Abstract
Arbuscular mycorrhizal (AM) symbiosis, a mutually beneficial interaction between plant roots and AM fungi, plays a key role in plant growth, nutrient acquisition, and stress tolerance, which make it a major focus for sustainable agricultural strategies. This intricate association involves extensive transcriptional reprogramming in host plant cells during the formation of arbuscules, which are specialized fungal structures for nutrient exchange. The symbiosis is initiated by molecular signaling pathways triggered by fungal chitooligosaccharides and strigolactones released by plant roots, which act as chemoattractants and signaling molecules to promote fungal spore germination, colonization, and arbuscule development. Calcium spiking, mediated by LysM domain receptor kinases, serves as a critical second messenger in coordinating fungal infection and intracellular accommodation. GRAS transcription factors are key components that regulate the transcriptional networks necessary for arbuscule development and maintenance, while small RNAs (sRNAs) from both plant and fungi, contribute to modifications in gene expression, including potential bidirectional sRNA exchange to modulate symbiosis. Understanding the molecular mechanisms related to AM symbiosis may provide valuable insights for implementation of strategies related to enhancing plant productivity and resilience.
1. Introduction
The intricate ecological interplay between plants and microorganisms, specifically rhizospheric microorganisms, has a substantial impact on plant physiology and agricultural outcomes. These microorganisms can boost plant performance by enhancing nutrient acquisition, stress tolerance, and overall growth, thereby contributing to more sustainable and productive agricultural systems [1,2].
Arbuscular mycorrhizal fungi (AMF) are obligate biotrophic organisms that belong to the Glomeromycota phylum. These fungi form one of the most widespread symbiotic associations with plant roots [3]. This ancient symbiosis is characterized by a complex molecular dialogue between the plant and the fungus, leading to the formation of specialized structures within plant roots. These structures, known as arbuscules, serve as the primary site for nutrient exchange [4]. AMF enhance plant nutrient uptake, particularly phosphorus and nitrogen, and improve plant tolerance to various biotic and abiotic stresses. In return, plants provide carbon-based compounds to the fungi (Figure 1).
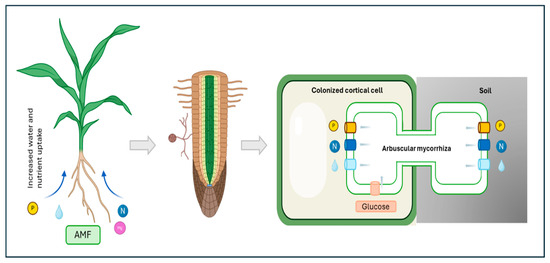
Figure 1.
Nutrient exchange between plants and arbuscular mycorrhizal fungi. The mycorrhizal fungus provides minerals and water to plants, which in exchange provide a fixed carbon source to the fungus.
The establishment of the AM symbiosis requires a complex chemical crosstalk between the plant host and its fungal partner. During the pre-symbiotic phase, plant roots release signaling molecules called strigolactones into the rhizosphere, which induce fungal spore germination and hyphal branching. In response, AM fungi release chitooligosaccharides (COs) and lipochitooligosaccharides (LCOs), signaling molecules that are recognized at the plant plasma membrane [5]. This recognition initiates a cascade of signaling events that promote fungal colonization of the root and induce extensive transcriptional reprogramming in the host plant. A central pathway involved in this signaling process is the common symbiosis signaling pathway, also known as the sym pathway, which is required for both mycorrhization and nodulation in legume plants [6]. Nodulation is a symbiotic process that occurs primarily in legume plants such as peas, beans, clover, and alfalfa. It involves a mutually beneficial relationship between the plant and nitrogen-fixing bacteria, mainly Rhizobium species. Medicago truncatula has been extensively studied as a model legume for investigating the molecular genetics of nitrogen-fixing root–nodule symbiosis [6]. One of the key components of the sym pathway is the induction of perinuclear calcium oscillations in the host cell. These calcium signals activate a calcium- and calmodulin-dependent serine/threonine protein kinase known as CCaMK, which, in turn, triggers the transcriptional changes necessary for the establishment of the symbiotic interface [7]. This interface, called the peri-arbuscular membrane, envelops the arbuscule and provides a site for bidirectional nutrient and signal exchange. The formation and maintenance of the peri-arbuscular membrane require significant structural and functional modifications in the host root cells, indicating a high degree of coordination between plant and fungal gene expression. The composition of the peri-arbuscular membrane is distinct from the typical plasma membrane, containing specific proteins and lipids that facilitate the efficient transfer of nutrients, such as phosphate transporters and ammonium channels. These molecular components ensure that the symbiotic relationship remains beneficial and controlled, preventing any overexploitation by the fungal partner [8,9].
AM colonization not only affects local root physiology but also induces systemic changes in the plant, impacting nutrition, development, and responses to both biotic and abiotic stresses. These systemic effects imply a root-to-shoot signaling axis that influences multiple aspects of plant biology. Metabolic changes in shoots are often observed in plants undergoing AM colonization, suggesting a coordinated response across the entire plant [10]. Additionally, systemic changes influence the plant’s ability to cope with environmental stresses, indicating that AM symbiosis has extensive benefits beyond the root system. For example, AM colonization can improve drought tolerance by enhancing water uptake and increasing the production of stress-related proteins and metabolites. It also aids in the uptake of less-mobile nutrients, such as zinc and copper, which are crucial for various plant metabolic processes [11,12]. Recent research has also highlighted the role of small RNAs (sRNAs) in regulating the AM symbiosis. These molecules are involved in regulating gene expression in both the plant and fungal partners. Some sRNAs have been found to act as mobile signals that can move between the plant and the fungus, influencing the symbiotic relationship at both local and systemic levels (Figure 2). This cross-kingdom RNA interference is a promising area of research that could unlock new strategies for optimizing AM symbiosis for agricultural benefit. The aim of this review was to delve deeper into the key molecular processes involved in the AMF–plant interaction. By understanding the molecular mechanisms underlying this sophisticated symbiotic relationship, different strategies to enhance plant growth and productivity may be developed, especially in challenging environmental conditions.
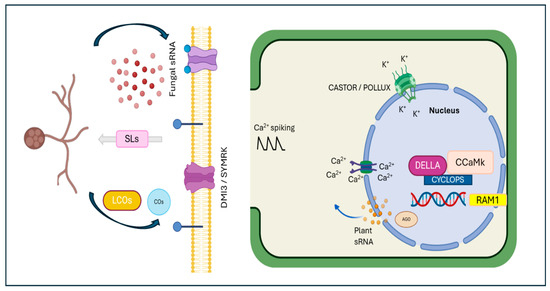
Figure 2.
Establishment of arbuscular mycorrhizal interaction. Plant roots release strigolactones, which induce arbuscular mycorrhizal spore germination and hyphal branching. The mycorrhizal fungi produce Myc factors, which are recognized by the DMI3/SYMRK receptor complex. This stimulus activates perinuclear calcium spiking, involving the CASTOR and POLLUX channels. Required for arbuscular mycorrhization 1 (RAM1) is essential during the early colonization stage, while the DELLA/CYCLOPS/CCaMK complex is necessary for arbuscule development. Additionally, both plant and fungal sRNAs are involved in mycorrhizal symbiosis, targeting various genes and regulating their expression—a crucial process for maintaining symbiosis.
