Phenotypic Characterization and Transcriptome Analysis of the Dwarf Mutant zmbrd1 in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Identification of Genotype of Mutant Strains
2.3. Bioinformatics Analysis of ZmBRD1 Gene
2.4. Transcriptome Sequencing
2.5. Observation of Tissue Paraffin Sections
2.6. Gene Expression Level Analysis
3. Results
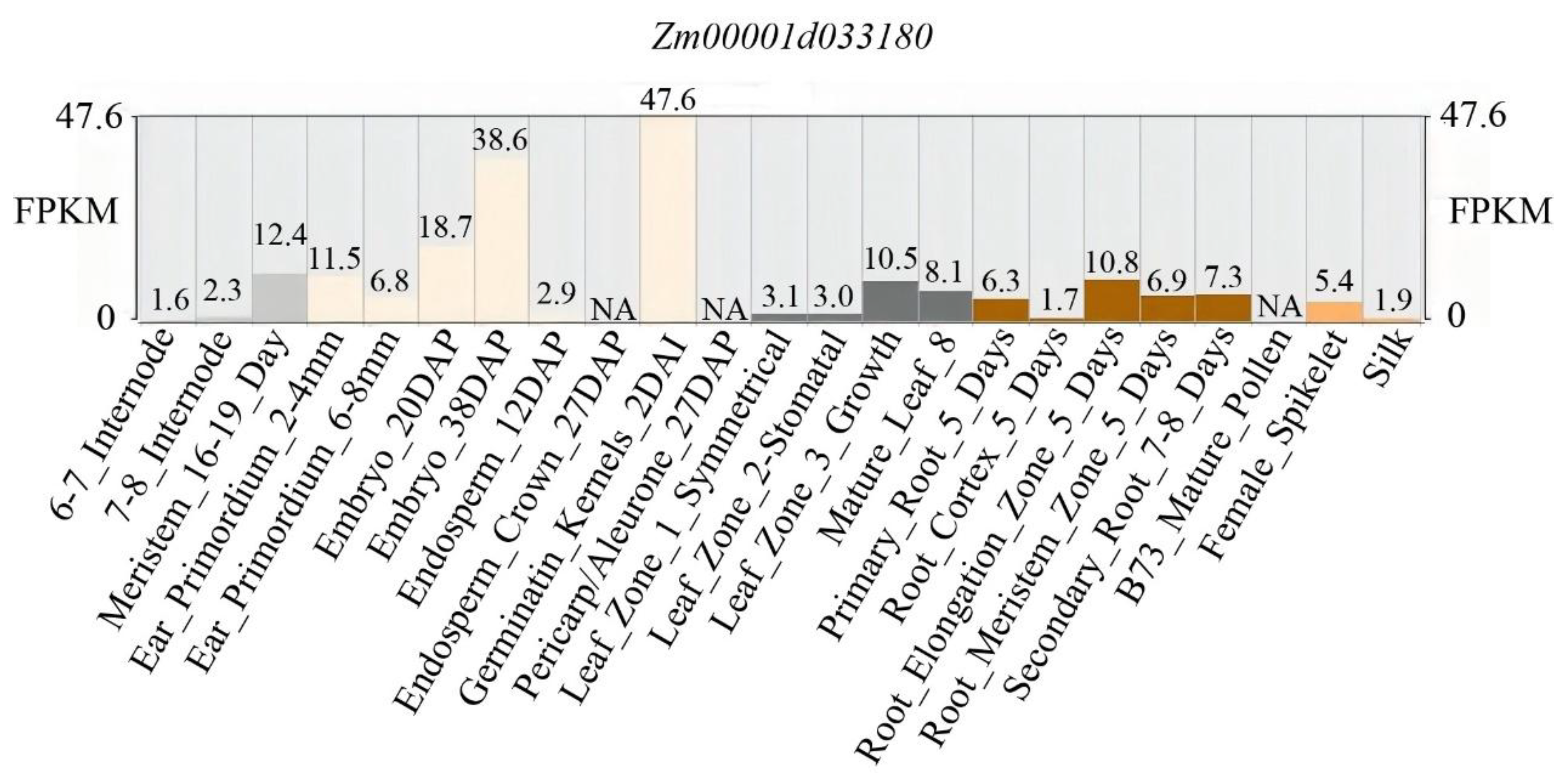
3.1. Identification of ZmBRD1 Mutant
3.2. Evolutionary Tree Analysis of ZmBRD1
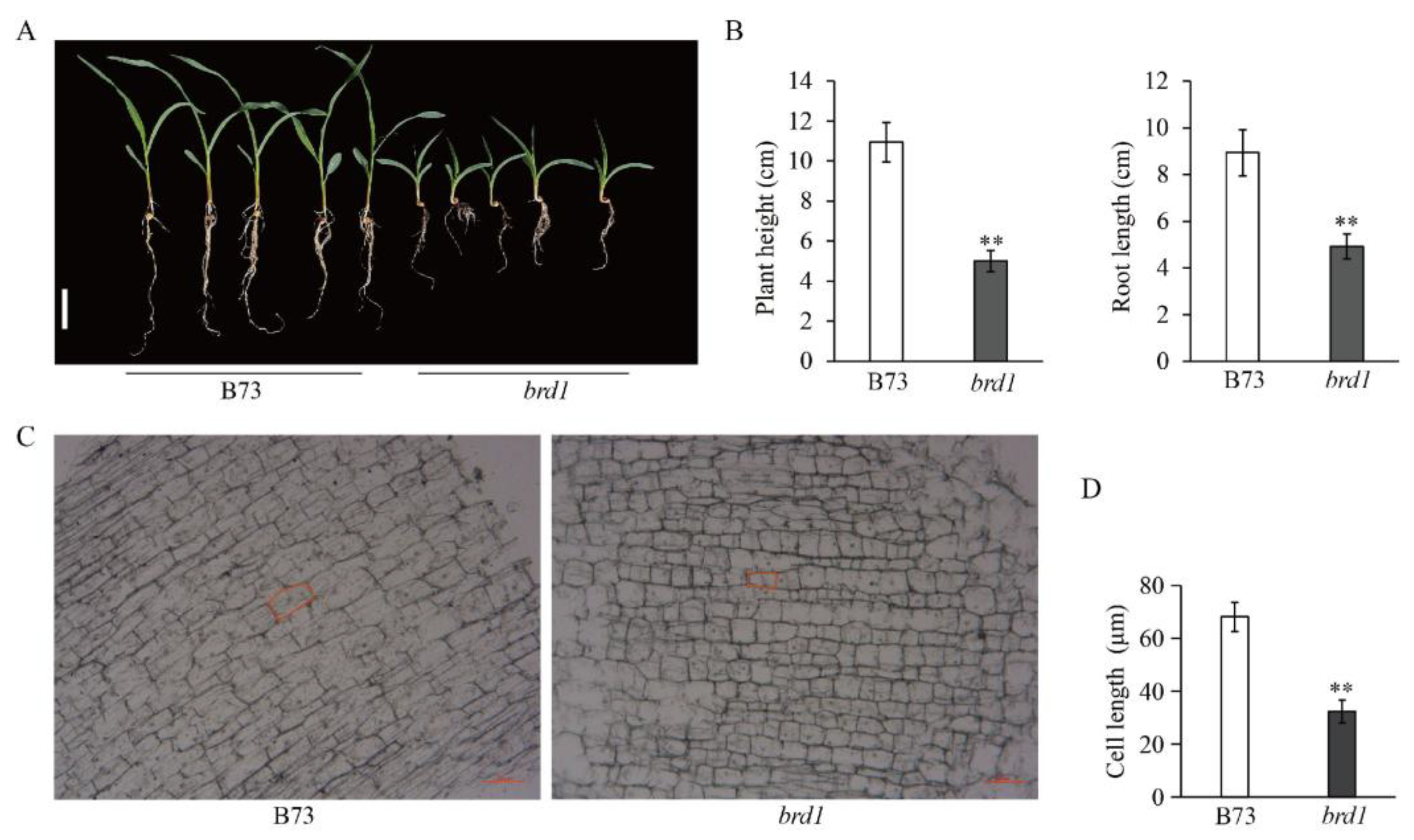
3.3. The Impact of ZmBRD1 on Plant Architecture in Maize
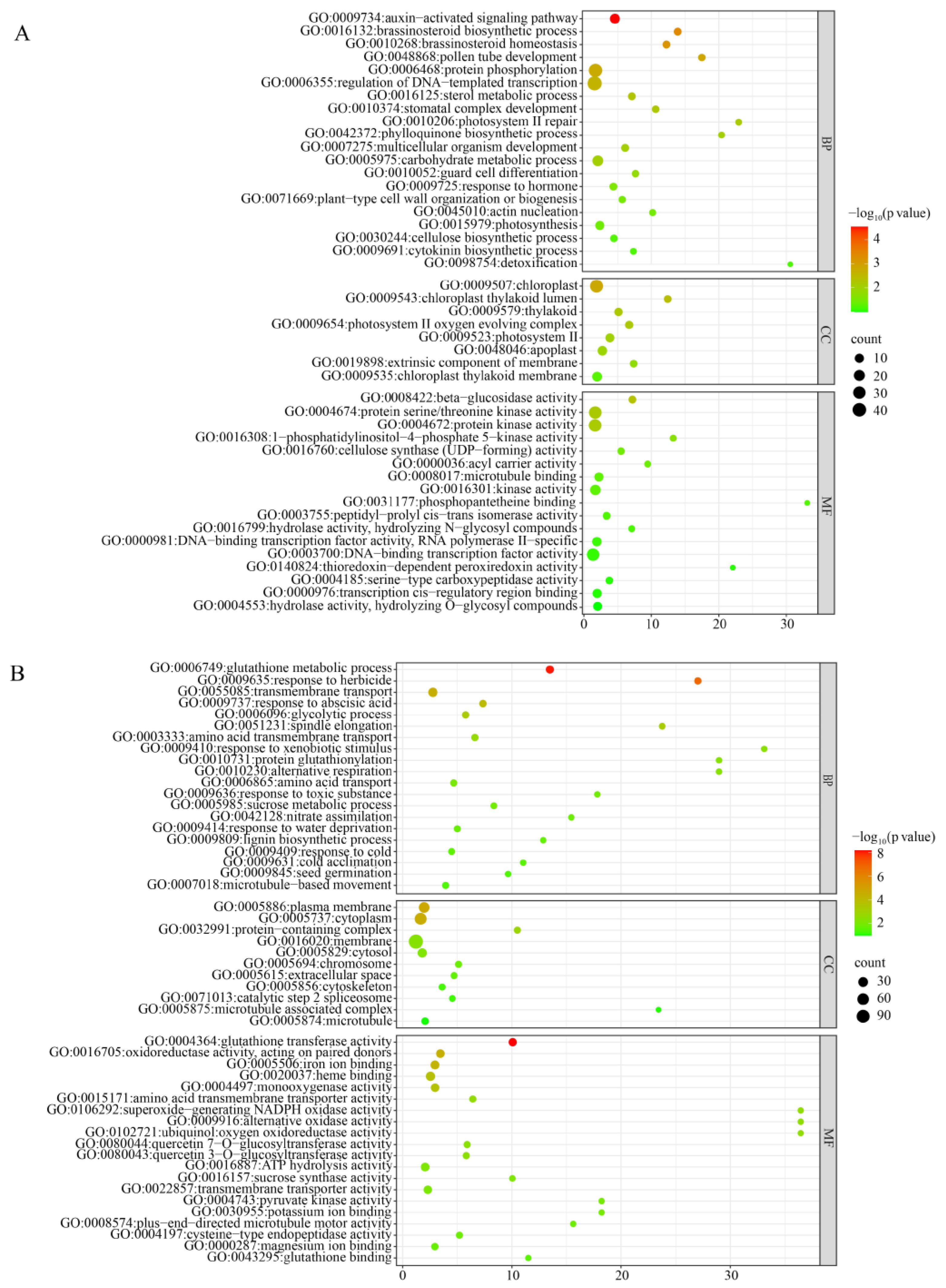
3.4. Transcriptome Analysis of zmbrd1 Mutant Leaf Tissue
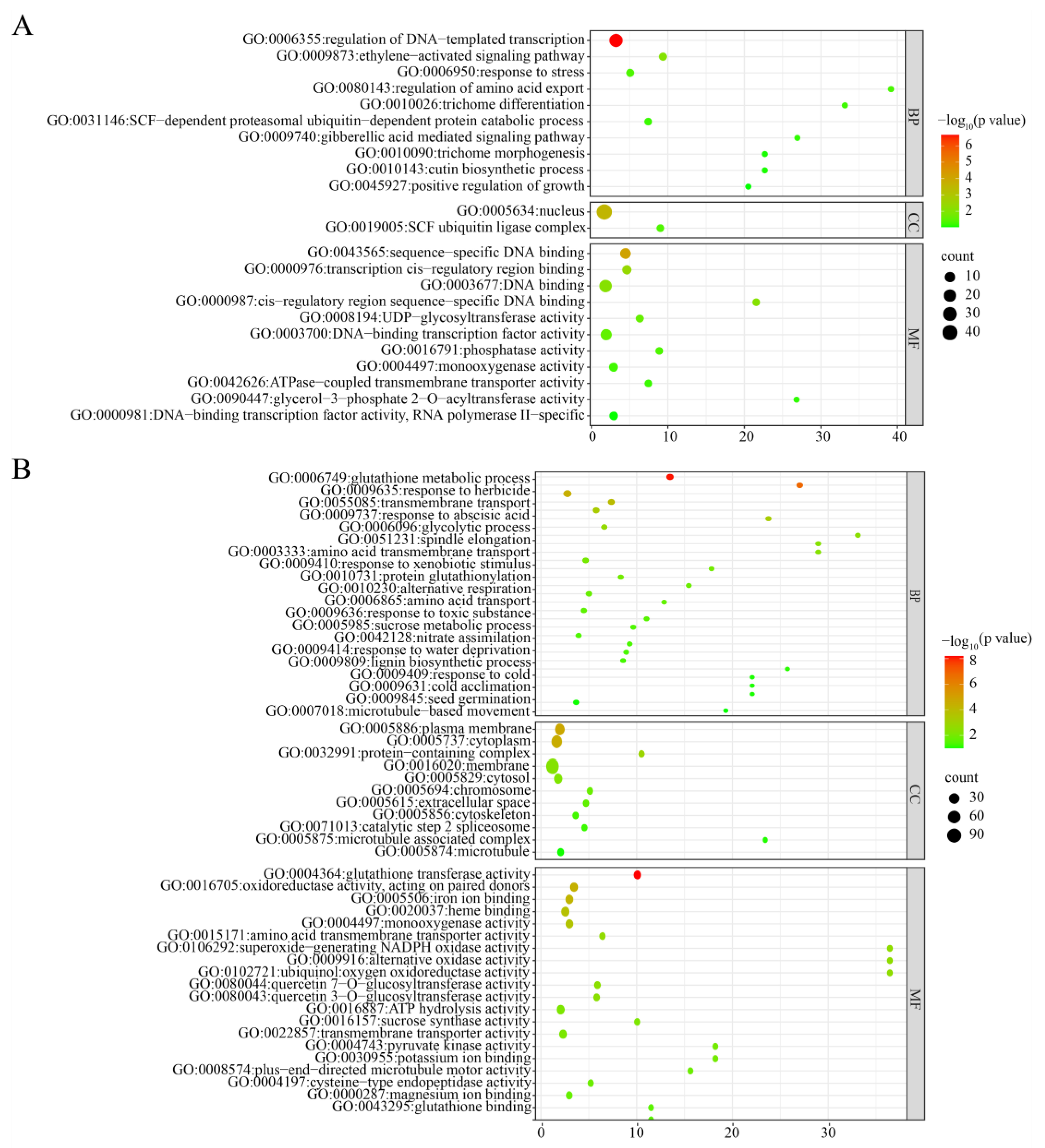
3.5. Transcriptome Analysis of Root Tissue of zmbrd1 Mutant
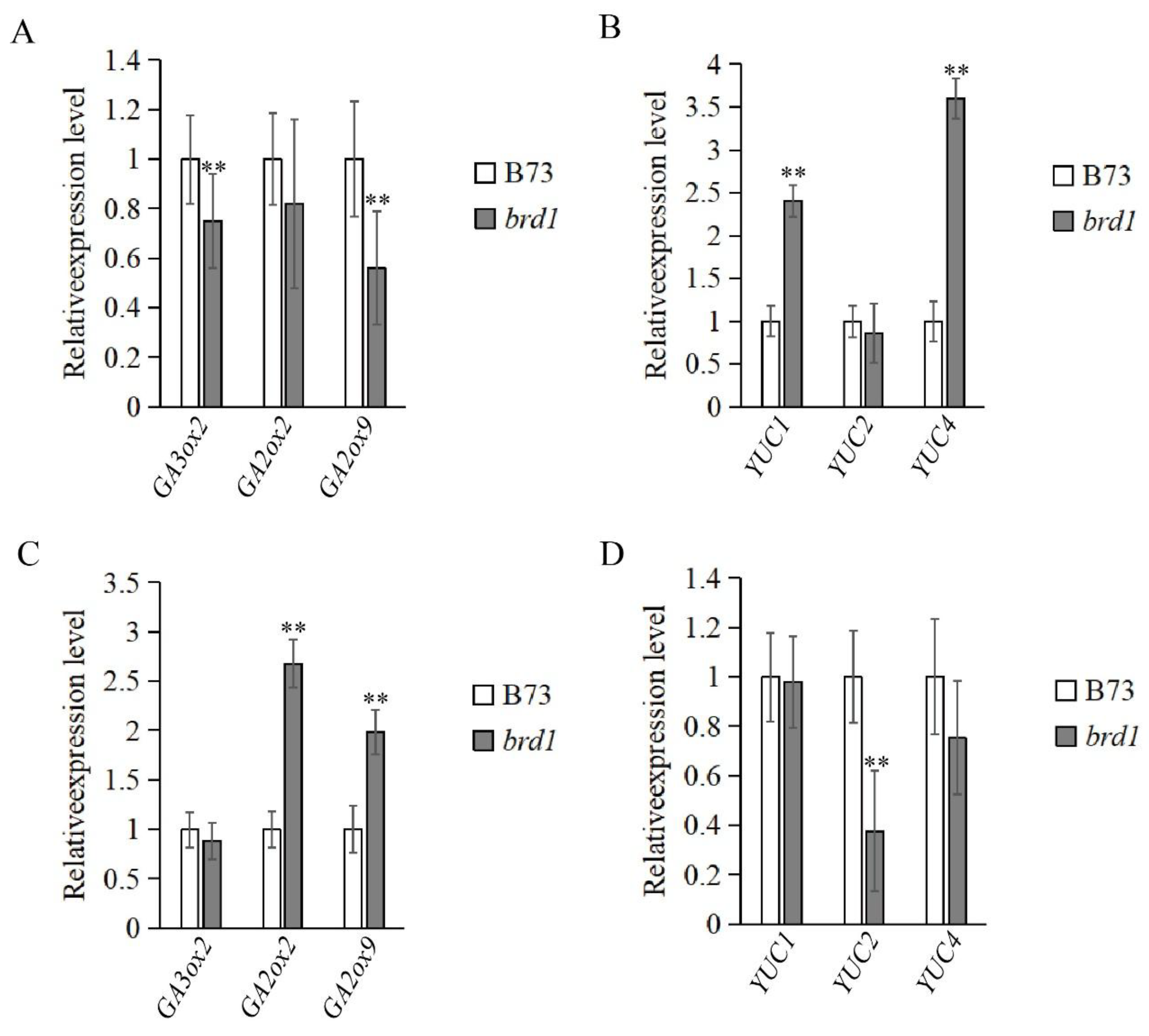
3.6. qRT-PCR Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, X.; Feng, F.; Li, Y.; Xu, S.; Kadambot, H.M.S.; Liao, Y. Maize yield improvements in China: Past trends and future directions. Plant Breed. 2016, 135, 166–176. [Google Scholar] [CrossRef]
- Yin, X.H.; Mc Clure, M.A.; Jaja, N.; Tyler, D.; Hayes, R. In-season prediction of corn yield using plant height under major production systems. Agron. J. 2011, 103, 923. [Google Scholar] [CrossRef]
