Differentially Expressed Genes in Cardiomyocytes of the First Camelized Mouse Model, Nrapc.255ins78 Mouse
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA-Seq
2.3. Differentially Expressed Gene (DEG) Analyses
2.4. Gene Ontology (GO) Functional Enrichment Analysis
2.5. Interaction Network Construction
2.6. RT-qPCR Validation
2.7. Statistics
3. Results
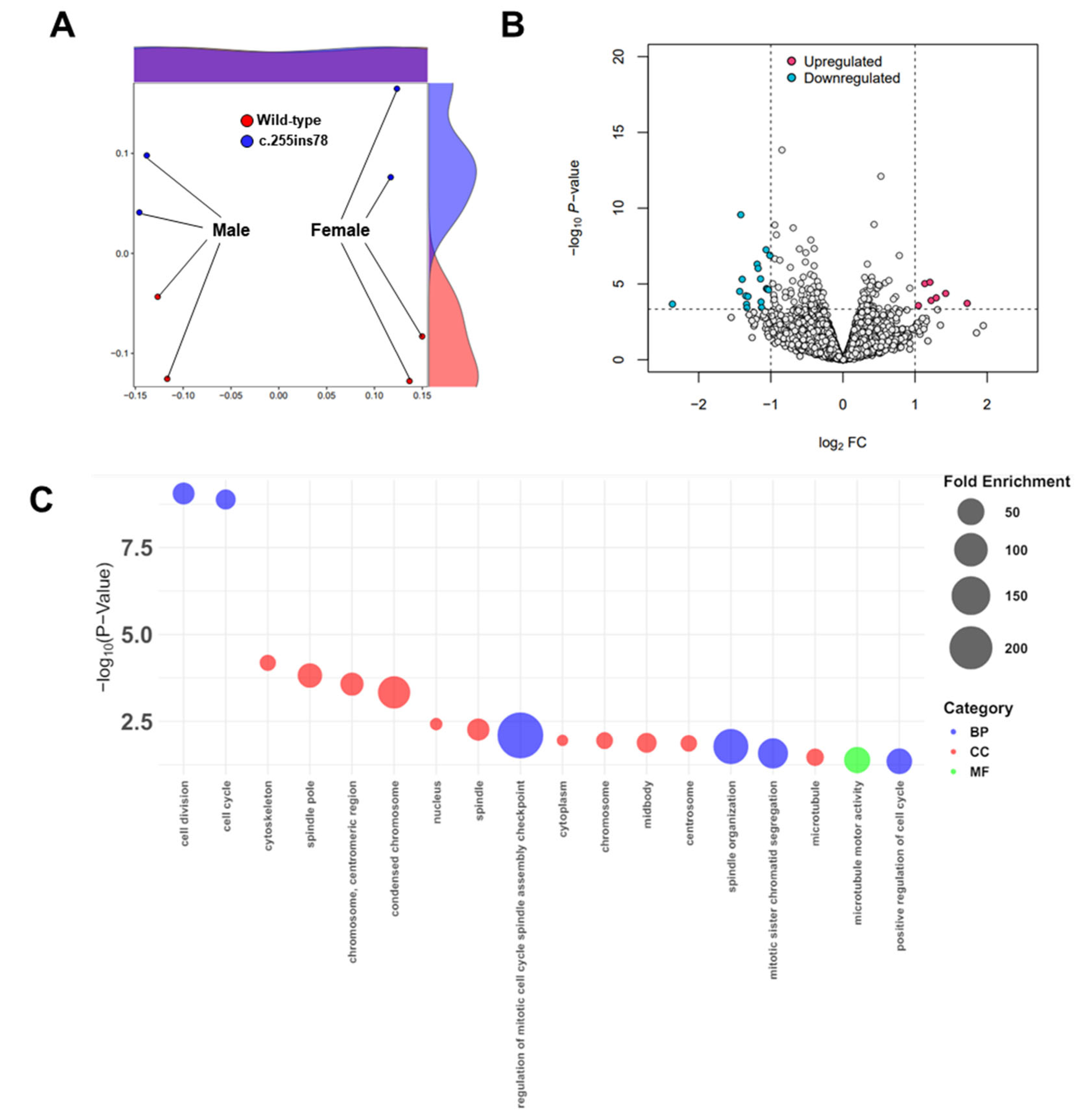
3.1. Data Processing and Transcriptomes
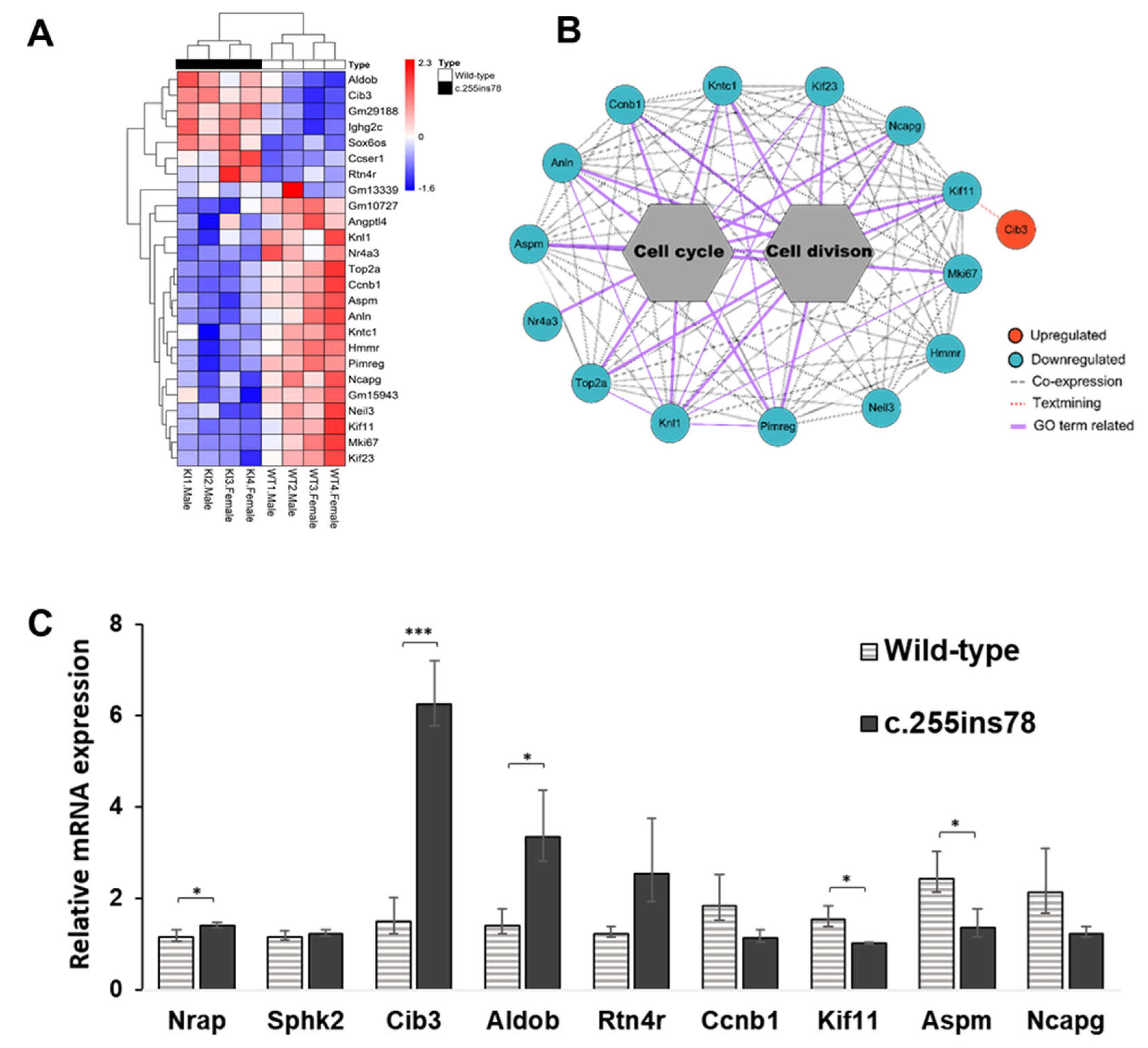
3.2. Functional Annotations
3.3. Expression Pattern and Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kandeel, M.; Al-Taher, A.; Venugopala, K.N.; Marzok, M.; Morsy, M.; Nagaraja, S. Camel proteins and enzymes: A growing resource for functional evolution and environmental adaptation. Front. Vet. Sci. 2022, 9, 911511. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef] [PubMed]
- Alvira-Iraizoz, F.; Gillard, B.T.; Lin, P.; Paterson, A.; Pauža, A.G.; Ali, M.A.; Alabsi, A.H.; Burger, P.A.; Hamadi, N.; Adem, A.; et al. Multiomic analysis of the Arabian camel (Camelus dromedarius) kidney reveals a role for cholesterol in water conservation. Commun. Biol. 2021, 4, 779. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, B.; Lim, B.; Uzzaman, R.; Jang, G.; Kim, K. Exploring the importance of predicted camel NRAP exon 4 for environmental adaptation using a mouse model. Anim. Genet. 2025, 56, e13490. [Google Scholar] [CrossRef] [PubMed]
- Buggiotti, L.; Yurchenko, A.A.; Yudin, N.S.; Vander Jagt, C.J.; Vorobieva, N.V.; Kusliy, M.A.; Vasiliev, S.K.; Rodionov, A.N.; Boronetskaya, O.I.; Zinovieva, N.A.; et al. Demographic History, Adaptation, and NRAP Convergent Evolution at Amino Acid Residue 100 in the World Northernmost Cattle from Siberia. Mol. Biol. Evol. 2021, 38, 3093–3110. [Google Scholar] [CrossRef]
- Arias, N.; Velapatiño, B.; Hung, A.; Cok, J. Cytokines expression in alpacas and llamas exposed to cold stress. Small Rumin. Res. 2016, 141, 135–140. [Google Scholar] [CrossRef]
- Leon, L.R. Molecular Biology of Thermoregulation Invited Review: Cytokine regulation of fever: Studies using gene knockout mice. J. Appl. Physiol. 2002, 92, 2648–2655. [Google Scholar] [CrossRef]