2. Signaling Molecules in the Establishment of Symbiotic Associations
Chitin, a polymer of N-acetylglucosamine, and its derivative chitosan are essential components of fungal cell walls and central to plant–fungal interactions. In rhizobium–legume associations, flavonoids, isoflavonoids, and betaines produced by legumes induce the expression of nod genes in rhizobia, leading to the synthesis of LCOs, which play a major role in symbiotic association [13,14]. Plants also recognize fragments of chitin, called COs, as microbe-associated molecular patterns, triggering immune responses such as gene expression, activation of mitogen-activated protein kinases, production of reactive oxygen species (ROS), and calcium influx. Eight-residue COs are particularly effective in activating these immune responses, and their detection involves receptor-like kinases containing LysM domains [15]. For instance, in M. truncatula, the MtLYK9/MtCERK1 and MtLYR4 receptor complex binds eight-residue COs, initiating immune signaling [16,17]. Similarly, other plants like rice and Arabidopsis thaliana utilize analogous receptor complexes to recognize COs and activate defense mechanisms [18]. Nevertheless, COs are also produced by beneficial symbionts, such as AMF, compelling a balance between immune responses and symbiotic signaling [18]. The original paradigm proposed that long-chain COs stimulate immunity, whereas short COs initiate symbiotic pathways. However, recent studies have revealed a more complex interaction, demonstrating that long COs can activate both immune and symbiotic responses. In M. truncatula, eight-residue COs can induce calcium spiking and upregulate symbiosis-related genes, mediated by the same receptor complex that detects immune signals, highlighting the important role of COs recognition in host discrimination [17].
AMF synthesize a mixture of COs and LCOs, both of which contribute to symbiotic signaling. Sulfated LCOs are particularly effective, surpassing non-sulfated LCOs and COs in triggering calcium spiking and promoting fungal colonization in various plant species. For example, in M. truncatula, sulfated LCOs stimulate calcium spiking in root hairs and atrichoblasts of lateral roots, which are the main sites for fungal colonization [19]. In non-leguminous plants like French marigold and organ culture carrot, treatments combining sulfated and non-sulfated LCOs enhance root colonization and calcium spiking, underscoring the importance of LCOs in establishing AM symbiosis [19,20]. Moreover, AM fungal LCOs suppress defense responses to COs, facilitating symbiotic interactions. Interestingly, LCOs from non-mycorrhizal fungi can also induce early symbiotic responses, such as root hair branching in M. truncatula and Vicia sativa, suggesting that LCO synthesis is a conserved fungal trait [14,19]. Mass spectrometry analyses of fungal exudates reveal that fungal LCOs share structural similarities with those of AMF, indicating a conserved and versatile role in plant–fungal communication. These findings highlight the dual nature of COs and LCOs in modulating plant immunity and symbiosis, emphasizing the intricacies of their molecular interplay.
The interaction between COs and LCOs illustrates a sophisticated mechanism by which plants discern between pathogenic and symbiotic fungi, fine-tuning their responses to optimize both defense and symbiotic efficiency. For instance, while eight-residue COs trigger immune signaling, the presence of LCOs can suppress these defenses, allowing AMF to establish colonization and promote nutrient exchange [17]. This delicate balance between immunity and symbiosis is mediated by calcium spiking, a pivotal second messenger in both pathways, and receptor complexes capable of integrating diverse molecular signals. Understanding these dynamics provides valuable insights into the evolutionary and functional significance of chitin derivatives in plant–fungal interactions [14]. The ability of AMF to modulate plant responses through COs and LCOs underscores their critical role in promoting plant health and productivity. These molecules not only enhance nutrient acquisition by facilitating AM symbiosis but also influence root development, such as lateral root formation, further supporting fungal colonization [21].
While fungi produce LCOs and COs to establish symbiotic associations, plants produce key molecules called strigolactones (SLs). SLs are a group of carotenoid-derived phytohormones exuded by plant roots. These molecules serve dual roles as developmental regulators within plants and as signaling compounds that mediate interactions with other organisms in the soil. Initially discovered as germination stimulants for parasitic weeds, such as those in the genus Striga [22], SLs have been then recognized as key players in plant architecture regulation and facilitators of AMF development [23]. AMF rely on SLs to induce hyphal growth and branching, enabling them to establish symbiotic relationships with host plants. This interaction is particularly well-documented in members of the Gigasporaceae and Glomeraceae families, where SLs stimulate processes such as hyphal proliferation, mitochondrial enlargement, and increased ATP content [24,25,26]. Interestingly, other root-exuded compounds, like hydroxy fatty acids, can also induce hyphal branching, suggesting that SLs are part of a broader signaling network [27]. The structural composition of SLs is central to their bioactivity. These molecules consist of a tricyclic lactone connected to a methylbutenolide via an enol ether bond, with both components being essential for their function. Modifications to the A and B rings, such as the presence of hydroxyl, keto, or acetyl groups, significantly influence their activity [28]. Plant genetic studies have identified key components in SL biosynthesis and secretion. For instance, the G-subfamily ABC transporter PDR1 in Petunia mediates the exudation of orobanchol, which is a prominent SL. Mutations in PDR1 result in reduced SL release, impairing AMF colonization due to lower hyphopodium initiation and intraradical fungal spread [29]. The localized expression of PDR1 in hypodermal passage cells, which serve as entry points for AMF into the root cortex, underscores the targeted nature of SL signaling [30].
SL biosynthesis is tightly regulated and responsive to environmental stimuli, particularly phosphate availability. Under phosphate starvation, plants boost SL production to enhance AMF colonization, as the symbiosis aids in phosphate acquisition [31]. This process is coordinated by GRAS-type transcription factors, notably NSP1 and NSP2, which regulate the expression of DWARF27 (D27), encoding a β-carotene isomerase critical for the initial steps of SL biosynthesis [32,33]. Mutants deficient in NSP1 or NSP2 exhibit disrupted SL biosynthesis and exudation, leading to compromised AMF colonization. Notably, in Medicago, NSP2 is regulated by microRNA (miRNA) miR171h, which restricts colonization to specific root zones by locally reducing SL biosynthesis. Overexpressing a miR171h-resistant version of NSP2 enables colonization of typically uncolonized root elongation zones, revealing a fine-tuned mechanism of spatial regulation in SL-mediated interactions [34]. The perception of SLs involves the α/β-hydrolase receptor D14/DAD2/HTD2, which interacts with the F-box protein D3/MAX2/RMS4. Binding of SLs to D14 triggers a conformational change, facilitating its interaction with D3 and subsequent activation of an E3 ligase, which targets specific proteins for degradation to enable downstream signaling [35]. The F-box protein D3 is essential for AMF colonization, as evidenced in rice and pea mutants lacking this protein. These mutants fail to support AMF accommodation and expression of symbiosis-related genes, despite showing enhanced hyphal branching due to elevated SL exudation [36,37]. Intriguingly, SL signaling is not always required for AMF colonization, as demonstrated in rice D14 mutants, which exhibit higher colonization levels likely due to increased SL release. This suggests that D3 may participate in additional pathways that promote symbiosis, independent of SL signaling [37,38].