- Mansfield, B. Survey of plant density tolerance in US maize germplasm. Crop Sci. 2012, 54, 157–173. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Wang, T.; Wang, R.; Wang, X.; Zhang, R.; Xu, R.; Jiao, Y.; Sun, X.; Wang, J.; Song, W.; Zhao, J. Research in maize dwarf genes and dwarf breeding. Biotechnol. Bull. 2023, 39, 43–51. [Google Scholar]
- Chen, Y.; Hou, M.; Liu, L.; Wu, S.; Shen, Y.; Ishiyama, K.; Kobayashi, M.; McCarty, D.R.; Tan, B.C. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014, 166, 2028–2039. [Google Scholar] [CrossRef]
- Lv, H.; Zheng, J.; Wang, T.; Fu, J.; Huai, J.; Min, H.; Zhang, X.; Tian, B.; Shi, Y.; Wang, G. The maize d2003, a novel allele of VP8, is required for maize internode elongation. Plant Mol. Biol. 2014, 84, 243–257. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, W.; Chen, Y.; Deng, D.; Ding, H.; Bian, Y.; Yin, Z.; Zhu, Y.; Zhao, J. Revealing physiological and genetic properties of a dominant maize dwarf Dwarf11(D11) by integrative analysis. Mol. Breed. 2016, 36, 31. [Google Scholar] [CrossRef]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef]
- Winkler, R.G. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell 1995, 7, 1307–1317. [Google Scholar]
- Andersen, J.R.; Schrag, T.; Melchinger, A.E.; Zein, I.; Lübberstedt, T. Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor. Appl. Genet. 