Integrating WGCNA, TCN, and Alternative Splicing to Map Early Caste Programs in Day-2 Honeybee Larvae
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Mapping and Quality Control of RNA-Seq Data
2.3. PCA
2.4. GO Enrichment Analysis of Differentially Expressed and Alternatively Spliced Genes
2.5. Alternative Splicing Analysis
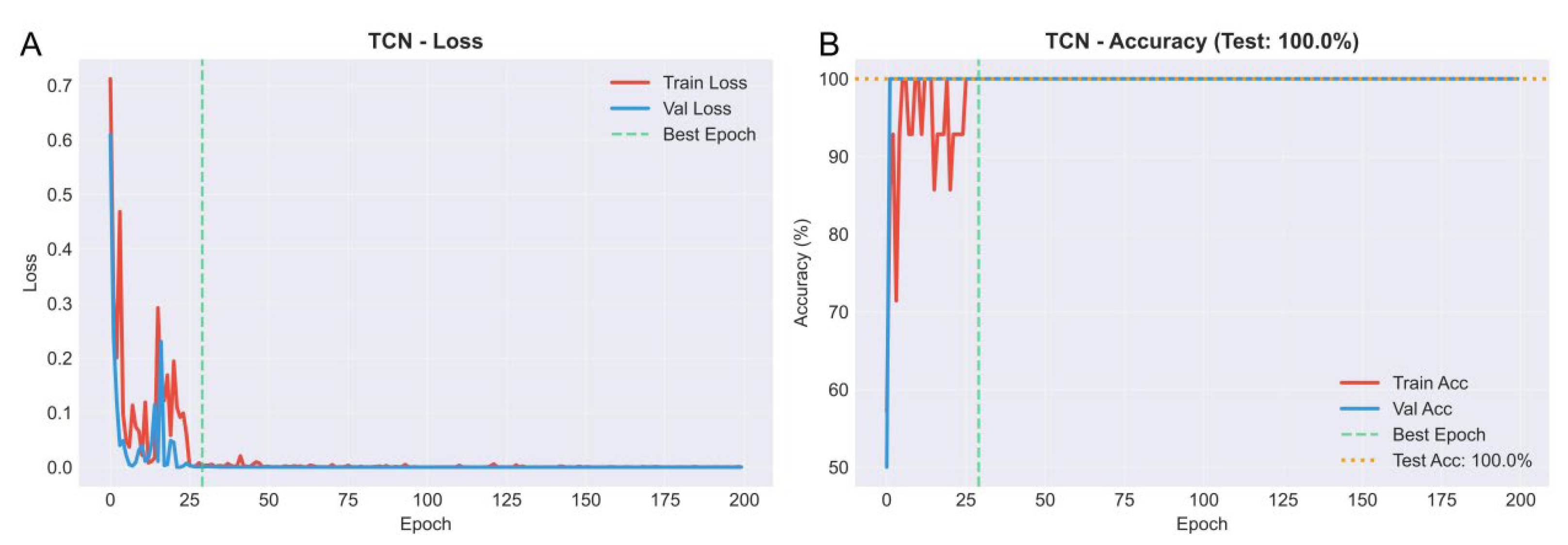
2.6. Weighted Gene Co-Expression Network Analysis (WGCNA) and TCN Modeling
3. Results
3.1. Overall RNA-Seq Results and Quality Control
3.2. Hierarchical Clustering and Principal Component Analysis of Samples
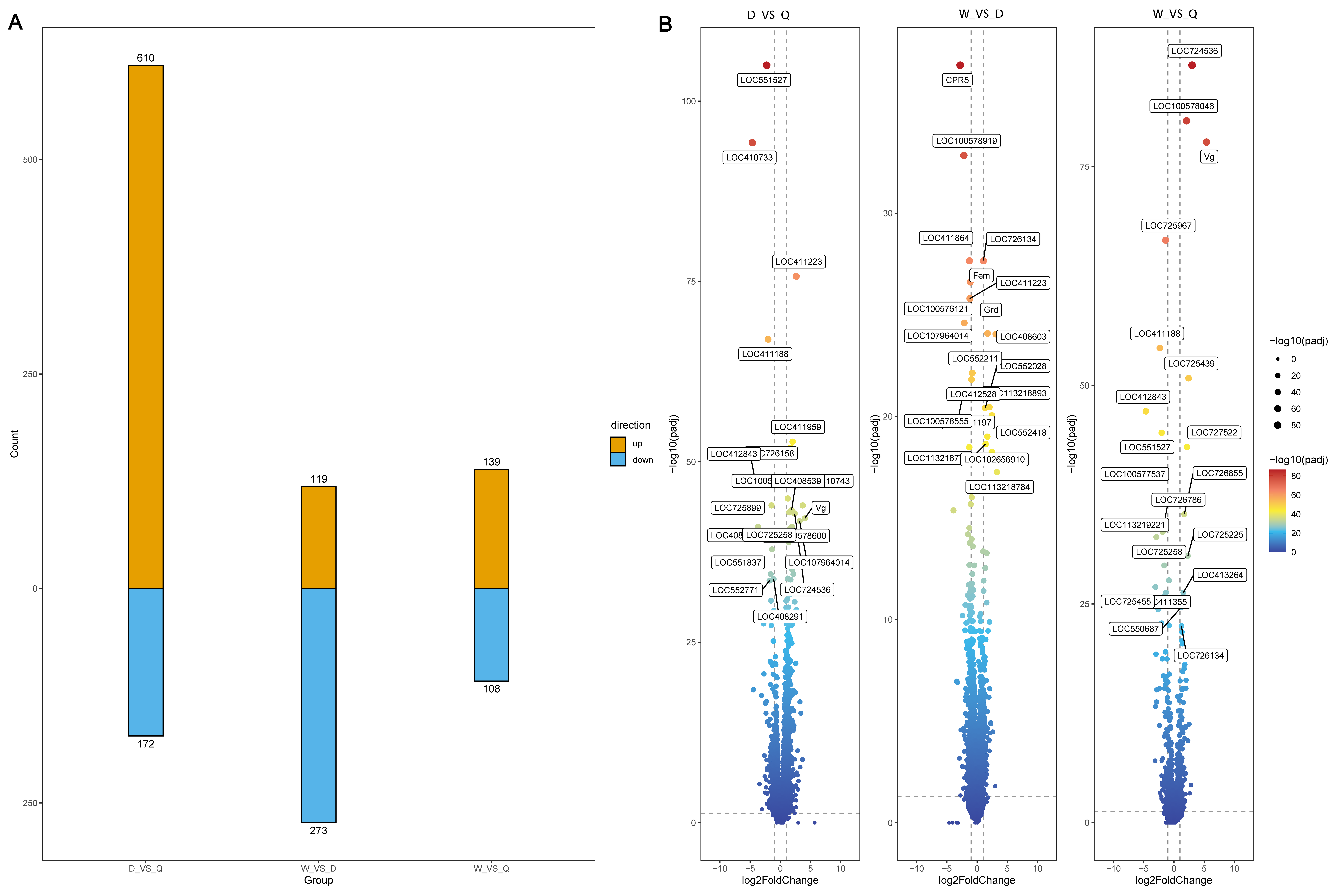
3.3. Analysis of Differentially Expressed Genes (DEGs)
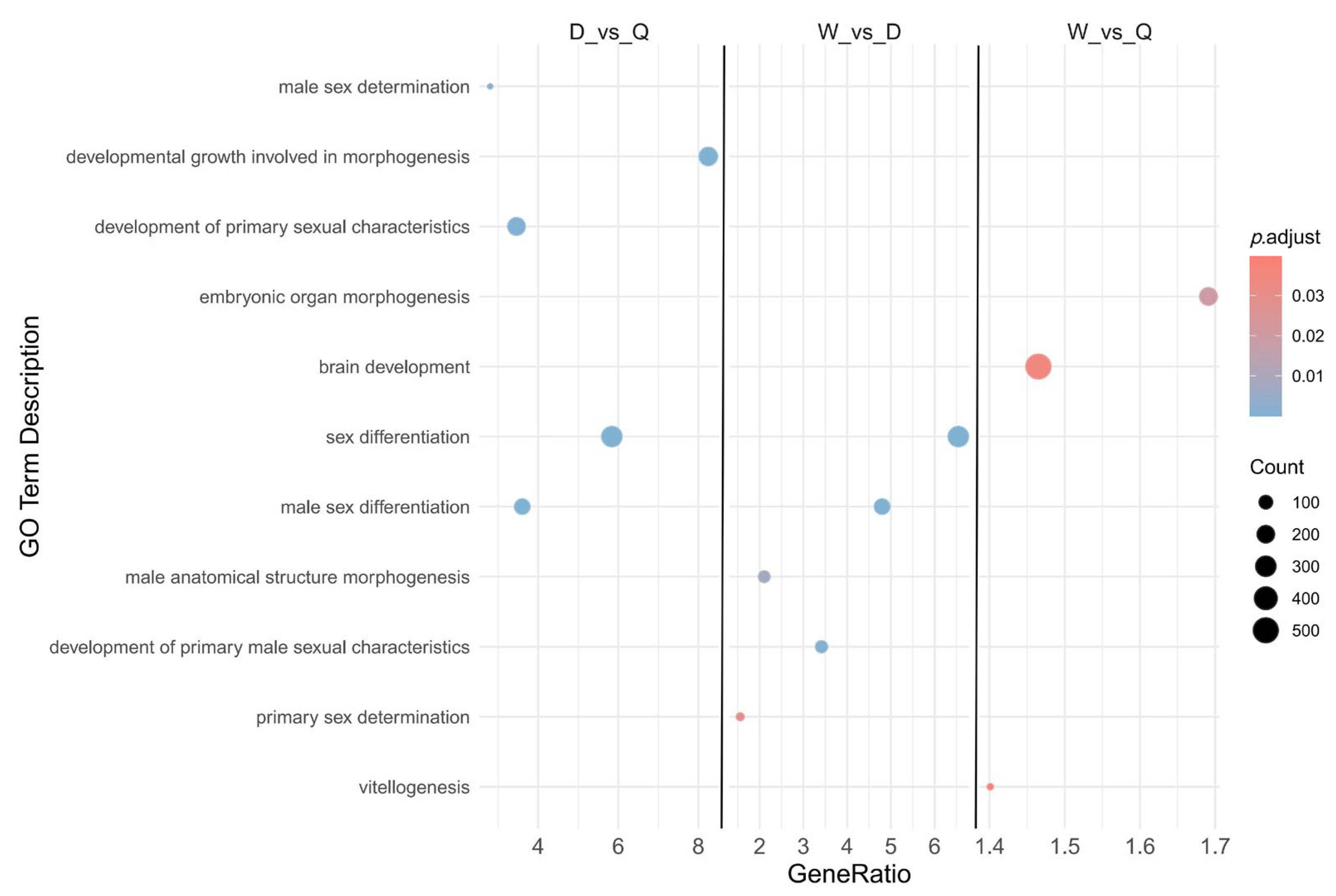
3.4. Gene Expression GO Analysis
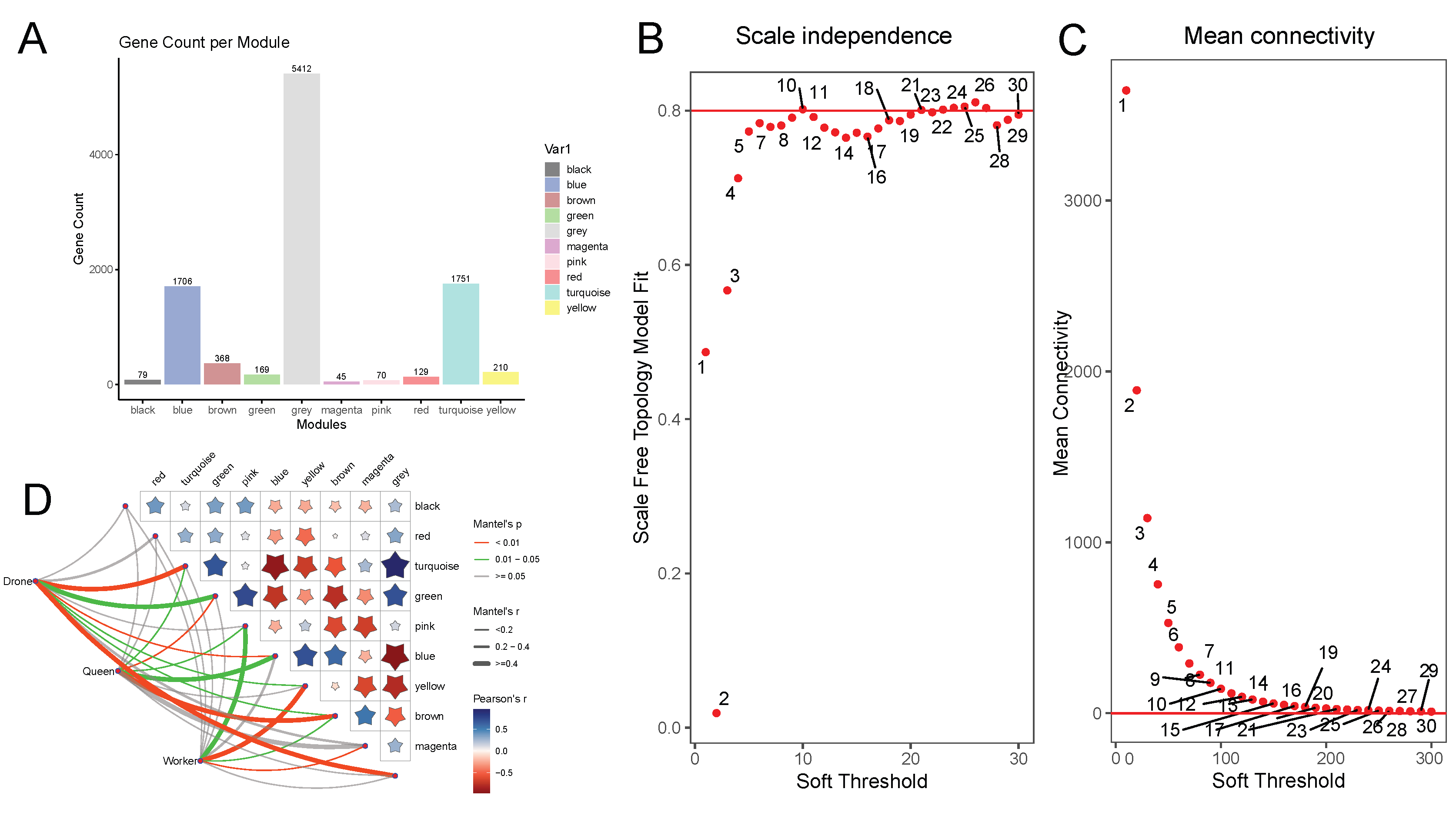
3.5. WGCNA
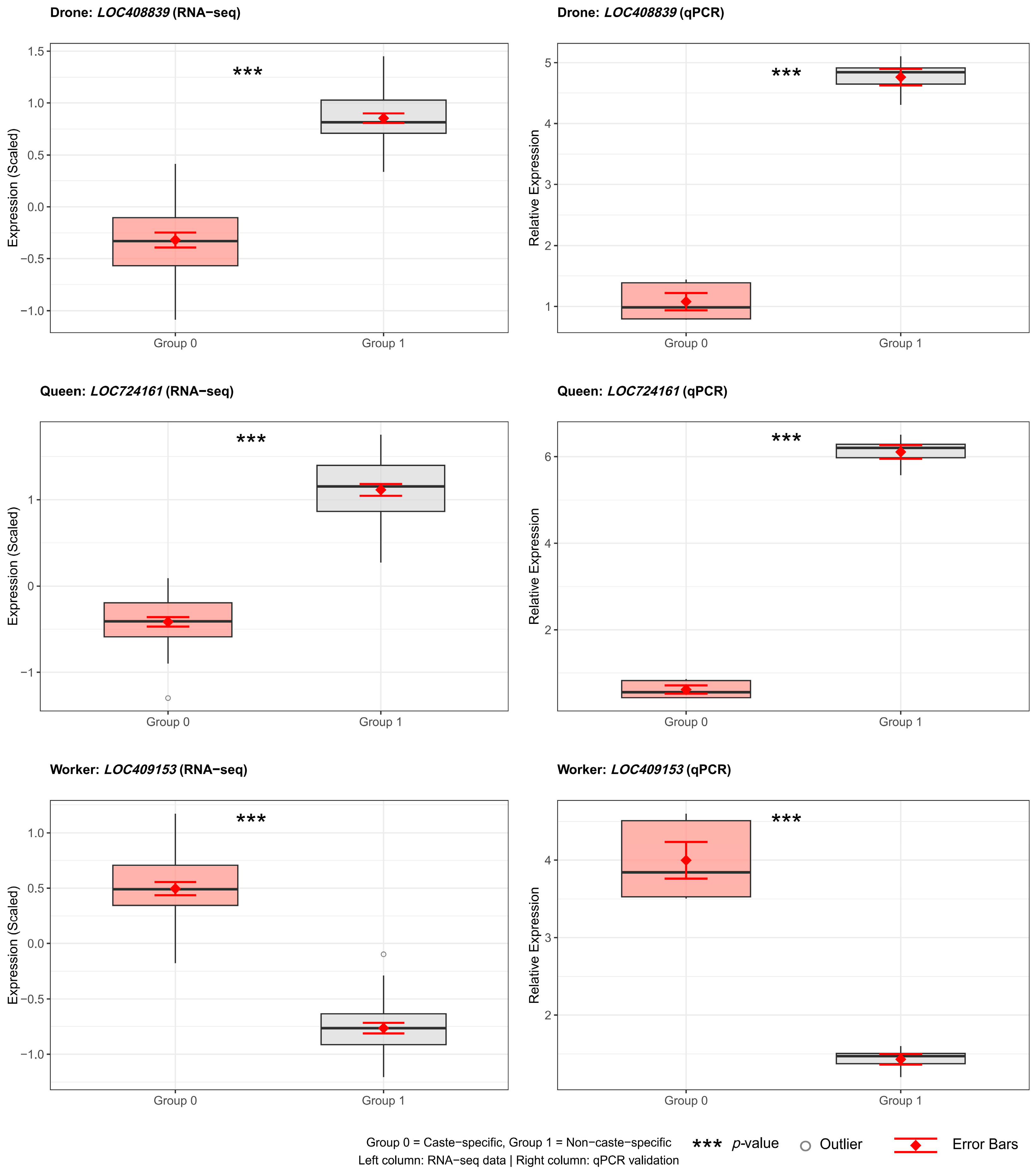
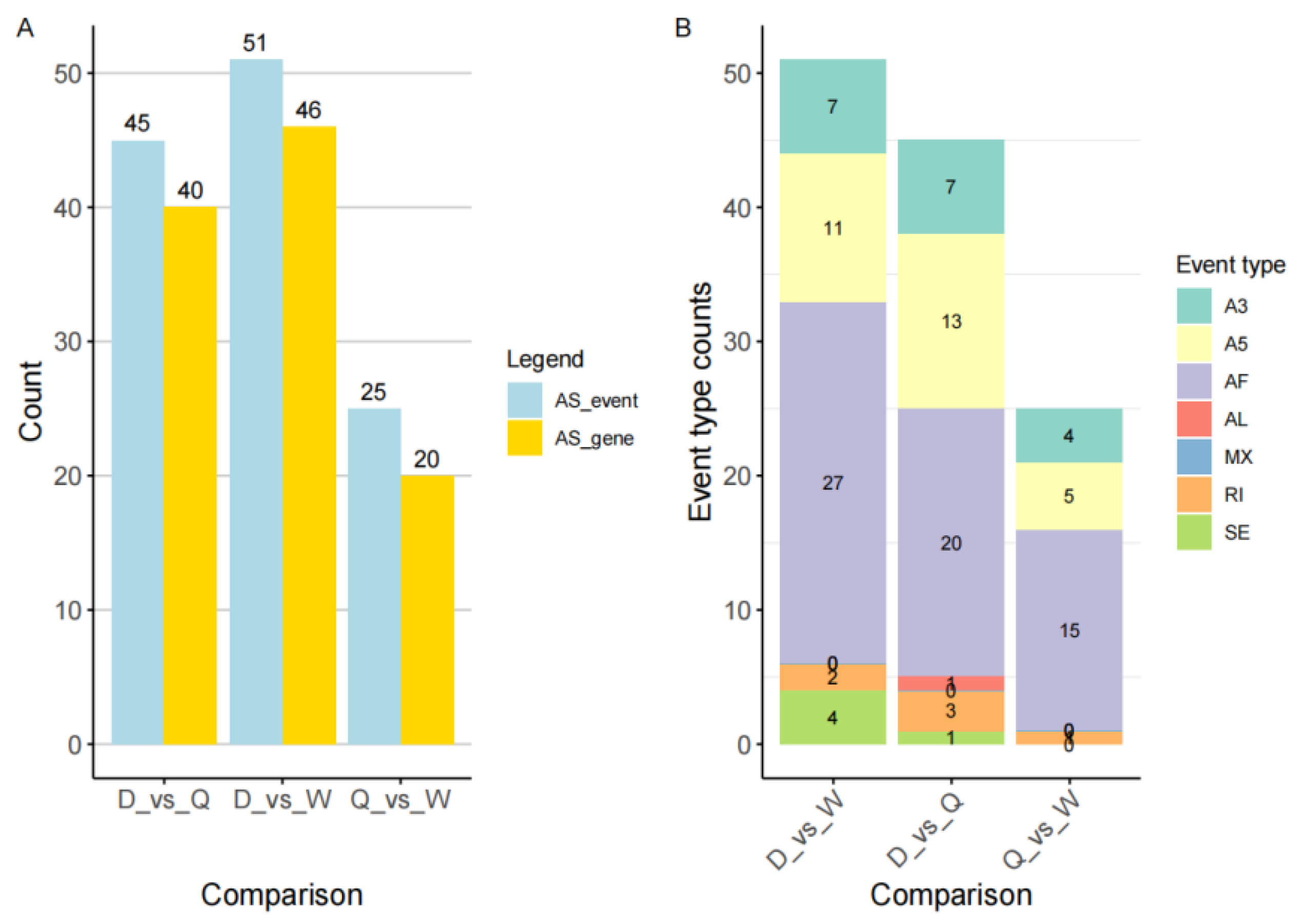
3.6. Statistical Analysis of Differential Genes and Events in Alternative Splicing Events
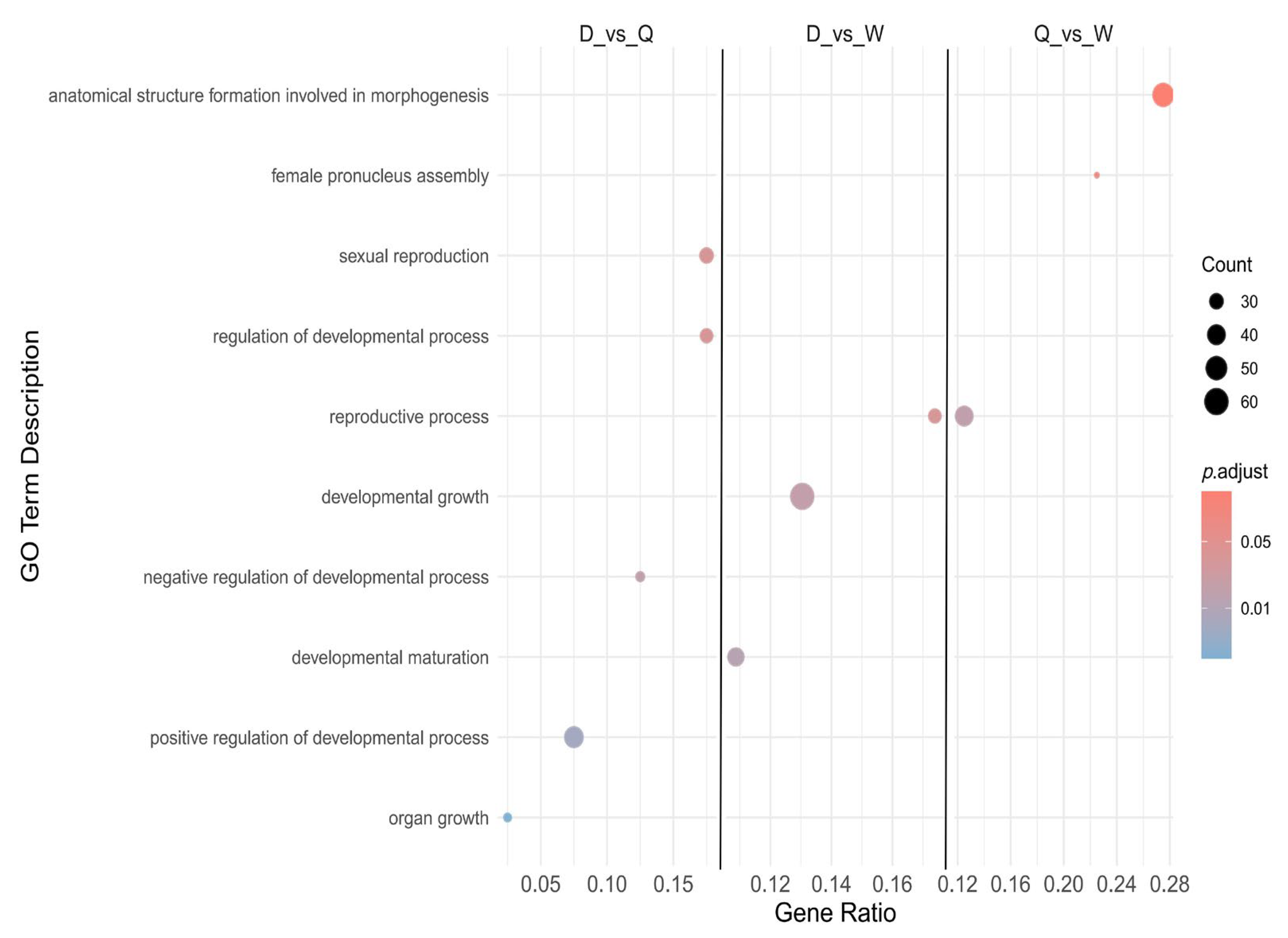
3.7. GO Enrichment Analysis of Alternative Splicing
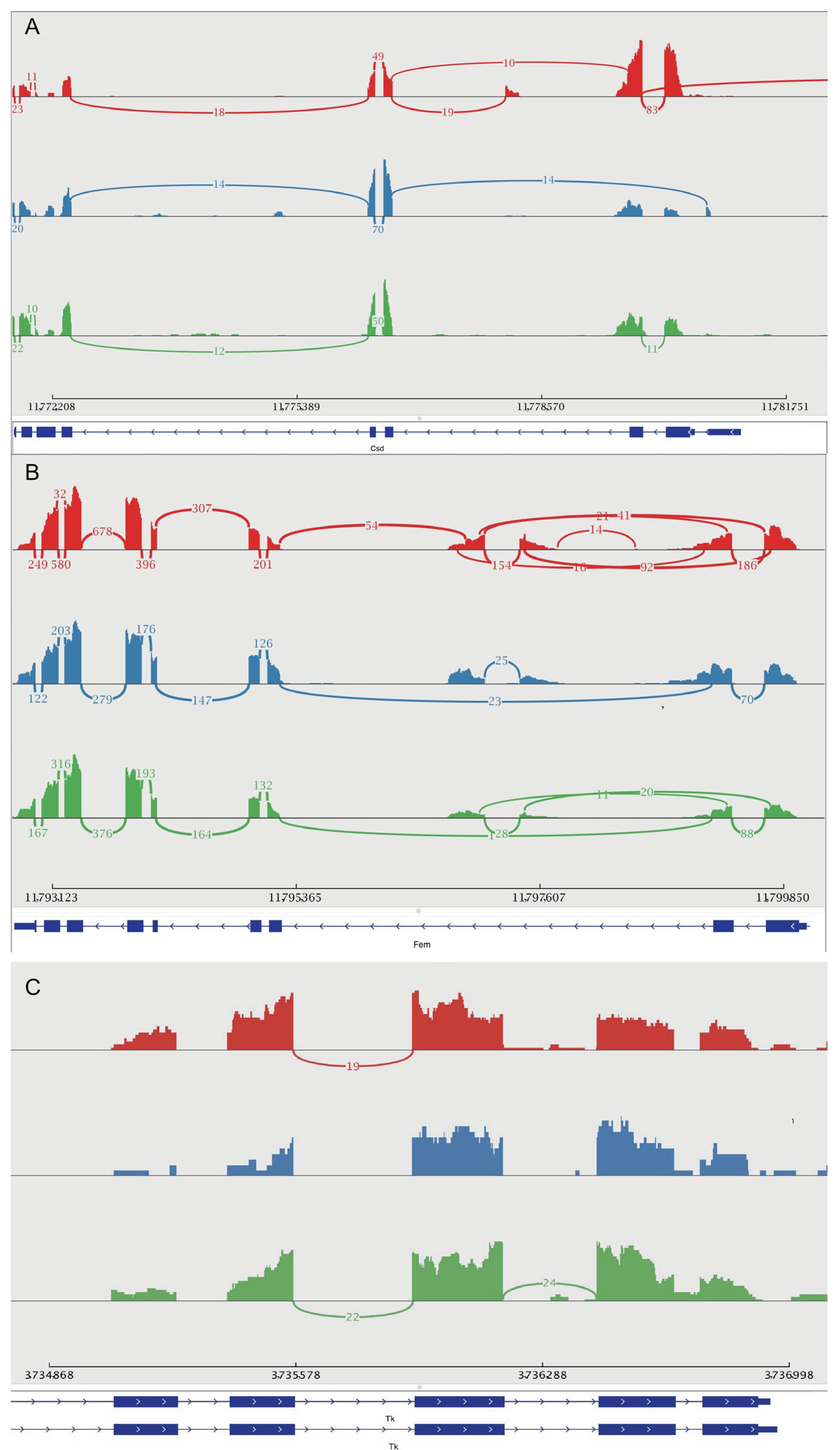
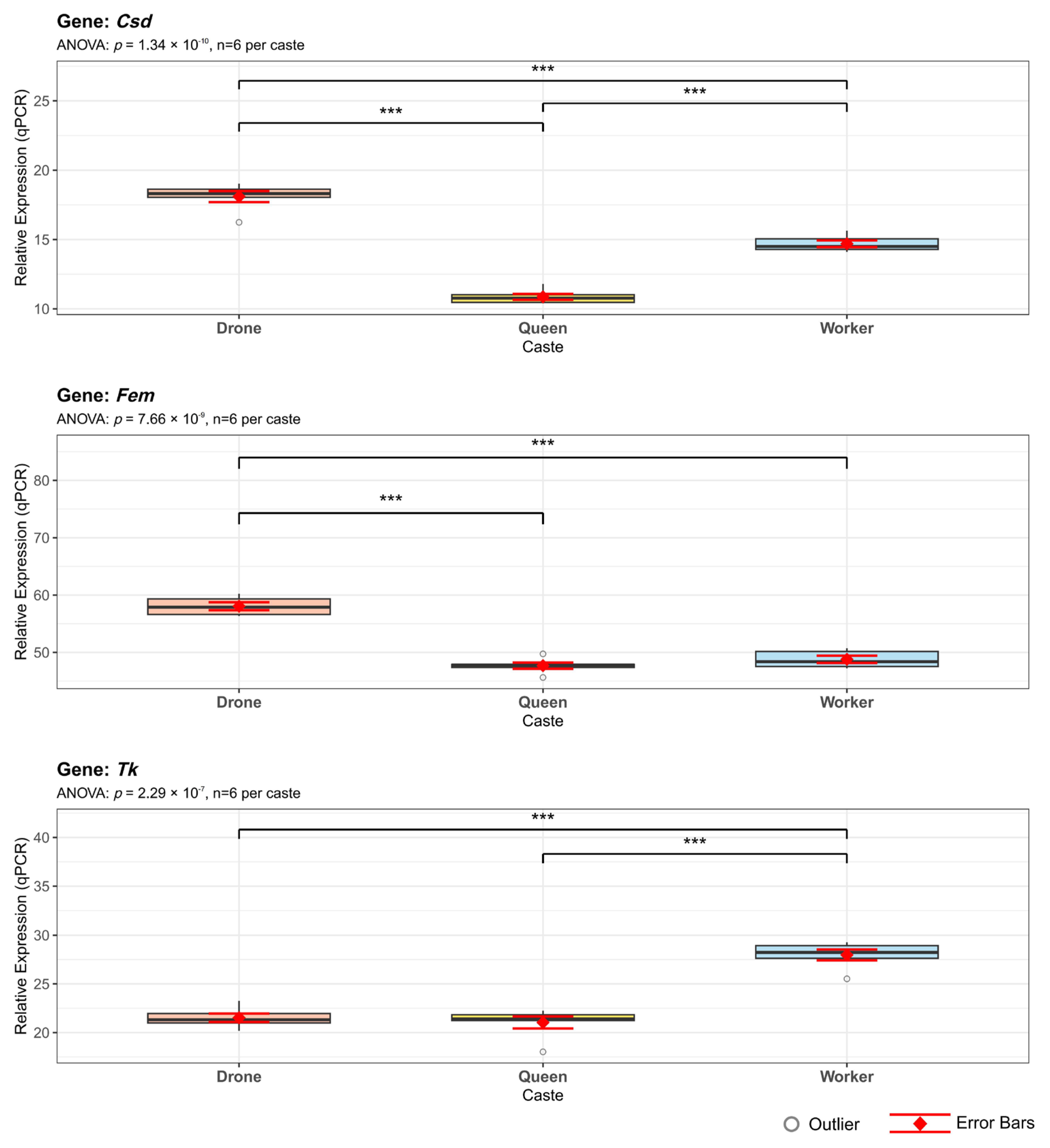
3.8. Alternative Splicing of Genes Related to Sex Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brouwers, E.V.M.; Ebert, R.; Beetsma, J. Behavioural and physiological aspects of nurse bees in relation to the composition of larval food during caste differentiation in the honeybee. J. Apic. Res. 1987, 26, 11–23. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Y.; Zheng, H.Q.; Cao, L.F.; Niu, D.F.; Yu, D.L.; Sun, Y.Q.; Hu, S.N.; Hu, F.L. Transcriptome comparison between honey bee queen-and worker-destined larvae. Insect Biochem. Mol. Biol. 2012, 42, 665–673. [Google Scholar] [CrossRef]
- Rangel, J.; Fisher, A. Factors affecting the reproductive health of honey bee (Apis mellifera) drones—A review. Apidologie 2019, 50, 759–778. [Google Scholar] [CrossRef]
- He, X.J.; Zhou, L.B.; Pan, Q.Z.; Barron, A.B.; Yan, W.Y.; Zeng, Z.J. Making a queen: An epigenetic analysis of the robustness of the honeybee (Apis mellifera) queen developmental pathway. Mol. Ecol. 2017, 26, 1598–1607. [Google Scholar] [CrossRef]
- Cnaani, J.; Robinson, G.E.; Hefetz, A. The critical period for caste determination in Bombus terrestris and its juvenile hormone correlates. J. Comp. Physiol. A 2000, 186, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, X.J.; Barron, A.B.; Li, Z.; Jin, M.J.; Wang, Z.L.; Huang, Q.; Zhang, L.Z.; Wu, X.B.; Yan, W.Y.; et al. The diverging epigenomic landscapes of honeybee queens and workers revealed by multiomic sequencing. Insect Biochem. Mol. Biol. 2023, 155, 103929. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.-Z.; et al. RNA m6A modification functions in larval development and caste differentiation in honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef]