3. Calcium Spiking in Response to Symbiosis
Calcium spiking is a critical component of plant cellular signaling, which involves dynamic and repetitive increases in calcium concentration within the nucleus and perinuclear cytoplasm. These calcium oscillations originate from calcium stores located near the nuclear envelope, likely involving the endoplasmic reticulum [7]. To facilitate calcium spikes, it is hypothesized that a core molecular machinery is necessary, comprising calcium channels for release, counter-ion channels such as potassium or anion channels to balance charges during release, and calcium ATPases that pump calcium back into the store for replenishment [39]. Among these key components, the CASTOR and POLLUX channels, also known as MtDMI1, has been identified to be implicated as counter-ion channels. Similarly, the SERCA-type ATPase MCA8 is proposed as a nuclear-localized calcium pump responsible for restoring calcium levels in these stores [40]. Additional insights reveal that mutations in nucleoporin genes—NUP85, NUP133, and NENA—disrupt calcium spiking. These genes belong to the NUP107 subcomplex, a group of nuclear pore proteins potentially involved in trafficking essential components of the spiking machinery, such as channels and pumps, to their functional sites in the inner nuclear envelope [39]. However, the precise mechanisms regulating this transport remain unclear.
Calcium signaling plays a pivotal role in symbiotic interactions between plants and microorganisms, such as rhizobia and AMF. Upon recognizing these symbiotic partners, plants initiate calcium oscillations in the nucleus that is one of the earliest detectable responses [27]. For rhizobia, this process begins with the perception of Nod factors by the plant, triggering calcium spikes within minutes [41]. The involvement of plasma membrane-bound receptor complexes, as well as additional components associated with the nuclear envelope, underscores the complexity of this signaling pathway. Similarly, calcium spiking is observed in plant roots responding to AMF, even before direct contact with fungal hyphae [40]. This suggests that diffusible fungal signals, such as Myc factors, play a role in activating the calcium response. Among these signals are LCOs and COs which are considered as key molecules. Fungal-derived sulfated and non-sulfated LCOs, along with short-chain COs, have been shown to induce calcium oscillations in plant root epidermal cells. Interestingly, the calcium responses elicited by AMF and rhizobia exhibit notable similarities, with subtle differences in spiking frequency and duration [42]. While fungal and short-chain CO-induced oscillations tend to have more variability and longer durations compared to those induced by Nod factors or LCOs, their overall structural profiles are largely similar. These findings suggest that the specific composition and relative concentrations of these signaling molecules determine the calcium response’s characteristics. Moreover, calcium spiking patterns evolve during symbiotic colonization. For instance, low-frequency oscillations are observed in the outer cortical cells before infection, whereas higher-frequency spiking occurs during the entry of symbionts into host cells [43]. This progression implies that calcium signaling is tightly regulated and adjusts to the stages of symbiotic development, coordinating cellular reprogramming in the host plant to accommodate the symbiotic process.
These responses underscore the importance of calcium signaling in plant ability to distinguish between different symbiotic microorganisms at early interaction stages. While it was initially hypothesized that rhizobia and AMF might induce distinct calcium spiking signatures, subsequent studies revealed that these responses are remarkably similar, particularly when induced by LCOs [39,44]. However, the variability and longer duration of fungal-induced calcium spiking suggest a degree of signaling specificity. Additionally, short-chain COs and LCOs exhibit cell-type-specific activation patterns, with responses differing between trichoblasts and atrichoblasts depending on their combinations and concentrations. For instance, in rice, calcium spiking in trichoblasts is only observed when both COs and LCOs are present, emphasizing the combinatorial character of these signals [42,45].
4. Role of GRAS-Domain Proteins in Arbuscular Mycorrhizae
GRAS proteins form a diverse family of plant-specific transcription factors (TFs) with essential roles in a wide range of developmental processes, signaling pathways, and environmental responses. These proteins typically consist of 360 to 850 amino acids and are characterized by a conserved GRAS domain at the C-terminal region, which includes motifs such as VHIID, SAW, and PFYRE, as well as two leucine heptad repeats (LHR). This conserved structure, first detailed by [46], contrasts with the N-terminal region, which is variable and often intrinsically disordered, allowing GRAS proteins to interact with a wide variety of other proteins [47]. One notable feature of some GRAS proteins is the DELLA motif in their N-terminal region, which modulates interactions with other transcription factors, as demonstrated by [48]. These structural characteristics contribute to the multifaceted roles of GRAS proteins in plant biology. The acronym GRAS derives from the first identified members of this family: gibberellin acid insensitive (GAI), repressor of GA1 (RGA), and scarecrow-like (SCL) proteins [46]. These proteins serve as key regulators in critical processes such as gibberellin signaling, root and shoot development, light and stress responses, and symbiotic interactions with microorganisms [49,50,51]. The involvement of GRAS proteins in gibberellin acid signaling, for instance, is exemplified by DELLA proteins, which act as repressors in the GA pathway by interacting with other proteins, including those from the indeterminate domain (IDD) family [52].
Genome-wide analyses have revealed 29 orthologous groups across several species [53]. These groups have been classified into 17 subfamilies, with 5 newly established ones, including RAD1 and RAM1, which are implicated in mycorrhizal signaling [54,55]. Interestingly, these subfamilies exhibit taxonomic specificity, as demonstrated by the absence of some GRAS subfamilies in Brassicales. Such findings underscore the evolutionary diversification of GRAS proteins and their specialized roles in distinct plant lineages. The roles of GRAS proteins extend beyond individual pathways; they often function within complex regulatory networks. For example, the interplay between SCARECROW (SCR) and SHORT-ROOT (SHR) transcription factors in radial root organization has been well-studied [56]. Additionally, SCL3 mediates gibberellin-promoted cell elongation in roots, acting as a bridge between the SCR/SHR and GA/DELLA pathways [57,58]. These interactions illustrate the ability of GRAS proteins to integrate hormonal and developmental signals, ensuring coordinated growth and differentiation. One of the most intriguing aspects of GRAS protein functionality is their role in symbiotic processes, particularly in mycorrhizal associations. GRAS proteins such as RAM1 and RAD1 have been identified as central regulators of arbuscular mycorrhiza [59,60]. These transcription factors are essential for the establishment and maintenance of AM symbiosis, facilitating the accommodation of fungal structures within host root cells. RAM1, for instance, activates the expression of genes involved in lipid and carbohydrate metabolism, as well as those encoding membrane proteins critical for arbuscule function [61,62]. Furthermore, RAM1 interacts with other GRAS-domain proteins, including RAD1, to form regulatory networks that modulate arbuscule development [63].