2005, 111, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhao, Y.J.; Wu, J.W.; Wei, Y.M.; Ren, R.C.; Zang, J.; Zhang, W.T.; Shen, Q.; Zhang, X.S.; Zhao, X.Y. Overexpression of Zm DWF4 improves major agronomic traits and enhances yield in maize. Mol. Breed. 2020, 40, 71. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, B.; Zhang, H.; Guo, X.; Wang, L.; Qiu, X.; Xie, Q.; Mu, L.; Ma, C.; Zhou, T.; et al. Phenotypic characterization and genetic mapping of the semi-dwarf mutant sdw9 in maize. Theor. Appl. Genet. 2024, 137, 253. [Google Scholar] [CrossRef]
- Thilakarathne, A.S.; Liu, F.; Zou, Z. Plant Signaling Hormones and Transcription Factors: Key Regulators of Plant Responses to Growth, Development, and Stress. Plants 2025, 14, 1070. [Google Scholar] [CrossRef]
- Rao, Y.; Jiao, R.; Ye, H.; Hu, J.; Lu, T.; Wu, X.; Fang, Y.; Li, S.; Lin, H.; Wang, S.; et al. Fine mapping and candidate gene analysis of leaf tip premature senescence and Dwarf Mutant dls-1 in Rice. Plant Growth Regul. 2021, 94, 275–285. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Zhang, D.; Ye, H.; Guo, H.; Johnson, A.; Zhang, M.; Lin, H.; Yin, Y. Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 2014, 77, 59–70. [Google Scholar] [CrossRef]
- Makarevitch, I.; Thompson, A.; Muehlbauer, G.J.; Springer, N.M. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 2012, 7, e30798. [Google Scholar] [CrossRef]