- Shi, Y.Y.; Yan, W.Y.; Huang, Z.Y.; Wang, Z.L.; Wu, X.B.; Zeng, Z.J. Genomewide analysis indicates that queen larvae have lower methylation levels in the honey bee (Apis mellifera). Sci. Nat. 2013, 100, 193–197. [Google Scholar] [CrossRef]
- Santos, D.E.; de Oliveira Souza, A.; Tibério, G.J.; Alberici, L.C.; Hartfelder, K. Differential expression of antioxidant system genes in honey bee (Apis mellifera L.) caste development mitigates ROS-mediated oxidative damage in queen larvae. Genet. Mol. Biol. 2020, 43, e20200173. [Google Scholar] [CrossRef] [PubMed]
- Corona, M.; Estrada, E.; Zurita, M. Differential expression of mitochondrial genes between queens and workers during caste determination in the honeybee Apis mellifera. J. Exp. Biol. 1999, 202, 929–938. [Google Scholar] [CrossRef]
- Cameron, R.C.; Duncan, E.J.; Dearden, P.K. Biased gene expression in early honeybee larval development. BMC Genom. 2013, 14, 903. [Google Scholar] [CrossRef] [PubMed]
- El Agoze, M.; Drezen, J.M.; Renault, S.; Periquet, G. Analysis of the reproductive potential of diploid males in the wasp Diadromus pulchellus (Hymenoptera: Ichneumonidae). Bull. Entomol. Res. 1994, 84, 213–218. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yamada, T.; Suzuki, M.G. In vitro comparison of sex-specific splicing efficiencies of Fem. pre-mRNA under monoallelic and heteroallelic conditions of Csd, a master sex-determining gene in the honeybee. J. Dev. Biol. 