022, 15, 128. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhao, Y.; Chen, Q.; Zhu, S.; Niu, Y.; Ye, Z.; Hu, P.; Chen, D.; Xu, P.; Chen, J.; et al. Hypoxic exosomal HIF-1alpha-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 2021, 40, 5505–5517. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, J.Y.; Gong, L.P.; Feng, Z.Y.; Wang, D.; Pan, Y.H.; Sun, L.P.; Wen, J.Y.; Chen, G.F.; Liang, J.; et al. Hypoxia-induced ebv-circLMP2A promotes angiogenesis in EBV-associated gastric carcinoma through the KHSRP/VHL/HIF1alpha/VEGFA pathway. Cancer Lett. 2022, 526, 259–272. [Google Scholar] [CrossRef]
- Li, H.; Cao, B.; Zhao, R.; Li, T.; Xu, X.; Cui, H.; Deng, H.; Gao, J.; Wei, B. circDNMT1 Promotes Malignant Progression of Gastric Cancer Through Targeting miR-576-3p/Hypoxia Inducible Factor-1 Alpha Axis. Front. Oncol. 2022, 12, 817192. [Google Scholar] [CrossRef]
- Chen, W.; Ji, Y. CircC6orf132 Facilitates Proliferation, Migration, Invasion, and Glycolysis of Gastric Cancer Cells Under Hypoxia by Acting on the miR-873-5p/PRKAA1 Axis. Front. Genet. 2021, 12, 636392. [Google Scholar] [CrossRef]
- Liang, L.; Zhu, Y.; Li, J.; Zeng, J.; Wu, L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J. Exp. Clin. Cancer Res. 2022, 41, 261. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Zhang, Y.; Zheng, J.; Wang, S.; Cao, G. Circular RNA hsa_circ_0004543 Aggravates Cervical Cancer Development by Sponging MicroRNA hsa-miR-217 to Upregulate Hypoxia-Inducible Factor. J. Oncol. 2022, 2022, 4031403. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Huang, T.; Feng, W. Circular RNA HIPK3 Promotes EMT of Cervical Cancer Through Sponging miR-338-3p to Up-Regulate HIF-1alpha. Cancer Manag. Res. 2020, 12, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zou, D.; Sun, Y.; Dai, Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol. Biol. Lett. 2019, 24, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, B.; Zhao, R.; Pan, Z.; Zhang, S.; Qiu, W.; Guo, Q.; Qi, Y.; Gao, Z.; Fan, Y.; et al. Hypoxia-induced circADAMTS6 in a TDP43-dependent manner accelerates glioblastoma progression via ANXA2/ NF-kappaB pathway. Oncogene 2023, 42, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Song, Y.C.; Qiu, F.; Wang, Z.C.; Li, N.; Zhao, F.B. Hypoxic glioma cell-secreted exosomal circ101491 promotes the progression of glioma by regulating miR-125b-5p/EDN1. Brain Res. Bull. 2023, 195, 55–65. [Google Scholar] [CrossRef]
- Xi, Y.; Shen, Y.; Wu, D.; Zhang, J.; Lin, C.; Wang, L.; Yu, C.; Yu, B.; Shen, W. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol. Cancer 2022, 21, 145. [Google Scholar] [CrossRef]
- Gong, W.; Xu, J.; Wang, Y.; Min, Q.; Chen, X.; Zhang, W.; Chen, J.; Zhan, Q. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 2022, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yang, W.; Jiang, N.; Shi, J.; Chen, L.; Zhong, G.; Bi, J.; Dong, W.; Wang, Q.; Wang, C.; et al. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int. J. Biol. Sci. 2019, 15, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Z.; Liu, B.; Sun, L. Silencing of circCYP51A1 represses cell progression and glycolysis by regulating miR-490-3p/KLF12 axis in osteosarcoma under hypoxia. J. Bone Oncol. 2022, 37, 100455. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, Y.; Wu, P.; Li, D.; Lu, Y.; Shen, P.; Yang, T.; Shi, G.; Chen, Q.; Yuan, H.; et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol. Cancer 2022, 21, 121. [Google Scholar] [CrossRef]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019, 12, 3869–3879. [Google Scholar] [CrossRef]
- Tang, J.; Wang, R.; Tang, R.; Gu, P.; Han, J.; Huang, W. CircRTN4IP1 regulates the malignant progression of intrahepatic cholangiocarcinoma by sponging miR-541-5p to induce HIF1A production. Pathol. Res. Pract. 2022, 230, 153732. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ren, S.; Liu, J.; Feng, Y.; Li, Z.; He, L.; Li, L.; Cao, X.; Wang, Z.; Zhang, Y. Knockdown of circDENND4C inhibits glycolysis, migration and invasion by up-regulating miR-200b/c in breast cancer under hypoxia. J. Exp. Clin. Cancer Res. 2019, 38, 388. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef]
- Noguchi, M.; Morikawa, A.; Kawasaki, M.; Matsuno, Y.; Yamada, T.; Hirohashi, S.; Kondo, H.; Shimosato, Y. Small adenocarcinoma of the lung. Histologic characteristics and prognosis. Cancer 1995, 75, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, H.; Wang, J.; Wang, X.; Qian, L.; Xu, F.; Song, W.; Wu, D.; Shen, Z.; Feng, D.; et al. DeltaNp63alpha exerts antitumor functions in cervical squamous cell carcinoma. Oncogene 2020, 39, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef]
- Weng, X.; Liu, H.; Ruan, J.; Du, M.; Wang, L.; Mao, J.; Cai, Y.; Lu, X.; Chen, W.; Huang, Y.; et al. HOTAIR/miR-1277-5p/ZEB1 axis mediates hypoxia-induced oxaliplatin resistance via regulating epithelial-mesenchymal transition in colorectal cancer. Cell Death Discov. 2022, 8, 310. [Google Scholar] [CrossRef]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Barbier, A.J.; Jiang, A.Y.; Zhang, P.; Wooster, R.; Anderson, D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022, 40, 840–854. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e1716. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).