- Kir, G.; Ye, H.; Nelissen, H.; Neelakandan, A.K.; Kusnandar, A.S.; Luo, A.; Inzé, D.; Sylvester, A.W.; Yin, Y.; Becraft, P.W. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 2015, 169, 826–839. [Google Scholar] [CrossRef]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [CrossRef]
- Best, N.B.; Hartwig, T.; Budka, J.; Fujioka, S.; Johal, G.; Schulz, B.; Dilkes, B.P. nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol. 2016, 171, 2633–2647. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, J.; Ren, W.; Yang, Q.; Chai, Z.; Chen, R.; Wang, L.; Zhao, J.; Lang, Z.; Wang, H.; et al. Gene-Indexed Mutations in Maize. Mol. Plant 2018, 11, 496–504. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Lu, X. ZmRLCK1 modulates secondary cell wall deposition in maize. Plant J. 2025, 123, e70313. [Google Scholar] [CrossRef]
- Kenneth, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Miao, L.; Wang, X.; Yu, C.; Ye, C.; Yan, Y.; Wang, H. What factors control plant height? J. Integr. Agric. 2024, 23, 1803–1824. [Google Scholar] [CrossRef]
- Wang, F.; Yoshida, H.; Matsuoka, M. Making the ‘green revolution’ truly green: Improving crop nitrogen use efficiency. Plant Cell Physiol. 2021, 62, 942–947. [Google Scholar] [CrossRef]
- Wang, F.; Yu, Z.; Zhang, M.; Wang, M.; Lu, X.; Liu, X.; Li, Y.; Zhang, X.; Tan, B.C.; Li, C.; et al. ZmTE1 promotes plant height by regulating intercalary meristem formation and internode cell elongation in maize. Plant Biotechnol. J. 2022, 20, 526–537. [Google Scholar] [CrossRef]