2023, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Netschitailo, O.; Raub, S.; Kaftanoglu, O.; Beye, M. Sexual diversification of splicing regulation during embryonic development in honeybees (Apis mellifera), A haplodiploid system. Insect Mol. Biol. 2022, 31, 170–176. [Google Scholar] [CrossRef]
- Cheng, F.-P.; Hu, X.-F.; Pan, L.-X.; Gong, Z.-X.; Qin, K.-X.; Li, Z.; Wang, Z.-L. Transcriptome changes of Apis mellifera female embryos with Fem. gene knockout by CRISPR/Cas9. Int. J. Biol. Macromol. 2023, 229, 260–267. [Google Scholar] [CrossRef]
- Zhu, G.-Q.; Zhou, Y.-J.; Qiu, L.-X.; Wang, B.; Yang, Y.; Liao, W.-T.; Luo, Y.-H.; Shi, Y.-H.; Zhou, J.; Fan, J.; et al. Prognostic alternative mRNA splicing signature in hepatocellular carcinoma: A study based on large-scale sequencing data. Carcinogenesis 2019, 40, 1077–1085. [Google Scholar] [CrossRef]
- Wright, C.J.; Smith, C.W.J.; Jiggins, C.D. Alternative splicing as a source of phenotypic diversity. Nat. Rev. Genet. 2022, 23, 697–710. [Google Scholar] [CrossRef]
- Kalsotra, A.; Cooper, T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011, 12, 715–729. [Google Scholar] [CrossRef]
- Foret, S.; Kucharski, R.; Pellegrini, M.; Feng, S.H.; Jacobsen, S.E.; Robinson, G.E.; Maleszka, R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Nati. Acad. Sci. USA 2012, 109, 4968–4973. [Google Scholar] [CrossRef]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef]