- Dugue, B.; Leppaänen, E. Adaptation related to cytokines in man: Effects of regular swimming in ice-cold water. Clin. Physiol. 2000, 20, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Yan, Z.; Yan, W.; Xia, Q.; Zhang, Y. Cold exposure stimulates lipid metabolism, induces inflammatory response in the adipose tissue of mice and promotes the osteogenic differentiation of BMMSCs via the p38 MAPK pathway in vitro. Int. J. Clin. Exp. Pathol. 2015, 8, 10875–10886. [Google Scholar] [PubMed] [PubMed Central]
- Zhang, D.J.; Wang, L.; Ma, S.Z.; Ma, H.; Liu, D. Characterization of pig skeletal muscle transcriptomes in response to low temperature. Vet. Med. Sci. 2023, 9, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yu, X.; Portela Fontoura, A.B.; Javaid, A.; de la Maza-Escolà, V.S.; Salandy, N.S.; Fubini, S.L.; Grilli, E.; McFadden, J.W.; Duan, J.E. Transcriptomic regulations of heat stress response in the liver of lactating dairy cows. BMC Genom. 2023, 24, 410. [Google Scholar] [CrossRef]
- Jiao, D.; Ji, K.; Liu, H.; Wang, W.; Wu, X.; Zhou, J.; Zhang, Y.; Zhou, H.; Hickford, J.G.H.; Degen, A.A.; et al. Transcriptome analysis reveals genes involved in thermogenesis in two cold-exposed sheep breeds. Genes 2021, 12, 375. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, B.E.; Ryu, D.; Oh, S.W.; Oh, C.-M. Comparative Transcriptome Profiling of Young and Old Brown Adipose Tissue Thermogenesis. Int. J. Mol. Sci. 2021, 22, 13143. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Meijler, F.L.; Meijler, T.D. Archetype, Adaptation and the Mammalian Heart. Neth. Heart J. 2011, 19, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Torrent-Guasp, F.; Kocica, M.J.; Corno, A.F.; Komeda, M.; Carreras-Costa, F.; Flotats, A.; Cosin-Aguillar, J.; Wen, H. Towards new understanding of the heart structure and function. Eur. J. Cardio-Thorac. Surg. 2005, 27, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, J. Metabolic Shifts during Aging and Pathology. Compr. Physiol. 2015, 5, 667–686. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, C.; Koivumäki, J.; Pekkanen-Mattila, M.; Aalto-Setälä, K. Sex Differences in Heart: From Basics to Clinics. Eur. J. Med. Res. 2022, 27, 241. [Google Scholar] [CrossRef]