- van der Knaap, E.; Kim, J.H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef]
- Sauter, M.; Kende, H. Gibberellin-induced growth and regulation of the cell division cycle in deepwater rice. Planta 1992, 188, 362–368. [Google Scholar] [CrossRef]
- Bolduc, N.; Hake, S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, F.; Tan, L.; Xie, D.; Sun, C. Characterization of a novel high-tillering dwarf 3 mutant in rice. J. Genet. Genom. 2011, 38, 411–418. [Google Scholar] [CrossRef]
- Han, L.; Jiang, C.; Zhang, W.; Wang, H.; Li, K.; Liu, X.; Liu, Z.; Wu, Y.; Huang, C.; Hu, X. Morphological characterization and transcriptome analysis of new Dwarf and Narrow-Leaf (dnl2) mutant in maize. Int. J. Mol. Sci. 2022, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.L.; Li, B.Z.; Zhang, J.L.; Wang, H.L.; Wang, M.; Guo, S.Y.; Wang, P.T.; LI, Z.; Galbraith, D.W.; Li, D.; et al. GA Associated Dwarf 5 encodes an ent-kaurenoic acid oxidase required for maize gibberellin biosynthesis and morphogenesis. Crop J. 2023, 11, 1742–1751. [Google Scholar] [CrossRef]
- Yu, X.; Chen, G.; Tang, B.; Zhang, J.; Zhou, S.; Hu, Z. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2018, 267, 65–73. [Google Scholar] [CrossRef]