- He, X.J.; Jiang, W.J.; Zhou, M.; Barron, A.B.; Zeng, Z.J. A comparison of honeybee (Apis mellifera) queen, worker and drone larvae by RNA-Seq. Insect Sci. 2019, 26, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Bellora, N.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef]
- Vleurinck, C.; Raub, S.; Sturgill, D.; Oliver, B.; Beye, M. Linking genes and brain development of honeybee workers: A whole-transcriptome approach. PLoS ONE 2016, 11, e0157980. [Google Scholar] [CrossRef] [PubMed]
- Nunes, F.M.F.; Ihle, K.E.; Mutti, N.S.; Simões, Z.L.P.; Amdam, G.V. The gene vitellogenin affects microRNA regulation in honey bee (Apis mellifera) fat body and brain. J. Exp. Biol. 2013, 216, 3724–3732. [Google Scholar] [CrossRef]
- Seehuus, S.-C.; Norberg, K.; Gimsa, U.; Krekling, T.; Amdam, G.V. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. USA 2006, 103, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Gempe, T.; Hasselmann, M.; Schiøtt, M.; Hause, G.; Otte, M.; Beye, M. Sex determination in honeybees: Two separate mechanisms induce and maintain the female pathway. PLoS Biol. 2009, 7, e1000222. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Gou, X.; Qin, Z.Y.; Li, D.Q.; Wang, Y.; Ma, E.B.; Li, S.; Zhang, J.Z. Identification and expression of cuticular protein genes based on Locusta migratoria transcriptome. Sci. Rep. 2017, 7, 45462. [Google Scholar] [CrossRef]