DELLA proteins also play a dual role in AM symbiosis, promoting arbuscule formation while contributing to arbuscule degeneration through specific regulatory complexes. For instance, DELLA forms a complex with CYCLOPS and other proteins to activate RAM1 transcription, thereby initiating arbuscule formation [60]. Conversely, DELLA interacts with NSP1 and MYB1 to regulate arbuscule degeneration, highlighting its dynamic involvement in different stages of the symbiotic lifecycle [64]. These contrasting roles emphasize the complexity of GRAS-mediated regulation in symbiosis. In legumes, GRAS proteins such as NSP1 and NSP2 are integral to both nodulation and mycorrhization. These transcription factors form a complex that activates early nodulation genes and strigolactone biosynthesis, a process critical for AM fungal colonization [20,23]. Mutants lacking NSP1 or NSP2 exhibit reduced mycorrhizal colonization, demonstrating the essential role of these proteins in symbiotic signaling [65]. Additionally, NSP2 interacts with RAM1 to regulate mycorrhizal gene expression, providing further evidence of the interconnected roles of GRAS proteins in symbiotic pathways [66]. The regulatory versatility of GRAS proteins is further exemplified by their ability to form heterodimers, which are often essential for their functionality [66]. Genome analysis of the GRAS gene family highlights the combinatorial interactions between GRAS proteins and other transcriptional regulators, which enable them to participate in diverse processes without directly binding DNA [49]. This capacity for protein–protein interaction allows GRAS proteins to act as transcriptional regulators, forming complexes that modulate gene expression in a context-dependent manner. In addition to their established roles in GA signaling and symbiosis, GRAS proteins are implicated in stress responses and light signaling. For instance, they contribute to the regulation of root architecture under abiotic stress conditions, adapting plant growth to environmental challenges [49]. The involvement of GRAS proteins in light signaling further demonstrates their versatility, as they integrate external signals to optimize plant development and resource allocation. Nevertheless, the molecular mechanisms underlying their interactions with other proteins and their regulation by post-transcriptional and post-translational modifications are not fully understood. Recent studies have identified miRNA that target GRAS genes, such as miR171h, which regulates NSP2 expression during mycorrhization [34]. Such findings suggest additional layers of regulatory complexity that warrant further investigation.
5. Small RNAs in Mycorrhizal Symbiosis
RNA interference (RNAi) is a conserved regulatory mechanism across eukaryotes, playing a crucial role in development and stress responses [67]. This process is mediated by sRNAs, which guide gene silencing either transcriptionally (TGS) or post-transcriptionally (PTGS), depending on sequence complementarity. Plant sRNAs, typically 20–28 nucleotides long, are classified into miRNAs and short interfering RNAs (siRNAs) based on their biogenesis [68]. miRNAs are derived from single-stranded RNAs forming stable hairpin structures and are processed by the DICER-like enzyme DCL1. These miRNAs predominantly facilitate PTGS by cleaving target mRNAs or repressing their translation [69]. SiRNAs, in contrast, originate from double-stranded RNAs (dsRNAs) that may be endogenous or exogenous, processed by RNA-dependent RNA polymerases (RdRps). SiRNAs contribute to PTGS via 21–22 nucleotide siRNAs and TGS through 24 nucleotide heterochromatic siRNAs (hc-siRNAs), which guide RNA-directed DNA methylation [70]. Once synthesized, sRNA duplexes are stabilized through methylation by the HUA ENHANCER1 (HEN1) enzyme and incorporated into the RNA-induced silencing complex (RISC), where ARGONAUTE (AGO) proteins execute RNAi functions. AGO proteins, highly conserved across eukaryotes, vary in number and function among plant species. In A. thaliana, AGO proteins are grouped into three clades (AGO1/5/10, AGO2/3/7, and AGO4/6/8/9) and demonstrate functional specialization based on the sRNA length, 5′ nucleotide, and duplex structure [71,72]. Interestingly, sRNAs and AGO proteins often function beyond their originating cells, utilizing extracellular vesicles (EVs) or apoplastic spaces for transport. Unique RNAi components in certain plants, such as the monocot-specific DCL5 involved in temperature-dependent fertility or the unusually long sRNAs in Chlamydomonas reinhardtii AGO1, underscore evolutionary adaptations [71,73].
Plant sRNAs play a pivotal role in plant–biotic interactions, particularly in symbiosis and pathogen defense. They regulate endogenous genes, safeguard genome integrity from transposable elements, and defend against viral infections. Viral dsRNAs are processed into viral sRNAs by host RNAi machinery, which targets viral genomes to curtail infections or, in some cases, modulates host transcripts for viral persistence [74]. Beyond viruses, plant sRNAs respond to bacterial and fungal elicitors, such as flagellin and chitin, activating antimicrobial defenses [75,76]. Plant sRNAs can even translocate into pathogens, impacting their virulence; for example, miR159 and miR166 from cotton target Verticillium dahliae virulence genes [77]. This trans-kingdom RNAi phenomenon has inspired further exploration of plant-derived sRNAs for antimicrobial applications [78,79].
AMF colonization significantly reprograms sRNA expression in plants, a phenomenon conserved across species [63,80,81]. In M. truncatula, miRNAs affected by AM colonization target transcription factors that regulate AM development and immune suppression [82]. Similarly, in Nicotiana attenuata, AGO7 regulates AM-induced sRNAs, influencing colonization through phytohormone signaling and nutrient metabolism pathways [83]. Loss of AGO7 enhances mycorrhizal colonization, paralleling observations in M. truncatula mutants where nodulation is similarly increased [84], suggesting shared mechanisms between symbiotic interactions. Despite progress, mechanistic insights into miRNA roles in AM symbiosis are limited. For example, miR399, a key player in phosphate starvation signaling, modulates AM colonization by repressing PHO2, although its overexpression cannot overcome high-phosphate suppression of AM symbiosis [85,86]. Evidence points to a broader regulatory network involving phosphate starvation response components such as PHR in orchestrating AM colonization [87]. Unlike bacterial and fungal pathogen responses, where miRNA regulation suppresses interactions, auxin signaling positively regulates AM symbiosis. Downregulation of miR393, for instance, enhances auxin signaling essential for arbuscule formation [88], whereas miR396b overexpression reduces mycorrhizal colonization by targeting transcription factors in the GRF and bHLH families [89].
Certain miRNAs exhibit spatially localized expression patterns that restrict fungal colonization. In M. truncatula, miR171 isoforms like mtr-miR171h target NSP2, a GRAS transcription factor necessary for AM gene expression, limiting fungal colonization to specific root zones [90]. Similarly, miR171b in AM-compatible plants protects positive regulators like LOM1 from silencing, ensuring AM compatibility [91]. Populus trichocarpa also shows extensive miRNA reprogramming during AM colonization, with conserved expression patterns across mycorrhizal types. Systemic responses, such as miRNA changes in leaves, suggest a potential priming effect against foliar pathogens [91]. Future research integrating diverse RNA types, such as tiny and circular RNAs, alongside detailed studies of DCL and AGO family roles, could further elucidate the complex regulatory networks underlying AM symbiosis [92,93].