- Cheng, J.; Hill, C.; Han, Y.; He, T.; Ye, X.; Shabala, S.; Guo, G.; Zhou, M.; Wang, K.; Li, C. New semi-dwarfing alleles with increased coleoptile length by gene editing of gibberellin 3-oxidase 1 using CRISPR-Cas9 in barley (Hordeum vulgare L.). Plant Biotechnol. J. 2023, 21, 806–818. [Google Scholar] [CrossRef]
- Lan, D.; Cao, L.; Liu, M.; Ma, F.; Yan, P.; Zhang, X.; Hu, J.; Niu, F.; He, S.; Cui, J.; et al. The identification and characterization of a plant height and grain length related gene hfr131 in rice. Front. Plant Sci. 2023, 14, 1152196. [Google Scholar] [CrossRef]
- Wang, H.; Li, W.; Qin, Y.; Pan, Y.; Wang, X.; Weng, Y.; Chen, P.; Li, Y. The Cytochrome P450 gene CsCYP85A1 is a putative candidate for super compact-1 (Scp-1) plant architecture mutation in cucumber (Cucumis sativus L.). Front. Plant Sci. 2017, 8, 266. [Google Scholar] [CrossRef]
- Shafqat, A.; Abbas, S.; Ambreen, M.; Bhatti, A.S.; kausar, H.; Gull, K. Exploring the vital role of phytohormones and plant growth regulators in orchestrating plant immunity. Physiol. Mol. Plant Pathol. 2024, 133, 102359. [Google Scholar] [CrossRef]
- Nambara, E.; Van Wees, S.C.M. Plant hormone functions and interactions in biological systems. Plant J. 2021, 105, 287–289. [Google Scholar] [CrossRef]