- Hunziker, M.; Barandun, J.; Petfalski, E.; Tan, D.Y.; Delan-Forino, C.; Molloy, K.R.; Kim, K.H.; Dunn-Davies, H.; Shi, Y.; Chaker-Margot, M.; et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat. Commun. 2016, 7, 12090. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, L.T.; Zhang, W.X.; Cui, X.P.; Wang, H.F.; Xu, B.H. Comparison of the nutrient composition of royal jelly and worker jelly of honey bees (Apis mellifera). Apidologie 2016, 47, 48–56. [Google Scholar] [CrossRef]
- Lv, J.-H.; Hou, A.-J.; Zhang, S.-H.; Dong, J.-J.; Kuang, H.-X.; Yang, L.; Jiang, H. WGCNA combined with machine learning to find potential biomarkers of liver cancer. Medicine 2023, 102, e36536. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhu, H.X.; Ren, J.L.; Ren, M.H. Integrative machine learning models predict prostate cancer diagnosis and biochemical recurrence risk: Advancing precision oncology. NPJ Digit. Med. 2025, 8, 524. [Google Scholar] [CrossRef]
- Gómez-Pascual, A.; Rocamora-Pérez, G.; Ibanez, L.; Botía, J.A. Targeted co-expression networks for the study of traits. Sci. Rep. 2024, 14, 16675. [Google Scholar] [CrossRef]
- Gao, J.L.; Wang, Q.L.; Liu, J.; Zheng, S.Q.; Liu, J.H.; Gao, Z.Y.; Zhu, C. Integrating weighted gene co-expression network analysis and machine learning to elucidate neural characteristics in a mouse model of depression. Front. Psychiatry 2025, 16, 1564095. [Google Scholar] [CrossRef] [PubMed]