- Ma, W.; Jia, K.; Cheng, H.; Xu, H.; Li, Z.; Zhang, H.; Xie, H.; Sun, H.; Yi, L.; Chen, Z.; et al. Orphan Nuclear Receptor NR4A3 Promotes Vascular Calcification via Histone Lactylation. Circ. Res. 2024, 134, 1427–1447. [Google Scholar] [CrossRef] [PubMed]
- Yamniuk, A.P.; Vogel, H.J. Insights into the Structure and Function of Calcium- and Integrin-Binding Proteins. Calcium Bind. Proteins 2006, 1, 150–155. [Google Scholar]
- Huang, H.; Bogstie, J.N.; Vogel, H.J. Biophysical and structural studies of the human calcium-and integrin-binding protein family: Understanding their functional similarities and differences. Biochem. Cell Biol. 2012, 90, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Song, X.; Du, L.; Wang, C. Molecular Characterization of the Sheep CIB1 Gene. Mol. Biol. Rep. 2009, 36, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Qiu, X.; Dionne, G.; Cunningham, C.L.; Pucak, M.L.; Peng, G.; Kim, Y.H.; Lauer, A.; Shapiro, L.; Müller, U. CIB2 and CIB3 Are Auxiliary Subunits of the Mechanotransduction Channel of Hair Cells. Neuron 2021, 109, 2131–2149.e15. [Google Scholar] [CrossRef]
- Dal Cortivo, G.; Dell’orco, D. Calcium-and Integrin-Binding Protein 2 (CIB2) in Physiology and Disease: Bright and Dark Sides. Int. J. Mol. Sci. 2022, 23, 3552. [Google Scholar] [CrossRef]
- Olivieri, G.; Dal Cortivo, G.; Dal Conte, R.; Zanzoni, S.; Marino, V.; Dell’Orco, D.; Cantini, F. Structural Dynamics of Calcium and Integrin-Binding Protein 2 (CIB2) Reveal Uncommon Flexibility and Heterogeneous Calcium and Magnesium Loading. Int. J. Biol. Macromol. 2025, 286, 138003. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.U.; Naik, U.P. Calcium- and integrin-binding protein 1 regulates microtubule organization and centrosome segregation through polo like kinase 3 during cell cycle progression. Int. J. Biochem. Cell Biol. 2011, 43, 120–129. [Google Scholar] [CrossRef]
- Xu, Z.; Miyata, H.; Kaneda, Y.; Castaneda, J.M.; Lu, Y.; Morohoshi, A.; Yu, Z.; Matzuk, M.M.; Ikawa, M. CIB4 Is Essential for the Haploid Phase of Spermatogenesis in Mice. Biol. Reprod. 2020, 103, 235–243. [Google Scholar] [CrossRef]