Nearly all fungal lineages, including arbuscular mycorrhizal (AM) fungi, contain key components of RNA interference (RNAi) machinery, such as Dicer-like (DCL) proteins, Argonaute (AGO) proteins, and RNA-dependent RNA polymerases (RdRps). These components are often encoded by multiple genes per genome [94]. However, AMF exhibit unique adaptations in their RNAi-related gene repertoire, setting them apart from other fungi [95,96,97]. A particularly striking example is the extensive expansion of AGO genes in Rhizophagus irregularis, which possesses 26 full-length AGO genes and an additional 14 partial AGO sequences containing the PIWI domain but missing other domains. Furthermore, the RdRp family in R. irregularis includes 21 genes, some of which encode peptides with unusually short and unique RdRp domains [95]. These expansions coexist with the presence of one to two DCL genes and a unique prokaryotic-type class I ribonuclease III enzyme, likely acquired via horizontal gene transfer from cyanobacteria, which may assist in small RNA (sRNA) production [98]. Additionally, AMF retain the sRNA methyltransferase HEN1, which stabilizes sRNAs, a gene lost in several other fungal lineages [99]. Expression studies through transcriptomics and proteomics confirm that RNAi-related genes in AMF are actively expressed and regulated throughout their life cycle [95,96,99]. Sequencing of sRNAs has revealed that AMF, including R. irregularis and Gigaspora margarita, produce sRNAs with characteristic lengths of 24–26 nucleotides, consistent with the activity of a single DCL enzyme. These sRNAs exhibit a 5′ uracil bias, and some are 2′-O-methylated at the 3′ end—a modification mediated by HEN1 homologs [99]. Genomic analyses have identified two primary types of sRNA-generating loci: those associated with transposable elements (TEs), which produce 22–26 nt sRNAs, and others linked to protein-coding genes that generate sRNAs of variable lengths [95,96]. Moreover, a noncanonical, DCL-independent pathway, previously described in the non-mycorrhizal fungus Mucor circinelloides, might also function in AMF. This pathway relies on a prokaryotic-derived RNase III enzyme and could contribute to sRNA production in AMF [100,101].
TEs play a significant role in the genome dynamics of AMF, serving both as sources and targets of RNAi-mediated silencing. In R. irregularis, nearly half the genome comprises repetitive elements, with TEs contributing to 49% of the total sRNA pool. The remaining sRNAs originate from either unannotated genomic regions (41%) or protein-coding genes (10%), many of which are located near TEs, suggesting potential regulatory interactions [99]. The expansion of AGO genes in R. irregularis is thought to have co-evolved with TEs, reflecting their intertwined evolutionary dynamics. Besides RNAi, AMF also use DNA methylation to regulate TEs, with 5-methylcytosine (5mC) at CG sites serving as a key epigenetic mark. In addition, AMF exhibit a unique epigenetic feature: N6-methyladenine (6mA) at ApT motifs, which is primarily associated with increased transcription and genes related to symbiosis [102].
At the chromosomal level, R. irregularis nuclei are organized into distinct euchromatic and heterochromatic compartments. Gene-rich euchromatic regions house core functions, while heterochromatic regions contain repetitive sequences and genes encoding secreted proteins that are highly expressed during symbiosis [103,104]. This chromatin organization underscores dynamic interactions between fungal genomes and host plants, with evidence that host plants can induce changes in fungal chromatin and DNA methylation. Conversely, AMF can influence host epigenetics, as demonstrated by the effector protein RiNEL1 from R. irregularis, which interacts with plant histone H2B to suppress defense gene expression and enhance symbiosis [105]. AMF also seem to employ RNAi as a defense mechanism against viruses, as evidenced by the presence of sRNAs mapping to viral genomes within these fungi. However, the roles of mycoviruses in AM fungal biology remain poorly understood [95]. Beyond self-defense, AMF may engage in cross-kingdom RNAi, where sRNAs are exchanged between species to regulate gene expression. In pathogenic systems, fungal sRNAs derived from TEs can suppress host defense genes to facilitate infection [106]. Mutualistic interactions may also involve cross-kingdom RNAi, as seen in ectomycorrhizal fungi, where fungal sRNAs target plant genes to sustain the symbiosis [107]. Although direct evidence for cross-kingdom RNAi in AM symbiosis is limited, related studies demonstrate its feasibility. For instance, host-induced gene silencing (HIGS) has been used to manipulate fungal gene expression in AMF. Silencing the R. irregularis monosaccharide transporter gene MST2 in M. truncatula roots reduced fungal colonization [108]. Similar methods have validated functions of other fungal genes related to phosphate transport, effectors, and MAPK cascades [109,110]. A computational and experimental study has also identified fungal sRNAs that target host genes, such as an R. irregularis sRNA that modulates M. truncatula WRKY transcription factors to regulate colonization [111]. Fungal sRNAs often target host immune genes like MAPK cascade components and WRKY transcription factors, while in mutualistic AM symbioses, fungal sRNAs may modulate host defense responses and membrane remodeling for arbuscule development. Recent studies suggest additional complexities, including the potential for mRNA transfer between interacting organisms, the involvement of host AGO4 proteins in driving transcriptional gene silencing via methylation, and the transport of sRNAs within extracellular vesicles, broadening the scope of inter-organismal communication beyond canonical cross-kingdom RNAi [111].
6. Conclusions and Future Prospects
The establishment and maintenance of AMF symbiosis relies on a complex network of molecular interactions coordinated by signaling molecules, calcium spiking, GRAS transcription factors, and small RNAs (sRNAs). Communication between plants and AM fungi hinges on the exchange of signaling molecules, including chitin derivatives (COs and LCOs) and SLs, which enable plants to discriminate between beneficial and pathogenic fungi. These molecules trigger calcium spiking, a crucial early signaling event characterized by dynamic calcium oscillations regulated by core machinery involving calcium channels, counter-ion channels, and ATPases. Notably, GRAS-domain proteins RAM1, RAD1, NSP1, and NSP2 play central roles in AM development and function, influencing fungal accommodation, arbuscule formation, and coordinating symbiotic signaling with other pathways, such as hormone signaling. Acting as key gene expression regulators, sRNAs mediate plant responses to AM colonization by influencing nutrient transport, hormone signaling, and defense pathways. Concurrently, AM fungi possess adapted RNAi machinery potentially involved in modulating host silencing and defending against mycoviruses. Future research should prioritize several key areas, including a deeper understanding of the crosstalk between CO/LCO and SL signaling pathways, as well as the role of other root exudates. Further investigation of the downstream signaling cascades triggered by these molecules, including calcium signaling and transcriptional regulation, is essential, particularly regarding the mechanisms regulating the transport of calcium spiking machinery components and the functional consequences of differing spiking dynamics between rhizobial and AM fungal interactions. Regarding GRAS-domain proteins, future research should address post-transcriptional and post-translational modifications, as well as the roles of different GRAS subfamilies in various plant species. Finally, functional characterization of diverse sRNA populations and RNAi components in both plants and AM fungi is crucial, with emphasis on the mechanisms of sRNA exchange (cross-kingdom RNAi) via extracellular vesicles, validation of in silico sRNA–mRNA interactions, and exploration of mycovirus-mediated modulation. These investigations will provide a more comprehensive understanding of AM symbiosis and provide a framework for developing strategies to enhance plant–AMF interactions for sustainable agriculture in a context of climate change.
Author Contributions
Conceptualization, V.D., A.V., and L.P.H.-S.; writing—original draft preparation, V.D., M.V. and A.V.; writing—review and editing, K.A., K.F. and L.P.H.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest that influence this study.