- Li, A.N.; Zhang, Y.R.; Zhao, Z.D.; Wang, M.M.; Zan, L.S. Molecular characterization and transcriptional regulation analysis of the bovine PDHB gene. PLoS ONE 2016, 11, e0157445. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo, S.V.; Hartfelder, K. The insulin signaling pathway in honey bee (Apis mellifera) caste development—Differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 2008, 54, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Campbell, P.M.; Weisman, S.; Trueman, H.E.; Sriskantha, A.; Wanjura, W.J.; Haritos, V.S. A highly divergent gene cluster in honey bees encodes a novel silk family. Genome Res. 2006, 16, 1414–1421. [Google Scholar] [CrossRef]
- Xu, R.M.; Ma, B.B.; Yang, Y.Y.; Dong, X.C.; Li, J.K.; Xu, X.; Fang, Y. Proteome-metabolome profiling of wax gland complex reveals functional changes in honeybee, Apis mellifera L. iScience 2024, 27, 109279. [Google Scholar] [CrossRef]
- Robinson, E.K.; Jagannatha, P.; Covarrubias, S.; Cattle, M.; Smaliy, V.; Safavi, R.; Shapleigh, B.; Abu-Shumays, R.; Jain, M.; Cloonan, S.M.; et al. Inflammation drives alternative first exon usage to regulate immune genes including a novel iron-regulated isoform of Aim2. Elife 2021, 10, e69431. [Google Scholar] [CrossRef]
- Hasselmann, M.; Gempe, T.; Schiøtt, M.; Nunes-Silva, C.G.; Otte, M.; Beye, M. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 2008, 454, 519–522. [Google Scholar] [CrossRef]
- Beye, M.; Hasselmann, M.; Fondrk, M.K.; Page, R.E.; Omholt, S.W. The gene Csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 2003, 114, 419–429. [Google Scholar] [CrossRef]