- Clark, A.L.; Kanekura, K.; Lavagnino, Z.; Spears, L.D.; Abreu, D.; Mahadevan, J.; Yagi, T.; Semenkovich, C.F.; Piston, D.W.; Urano, F. Targeting Cellular Calcium Homeostasis to Prevent Cytokine-Mediated beta Cell Death. Sci. Rep. 2017, 7, 5611. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, J.W.; Steiner, S.R.; O’Neill, C.M.; Nunemaker, C.S. The central role of calcium in the effects of cytokines on beta-cell function: Implications for type 1 and type 2 diabetes. Cell Calcium 2011, 50, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Bootman, M.D.; Bultynck, G. Fundamentals of Cellular Calcium Signaling: A Primer. Cold Spring Harb. Perspect. Biol. 2020, 12, a038802. [Google Scholar] [CrossRef]
- Giorgi, C.; Marchi, S.; Pinton, P. The Machineries, Regulation, and Cellular Functions of Mitochondrial Calcium. Nat. Rev. Mol. Cell Biol. 2018, 19, 713–730. [Google Scholar] [CrossRef] [PubMed]
- Boyman, L.; Karbowski, M.; Lederer, W.J. Regulation of Mitochondrial ATP Production: Ca2+ Signaling and Quality Control. Trends Mol. Med. 2020, 26, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Qiu, H. The Physiological and Pathological Roles of Mitochondrial Calcium Uptake in Heart. Int. J. Mol. Sci. 2020, 21, 7689. [Google Scholar] [CrossRef]
- Iamartino, L.; Brandi, M.L. The Calcium-Sensing Receptor in Inflammation: Recent Updates. Front. Physiol. 2022, 13, 1059369. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gehmlich, K.; Geier, C.; Osterziel, K.J.; Van der Ven, P.F.; Furst, D.O. Decreased Interactions of Mutant Muscle LIM Protein (MLP) with N-RAP and α-Actinin and Their Implication for Hypertrophic Cardiomyopathy. Cell Tissue Res. 2004, 317, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Vermij, S.H.; Abriel, H.; Van Veen, T.A.B. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc. Res. 2017, 113, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Strong, P.N.; Dubowitz, V.; Dunn, M.J. Calmodulin-binding profiles for nebulin and dystrophin in human skeletal muscle. FEBS Lett. 1988, 234, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Kahl, C.R.; Means, A.R. Regulation of Cell Cycle Progression by Calcium/Calmodulin-Dependent Pathways. Endocr. Rev. 2003, 24, 719–736. [Google Scholar] [CrossRef]