- Sharma, S.; Bennett, M.J.; Mehra, P. Roles of hormones in regulating root growth-water interactions. J. Exp. Bot. 2025, 76, 1987–1995. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wang, L.; Lyu, J.; Shi, W.; Yao, L.; Fan, M.; Bai, M.Y. Brassinosteroid signaling and molecular crosstalk with nutrients in plants. J. Genet. Genom. 2023, 50, 541–553. [Google Scholar] [CrossRef]
- Xiong, J.; Bonney, S.; Gonçalves, R.V.; Esposito, D. Brassinosteroids control the inflammation, oxidative stress and cell migration through the control of mitochondrial function on skin regeneration. Life Sci. 2022, 307, 120887. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Ramon, U.; Shohat, H.; Blum, S.; Livne, S.; Mendelson, D.; Weiss, D. Multiple gibberellin receptors contribute to phenotypic stability under changing environments. Plant Cell 2019, 31, 1506–1519. [Google Scholar] [CrossRef]
- Anbarasan, S.; Ramesh, S. The Role of Plant Roots in Nutrient Uptake and Soil Health. Plant Sci. Arch. 2021, 6, 5–8. [Google Scholar] [CrossRef]
- Hansen, A.; Gladala-Kostarz, A.; Hindhaugh, R.; Doonan, J.H.; Bosch, M. Mechanical stimulation in plants: Molecular insights, morphological adaptations, and agricultural applications in monocots. BMC Biol. 2025, 23, 58. [Google Scholar] [CrossRef]