References
- Vega-Celedón, P.; Bravo, G.; Velásquez, A.; Cid, F.P.; Valenzuela, M.; Ramírez, I.; Vasconez, I.-N.; Álvarez, I.; Jorquera, M.A.; Seeger, M. Microbial Diversity of Psychrotolerant Bacteria Isolated from Wild Flora of Andes Mountains and Patagonia of Chile towards the Selection of Plant Growth-Promoting Bacterial Consortia to Alleviate Cold Stress in Plants. Microorganisms 2021, 9, 538. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.; Bodenhausen, N.; Hess, J.; Valzano-Held, A.; Waelchli, J.; Deslandes-Hérold, G.; Schlaeppi, K.; van der Heijden, M.G.A. Soil microbiome indicators can predict crop growth response to large-scale inoculation with arbuscular mycorrhizal fungi. Nat. Microbiol. 2023, 8, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 4th ed.; Academic: London, UK, 2008. [Google Scholar]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Gough, C.; Bécard, G. Strigolactones and Lipo-Chitooligosaccharides as Molecular Communication Signals in the Arbuscular Mycorrhizal Symbiosis. In Molecular MycorrhizalSymbiosis; Martin, F., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. 107–124. [Google Scholar] [CrossRef]
- Debellé, F. The Common Symbiotic Signaling Pathway. In The Model Legume Medicago Truncatula; de Bruijn, F.J., Ed.; JohnWiley & Sons, Ltd.: New York, NY, USA, 2020; pp. 523–528. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient Exchange and Regulation in Arbuscular Mycorrhizal Symbiosis. Mol. Plant 2017, 10, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Banasiak, J.; Jamruszka, T.; Murray, J.D.; Jasiński, M. A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium symbioses. Plant Physiol. 2021, 187, 2071–2091. [Google Scholar] [CrossRef]
- Fiorilli, V.; Vannini, C.; Ortolani, F.; Garcia-Seco, D.; Chiapello, M.; Novero, M.; Domingo, G.; Terzi, V.; Morcia, C.; Bagnaresi, P.; et al. Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci. Rep. 2018, 8, 9625. [Google Scholar] [CrossRef] [PubMed]
- Founoune-Mboup, H.; Diallo, B.; Adigoun, R.F.R.; Kane, A.; Fall, A.F. Contribution of arbuscular mycorrhizal fungi to the bioavailability of micronutrients (iron and zinc) in millet accessions. Front. Plant Sci. 2024, 15, 1364469. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Sarkar, S. Arbuscular mycorrhizal fungal contribution towards plant resilience to drought conditions. Front. Fungal Biol. 2024, 5, 1355999. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Parniske, M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell. Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef] [PubMed]
- Rush, T.A.; Puech-Pagès, V.; Bascaules, A.; Jargeat, P.; Maillet, F.; Haouy, A.; Maës, A.Q.; Carriel, C.C.; Khokhani, D.; Keller-Pearson, M.; et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 2020, 11, 3897. [Google Scholar] [CrossRef] [PubMed]
- Cope, K.R.; Bascaules, A.; Irving, T.B.; Venkateshwaran, M.; Maeda, J.; Garcia, K.; Rush, T.A.; Ma, C.; Labbé, J.; Jawdy, S.; et al. The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize Populus roots. Plant Cell 2019, 31, 2386–2410. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Sun, J.; Radhakrishnan, G.V.; Lee, T.; Bozsóki, Z.; Fort, S.; Gavrin, A.; Gysel, K.; Thygesen, M.B.; Andersen, K.R.; et al. A combination of chitooligosaccha ride and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 2019, 10, 5047. [Google Scholar] [CrossRef] [PubMed]
- Desaki, Y.; Kohari, M.; Shibuya, N.; Kaku, H. MAMP-triggered plant immunity mediated by the LysM-receptor kinase CERK1. J. Gen. Plant Pathol. 2019, 85, 1–11. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pagès, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Bécard, G.; Bonfante, P.; et al. Short-chainchitinoligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New. Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Khan, W.; Costa, C.; Souleimanov, A.; Prithiviraj, B.; Smith, D.L. Response of Arabidopsis thaliana roots to lipo-chitooligosaccharide from Bradyrhizobium japonicum and other chitin-like compounds. Plant Growth Regul. 2011, 63, 243–249. [Google Scholar] [CrossRef]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Al-Babili, S.; van der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 2012, 18, 72–83. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Besserer, A.; Puech-Pagès, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.-C.; Roux, C.; Bécard, G.; Séjalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, 226. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Becard, G.; Jauneau, A.; Roux, C.; Sejalon-Delmas, N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008, 148, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, G.; Douds, D.D., Jr. The effects of hydroxy fatty acids on the hyphal branching of germinated spores of AM fungi. Fungal Biol. 2011, 115, 351–358. [Google Scholar] [CrossRef]
- Akiyama, K.; Ogasawara, S.; Ito, S.; Hayashi, H. Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 2010, 51, 1104–1117. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Sharda, J.N.; Koide, R.T. Can hypodermal passage cell distribution limit root penetration by mycorrhizal fungi? New Phytol. 2008, 180, 696–701. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone–like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kohlen, W.; Lillo, A.; Op den Camp, R.; Ivanov, S.; Hartog, M.; Limpens, E.; Jamil, M.; Smaczniak, C.; Kaufmann, K.; et al. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 2011, 23, 3853–3865. [Google Scholar] [CrossRef] [PubMed]
- Lauressergues, D.; Delaux, P.M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Drummond, R.S.M.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an α/βhydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef]
- Foo, E.; Yoneyama, K.; Hugill, C.; Quittenden, L.; Reid, J. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 2012, 6, 76–87. [Google Scholar] [CrossRef]
- Yoshida, S.; Kameoka, H.; Tempo, M.; Akiyama, K.; Umehara, M.; Yamaguchi, S.; Hayashi, H.; Kyozuka, J.; Shirasu, K. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 2012, 196, 1208–1216. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Morita, R.; Tanaka, K.; Kodama, K.; Kameoka, H.; Kyozuka, J.; Seto, Y.; Yamaguchi, S. Activation of Strigolactone Biosynthesis by the DWARF14-LIKE/KARRIKIN-INSENSITIVE2 Pathway in Mycorrhizal Angiosperms, but Not in Arabidopsis, a Non-mycorrhizal Plant. Plant Cell Physiol. 2023, 64, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.; Oldroyd, G.E. Calcium signaling and transcriptional regulation in arbuscular mycorrhizal symbiosis. Mol. Mycorrhizal Symbiosis 2016, 8, 125–140. [Google Scholar] [CrossRef]
- Chabaud, M.; Genre, A.; Sieberer, B.J.; Faccio, A.; Fournier, J.; Novero, M.; Barker, D.G.; Bonfante, P. Arbuscular Mycorrhizal Hyphopodia and Germinated Spore Exudates Trigger Ca2+ Spiking in the Legume and Nonlegume Root Epidermis. New Phytol. 2011, 189, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.W.; Wais, R.; Long, S.R. Calcium Spiking in Plant Root Hairs Responding to Rhizobium Nodulation Signals. Cell 1996, 85, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Miller, J.B.; Granqvist, E.; Wiley-Kalil, A.; Gobbato, E.; Maillet, F.; Cottaz, S.; Samain, E.; Venkateshwaran, M.; Fort, S.; et al. Activation of Symbiosis Signaling by Arbuscular Mycorrhizal Fungi in Legumes and Rice. Plant Cell 2015, 27, 823–838. [Google Scholar] [CrossRef]