- Yuen, M.; Ottenheijm, C.A.C. Nebulin: Big protein with big responsibilities. J. Muscle Res. Cell Motil. 2020, 41, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Brodehl, A.; Belke, D.D.; Garnett, L.; Martens, K.; Abdelfatah, N.; Rodriguez, M.; Diao, C.; Chen, Y.X.; Gordon, P.M.; Nygren, A.; et al. Transgenic Mice Overexpressing Desmocollin-2 (DSC2) Develop Cardiomyopathy Associated with Myocardial Inflammation and Fibrotic Remodeling. PLoS ONE 2017, 12, e0174019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cai, X.; Wang, K.C.; Meng, Z. Mechanoregulation of YAP and TAZ in Cellular Homeostasis and Disease Progression. Front. Cell Dev. Biol. 2021, 9, 673599. [Google Scholar] [CrossRef] [PubMed]
- Halder, G.; Dupont, S.; Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012, 13, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ Upstream Signals and Downstream Responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-Y.; Lim, B.; Lee, B.-Y.; Jang, G.; Choi, J.-S.; Cui, X.-S.; Kim, K.-S. Differentially Expressed Genes in Cardiomyocytes of the First Camelized Mouse Model, Nrapc.255ins78 Mouse. Genes 2025, 16, 142. https://doi.org/10.3390/genes16020142
Lee S-Y, Lim B, Lee B-Y, Jang G, Choi J-S, Cui X-S, Kim K-S. Differentially Expressed Genes in Cardiomyocytes of the First Camelized Mouse Model, Nrapc.255ins78 Mouse. Genes. 2025; 16(2):142. https://doi.org/10.3390/genes16020142
Chicago/Turabian StyleLee, Sung-Yeon, Byeonghwi Lim, Bo-Young Lee, Goo Jang, Jung-Seok Choi, Xiang-Shun Cui, and Kwan-Suk Kim. 2025. "Differentially Expressed Genes in Cardiomyocytes of the First Camelized Mouse Model, Nrapc.255ins78 Mouse" Genes 16, no. 2: 142. https://doi.org/10.3390/genes16020142
APA StyleLee, S.-Y., Lim, B., Lee, B.-Y., Jang, G., Choi, J.-S., Cui, X.-S., & Kim, K.-S. (2025). Differentially Expressed Genes in Cardiomyocytes of the First Camelized Mouse Model, Nrapc.255ins78 Mouse. Genes, 16(2), 142. https://doi.org/10.3390/genes16020142