- Li, J.; Zou, Y.; Yang, K.; Zhu, Y.; Zhou, Q.; Shao, L.; Gong, J.; Peng, S.; Peng, G.; Qin, T.; et al. Well-developed root systems and a nitrogen-rich rhizosphere recruit key bacterial taxa to resist disease invasion of field crop. Agric. Ecosyst. Environ. 2025, 378, 109279. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839. [Google Scholar] [CrossRef]
- Lu, X.; Shi, H.; Ou, Y.; Cui, Y.; Chang, J.; Peng, L.; Gou, X.; He, K.; Li, J. RGF1-RGI1, a Peptide-Receptor Complex, Regulates Arabidopsis Root Meristem Development via a MAPK Signaling Cascade. Mol. Plant 2020, 13, 1594–1607. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Bao, Y.; Du, C.; Guo, X.; Lu, X.; Xie, F. Phenotypic Characterization and Transcriptome Analysis of the Dwarf Mutant zmbrd1 in Maize. Genes 2025, 16, 1410. https://doi.org/10.3390/genes16121410
Qin L, Bao Y, Du C, Guo X, Lu X, Xie F. Phenotypic Characterization and Transcriptome Analysis of the Dwarf Mutant zmbrd1 in Maize. Genes. 2025; 16(12):1410. https://doi.org/10.3390/genes16121410
Chicago/Turabian StyleQin, Li, Yu Bao, Chunlei Du, Xiaolong Guo, Xiaoduo Lu, and Fugui Xie. 2025. "Phenotypic Characterization and Transcriptome Analysis of the Dwarf Mutant zmbrd1 in Maize" Genes 16, no. 12: 1410. https://doi.org/10.3390/genes16121410
APA StyleQin, L., Bao, Y., Du, C., Guo, X., Lu, X., & Xie, F. (2025). Phenotypic Characterization and Transcriptome Analysis of the Dwarf Mutant zmbrd1 in Maize. Genes, 16(12), 1410. https://doi.org/10.3390/genes16121410