- Yuan, P.; Luo, F.; Gleason, C.; Poovaiah, B.W. Calcium/calmodulin-mediated microbial symbiotic interactions in plants. Front. Plant Sci. 2022, 13, 984909. [Google Scholar] [CrossRef] [PubMed]
- Czaja, L.F.; Hogekamp, C.; Lamm, P.; Maillet, F.; Martinez, E.A.; Samain, E.; Dénarié, J.; Küster, H.; Hohnjec, N. Transcriptional Responses toward Diffusible Signals from Symbiotic Microbes Reveal MtNFP- and MtDMI3-Dependent Reprogram ming of Host Gene Expression by Arbuscular Mycorrhizal Fungal Lipochitooligosaccharides. Plant Physiol. 2012, 159, 1671–1685. [Google Scholar] [CrossRef]
- Sieberer, B.J.; Chabaud, M.; Fournier, J.; Timmers, A.C.J.; Barker, D.G. A Switch in Ca2+ Spiking Signature Is Concomitant with Endosymbiotic Microbe Entry into Cortical Root Cells of Medicago truncatula. Plant J. 2012, 69, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef]
- la Rosa, N.M.-D.; Sotillo, B.; Miskolczi, P.; Gibbs, D.J.; Vicente, J.; Carbonero, P.; Oñate-Sánchez, L.; Holdsworth, M.J.; Bhalerao, R.; Alabadí, D.; et al. Large-Scale Identification of Gibberellin-Related Transcription Factors Defines Group VII ETHYLENE RESPONSE FACTORS as Functional DELLA Partners. Plant Physiol. 2014, 166, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Kakkar, M.; Kumari, P.; Zinta, G.; Gahlaut, V.; Kumar, S. Multifaceted roles of GRAS transcription factors in growth and stress responses in plants. iScience 2022, 25, 105026. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Sawers, R.J.H.; Marti, G.; Andrés-Hernández, L.; Yang, S.-Y.; Casieri, L.; Angliker, H.; Oakeley, E.J.; Wolfender, J.-L.; Abreu-Goodger, C.; et al. Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA 2015, 112, 6754–6759. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef]
- Cenci, A.; Rouard, M. Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef]
- Park, H.J.; Floss, D.S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M.J. Hyphal Branching during Arbuscule Development Requires RAM1. Plant Physiol. 2015, 169, 2774–2788. [Google Scholar] [CrossRef]
- Xue, L.; Cui, H.; Buer, B.; Vijayakumar, V.; Delaux, P.-M.; Junkermann, S.; Bucher, M. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 2015, 167, 854–871. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Kong, D.; Liu, X.; Hao, Y. SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 2014, 78, 319–327. [Google Scholar] [CrossRef]
- Heo, J.-O.; Chang, K.S.; A Kim, I.; Lee, M.-H.; Lee, S.A.; Song, S.-K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef]
- Ito, T.; Fukazawa, J. SCARECROW-LIKE3 regulates the transcription of gibberellin-related genes by acting as a transcriptional co-repressor of GAI-ASSOCIATED FACTOR1. Plant Mol. Biol. 2021, 105, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Levy, J.G.; Lévesque-Tremblay, V.; Pumplin, N.; Harrison, M.J. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, E5025–E5034. [Google Scholar] [CrossRef]
- Pimprikar, P.; Carbonnel, S.; Paries, M.; Katzer, K.; Klingl, V.; Bohmer, M.J.; Karl, L.; Floss, D.S.; Harrison, M.J.; Parniske, M.; et al. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016, 26, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Brands, M.; Wewer, V.; Dörmann, P.; Harrison, M.J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 2017, 214, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, S.; Liu, F.; Wang, W.; Wang, X.; Han, G.; Cheng, B. Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef]
- Floss, D.S.; Gomez, S.K.; Park, H.-J.; MacLean, A.M.; Müller, L.M.; Bhattarai, K.K.; Lévesque-Tremblay, V.; Maldonado-Mendoza, I.E.; Harrison, M.J. A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol. 2017, 27, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Delaux, P.M.; Bécard, G.; Combier, J.P. NSP1 is a component of the Myc signaling pathway. New Phytol. 2013, 199, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gobbato, E.; Marsh, J.F.; Vernié, T.; Wang, E.; Maillet, F.; Kim, J.; Miller, J.B.; Sun, J.; Bano, S.A.; Ratet, P.; et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012, 22, 2236–2241. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.; Martienssen, R. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, P.; Zhai, J.; Zhou, M.; Ma, L.; Liu, B.; Jeong, D.H.; Nakano, M.; Cao, S.; Liu, C.; et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. Cell Mol. Biol. 2012, 69, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Bajczyk, M.; Jarmolowski, A.; Jozwiak, M.; Pacak, A.; Pietrykowska, H.; Sierocka, I.; Swida-Barteczka, A.; Szewc, L.; Szweykowska-Kulinska, Z. Recent Insights into Plant miRNA Biogenesis: Multiple Layers of miRNA Level Regulation. Plants 2023, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Guo, M.; Liu, S.; Liu, L.; Yu, Y.; Mo, B.; Chen, X.; Gao, L. Origin, evolution anddiversification of plant ARGONAUTE proteins. Plant J. 2022, 109, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Belanger, S.; Zhan, J.; Meyers, C. Phylogenetic analyses of seven protein families refine the evolution of small RNA pathway singreen plants. Plant Physiol. 2023, 192, 1183–1203. [Google Scholar] [CrossRef]
- Teng, C.; Zhang, H.; Hammond, R.; Huang, K.; Meyers, B.C.; Walbot, V. Dicer like 5 deficiency confers temperature-sensitive male sterility in maize. Nat. Commun. 2020, 11, 2912. [Google Scholar] [CrossRef]
- Smith, N.A.; Eamens, A.L.; Wang, M.B. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011, 7, 1002022. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Marquez, D.; Del-Espino, A.; Lopez-Pagan, N.; Rodrıguez-Negrete, E.A.; Rubio-Somoza, I.; Ruiz-Albert, J.; Bejarano, E.R.; Beuzon, C.R. miR825-5p targets the TIR-NBS-LRR gene MIST1 and down-regulates basal immunity against Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 2021, 72, 7316–7334. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, F.; Baldrich, P.; Jimenez-Gongora, T.; Villar Martin, L.; Weigel, D.; Rubio Somoza, I. miR472 deficiency enhances Arabidopsis thaliana defence without reducing seed production. Mol. Plant-Microbe Interact. 2022, 37, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Liu, J.H.; Zhao, J.H.; Liu, T.; Chen, Y.Y.; Wang, C.H.; Zhang, Z.H.; Guo, H.S.; Duan, C.G. A fungal effector suppresses the nuclear export of AGO1-miRNA complex to promote infection in plants. Proc. Natl. Acad. Sci. USA 2022, 119, 2114583119. [Google Scholar] [CrossRef]
- Pandey, P.; Wang, M.; Baldwin, I.T.; Pandey, S.P.; Groten, K. Complex regulation of microRNAs in roots of competitively-grown isogenic Nicotiana attenuata plants with different capacities to interact with arbuscular mycorrhizal fungi. BMC Genom. 2018, 19, 937. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Soto, A.B.; Rodríguez-Corral, A.Z.; Bojórquez-López, A.; Cervantes-Rojo, M.; Castro-Martínez, C.; Lopez-Meyer, M. Arbuscular Mycorrhizal Symbiosis Leads to Differential Regulation of Genes and miRNAs Associated with the Cell Wall in Tomato Leaves. Biology 2022, 11, 854. [Google Scholar] [CrossRef] [PubMed]