- Graveley, B.R. Sorting out the complexity of SR protein functions. RNA 2000, 6, 1197–1211. [Google Scholar] [CrossRef]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Butler, B.; Pirrotta, V.; Irminger-Finger, I.; Nöthiger, R. The sex-determining gene tra of Drosophila: Molecular cloning and transformation studies. EMBO J. 1986, 5, 3607–3613. [Google Scholar] [CrossRef]
- Boggs, R.T.; Gregor, P.; Idriss, S.; Belote, J.M.; McKeown, M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell 1987, 50, 739–747. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; Li, J.; Yue, D.; Su, R. Integrating WGCNA, TCN, and Alternative Splicing to Map Early Caste Programs in Day-2 Honeybee Larvae. Genes 2025, 16, 1409. https://doi.org/10.3390/genes16121409
Ding X, Li J, Yue D, Su R. Integrating WGCNA, TCN, and Alternative Splicing to Map Early Caste Programs in Day-2 Honeybee Larvae. Genes. 2025; 16(12):1409. https://doi.org/10.3390/genes16121409
Chicago/Turabian StyleDing, Xiang, Jinyou Li, Dan Yue, and Runlang Su. 2025. "Integrating WGCNA, TCN, and Alternative Splicing to Map Early Caste Programs in Day-2 Honeybee Larvae" Genes 16, no. 12: 1409. https://doi.org/10.3390/genes16121409
APA StyleDing, X., Li, J., Yue, D., & Su, R. (2025). Integrating WGCNA, TCN, and Alternative Splicing to Map Early Caste Programs in Day-2 Honeybee Larvae. Genes, 16(12), 1409. https://doi.org/10.3390/genes16121409