- Devers, E.A.; Branscheid, A.; May, P.; Krajinski, F.; Bäumlein, H. Stars and symbiosis:microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011, 156, 1990–2010. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.; Baldwin, I.T.; Pandey, S.P. Argonaute7 (AGO7) optimizes arbuscular mycorrhizal fungal associations and enhances competitive growth in Nicotiana attenuata. New Phytol. 2023, 40, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Hobecker, K.V.; Reynoso, M.A.; Bustos-Sanmamed, P.; Wen, J.; Mysore, K.S.; Crespi, M.; Blanco, F.A.; Zanetti, M.E. The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 2017, 174, 2469–2486. [Google Scholar] [CrossRef] [PubMed]
- Branscheid, A.; Sieh, D.; Pant, B.D.; May, P.; Devers, E.A.; Elkrog, A.; Krajinski, F. Expression pattern suggests a role of miR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol. Plant–Microbe Interact. 2010, 23, 915–926. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F.; et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540. [Google Scholar] [CrossRef]
- Das, D.; Paries, M.; Hobecker, K.; Gigl, M.; Dawid, C.; Lam, H.; Zhang, J.; Chen, M.; Gutjahr, C. Phosphate starvation response transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 2022, 13, 477. [Google Scholar] [CrossRef] [PubMed]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Becard, G.; Combier, J.P. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M.; et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Hofferek, V.; Mendrinna, A.; Gaude, N.; Krajinski, F.; Devers, E.A. MiR171h restricts root symbioses and shows like its target NSP2 a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol. 2014, 14, 199. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Lauressergues, D.; Andre, O.; Gutjahr, C.; Guillotin, B.; Becard, G.; Combier, J.P. Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Rep. 2017, 20, 1339–1350. [Google Scholar] [CrossRef]
- Zeng, Z.; Liu, Y.; Feng, X.Y.; Li, S.X.; Jiang, X.M.; Chen, J.Q.; Shao, Z.Q. The RNAome landscape of tomato during arbuscular mycorrhizal symbiosis reveals an evolving RNA layer symbiotic regulatory network. Plant Commun. 2023, 4, 100429. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.; Requena, N. Distinguishing friends from foes: Can smRNAs modulate plant interactions with beneficial and pathogenic organisms? Curr. Opin. Plant Biol. 2022, 69, 102259. [Google Scholar] [CrossRef] [PubMed]
- Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Canovas-Marquez, J.T.; Perez-Ruiz, J.A.; Osorio Concepcion, M.; Navarro, E.; Calo, S. The evolutionary significance of RNAi in the fungal kingdom. Int. J. Mol. Sci. 2020, 21, 9348. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, A.; Fiorilli, V.; Miozzi, L.; Accotto, G.P.; Turina, M.; Lanfranco, L. Insilico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genom. 2019, 20, 169. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, A.; Turina, M.; Fiorilli, V.; Miozzi, L.; Venice, F.; Bonfante, P.; Lanfranco, L. Different genetic sources contribute to the small RNApopulation inthearbuscular mycorrhizal fungus Gigaspora margarita. Front. Microbiol. 2020, 11, 395. [Google Scholar] [CrossRef]
- Lanfranco, L.; Bonfante, P. Lessons from arbuscular mycorrhizal fungal genomes. Curr. Opin. Microbiol. 2023, 75, 102357. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kong, M.; Harrison, P.; Hijri, M. Conserved proteins of the RNA Interference system in the arbuscular mycorrhizal fungus Rhizoglomus irregulare provide new insight into the evolutionary history of Glomeromycota. Genome Biol. Evol. 2018, 10, 328–343. [Google Scholar] [CrossRef]
- Dallaire, A.; Manley, B.F.; Wilkens, M.; Bista, I.; Quan, C.; Evangelisti, E.; Bradshaw, C.R.; Ramakrishna, N.B.; Schornack, S.; Butter, F.; et al. Transcriptionalactivityand epigenetic regulation of transposable elements in the symbiotic fungus Rhizophagus irregularis. Genome Res. 2021, 31, 2290–2302. [Google Scholar] [CrossRef] [PubMed]
- Trieu, T.A.; Calo, S.; Nicolas, F.E.; Vila, A.; Moxon, S.; Dalmay, T.; Ruiz-Vazquez, R.M. Anon-canonical RNAsilencing pathway promotesmRNAdegradation in basal fungi. PLoS Genet. 2015, 11, 1005168. [Google Scholar] [CrossRef] [PubMed]
- Perez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Navarro, E.; Garre, V.; Nicolas, F.E. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet. 2020, 16, 1008611. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Cruz Corella, J.; Robbins, C.; Loha, A.; Menin, L.; Gasilova, N.; Masclaux, F.G.; Lee, S.J.; Sanders, I.R. The methylome of the model arbuscular mycorrhizal fungus, Rhizophagus irregularis, shares characteristics with early diverging fungi and Dikarya. Commun. Biol. 2021, 4, 901. [Google Scholar] [CrossRef] [PubMed]
- Yildirir, G.; Sperschneider, J.; Malar, C.M.; Chen, E.C.H.; Iwasaki, W.; Cornell, C.; Corradi, N. Long reads and Hi-C sequencing illuminate the two-compartment genome of the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 2022, 233, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Yildirir, G.; Rizzi, Y.; Malar, M.; Sorwar, E.; Chen, E.C.H.; Iwasaki, W.; Brauer, E.K.; Bosnich, W.; Gutjahr, C.; et al. Arbuscular mycorrhizal fungi heterokayons have two nuclear populations with distinct roles in host plant interactions. Nat. Microbiol. 2023, 8, 2142–2453. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Jiang, H.; Boeren, S.; Dings, H.; Kulikova, O.; Bisseling, T.; Limpens, E. Anuclear-targeted effector of Rhizophagus irregularis interferes with histone 2B mono-ubiquitination to promote arbuscular mycorrhisation. New Phytol. 2021, 230, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Wong-Bajracharya, J.; Singan, V.R.; Monti, R.; Plett, K.L.; Ng, V.; Grigoriev, I.V.; Martin, F.M.; Anderson, I.C.; Plett, J.M. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc. Natl. Acad. Sci. USA 2022, 119, 2103527119. [Google Scholar] [CrossRef] [PubMed]
- Helber, N.; Wippel, K.; Sauer, N.; Schaarschmidt, S.; Hause, B.; Requena, N. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp. is crucial for the symbiotic relationship with plants. Plant Cell 2011, 23, 3812–3823. [Google Scholar] [CrossRef]
- Tsuzuki, S.; Handa, Y.; Takeda, N.; Kawaguchi, M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol. Plant–Microbe Interact. 2016, 4, 277–286. [Google Scholar] [CrossRef]
- Wang, S.; Xie, X.; Che, X.; Lai, W.; Ren, Y.; Fan, X.; Hu, W.; Tang, M.; Chen, H. Host and virus-induced gene silencing of HOG1-MAPKcascade genes in Rhizophagus irregularis inhibit arbuscule development and reduce resistance of plants to drought stress. Plant Biotechnol. J. 2023, 4, 866–883. [Google Scholar] [CrossRef] [PubMed]
- Qiao, A.; Gao, Z.; Roth, R. Aperspective on cross-kingdom RNA interference in mutualistic symbioses. New Phytol. 2023, 240, 68–79. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).