Heat Shock Protein 104 (Hsp104) in the Marine Diatom Ditylum brightwellii: Identification and Transcriptional Responses to Environmental Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Extraction
2.3. cDNA Library Construction and Transcriptome Analysis
2.4. cDNA Synthesis, DbHsp104 Cloning, and Gene Expression Analysis
2.5. DbHsp104 Characterizations and Phylogeny
2.6. Thermal and Toxicant Treatments
| Primer Name | Sequence (5′→3′) | Source |
|---|---|---|
| Db-Hsp104-F144 | GGCACTTGTAGCAGGCGC | This study |
| Db-Hsp104-F736 | CGCAAGTTGGCACAGAACG | |
| Db-Hsp104-F1168 | GACAATGCTGTTAGAGAGGTG | |
| Db-Hsp104-F2002 | GAGGAGTATGAAATCGTGGGA | |
| Db-Hsp104-ClpN-R3 | CTCCAGGCTCACCCACTAA | |
| Db-HSP104-AAA-R1 | TGCACCCACCATACGAAG | |
| DbHSP104-N-R1 | GCTGCCTCATCAACCAAG | |
| DbHSP104-N-R2 | CGGAGAATAGAAATGGTC | |
| Db-TUA-F | CGGTATCCAGACTGGTAACGGC | [45] |
| Db-TUA-R | GAGGCACATGCTTTCCGTTTC | |
| B25 | GACTCTAGACGACATCGA | Universal primer |
| B26 | GACTCTAGACGACATCGA(T)18 |
2.7. DbHsp104 Expression Analysis
2.8. Statistical Analysis
3. Results
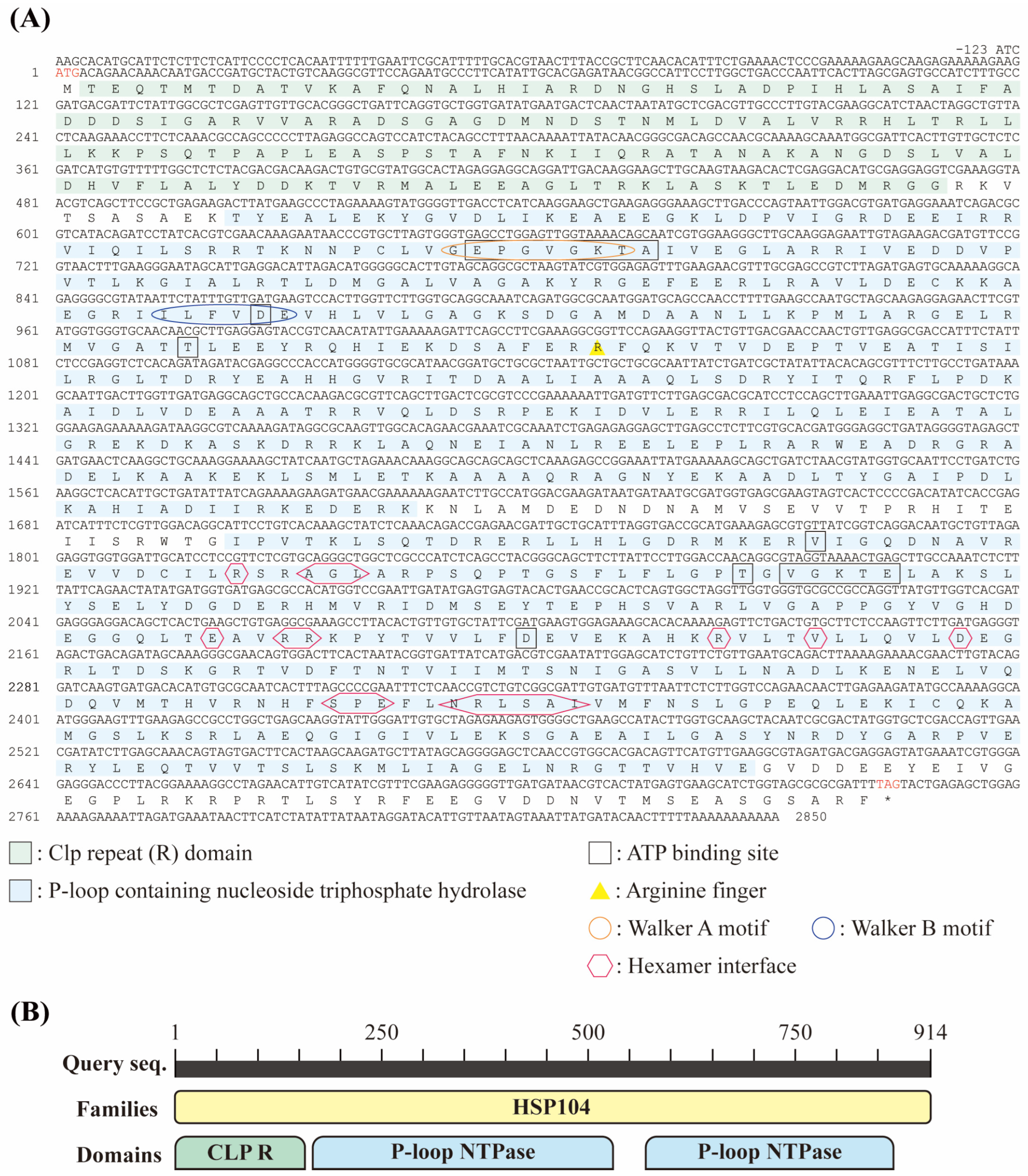
3.1. Sequence Identification of DbHsp104
3.2. Molecular Characterization of DbHsp104
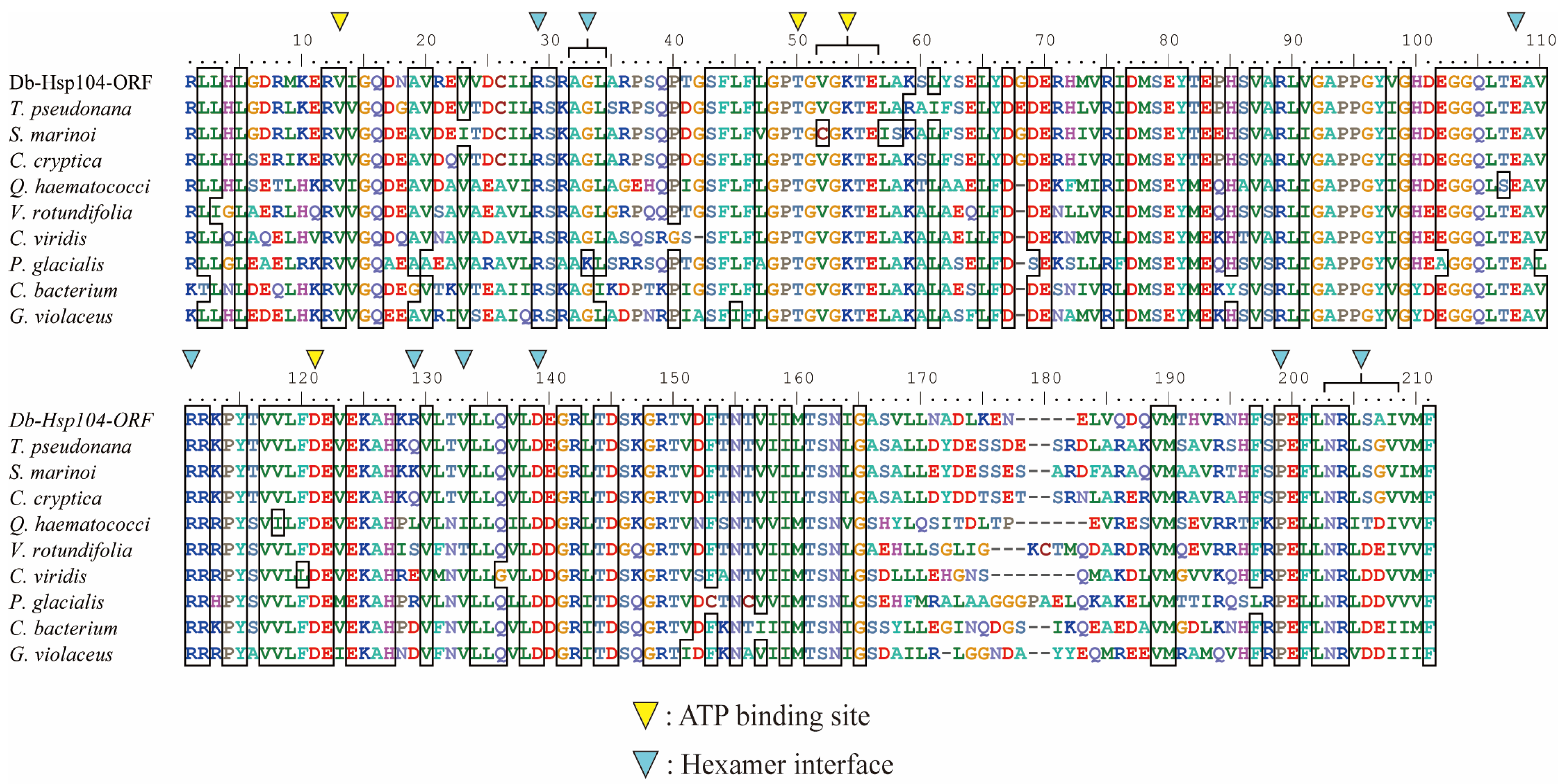
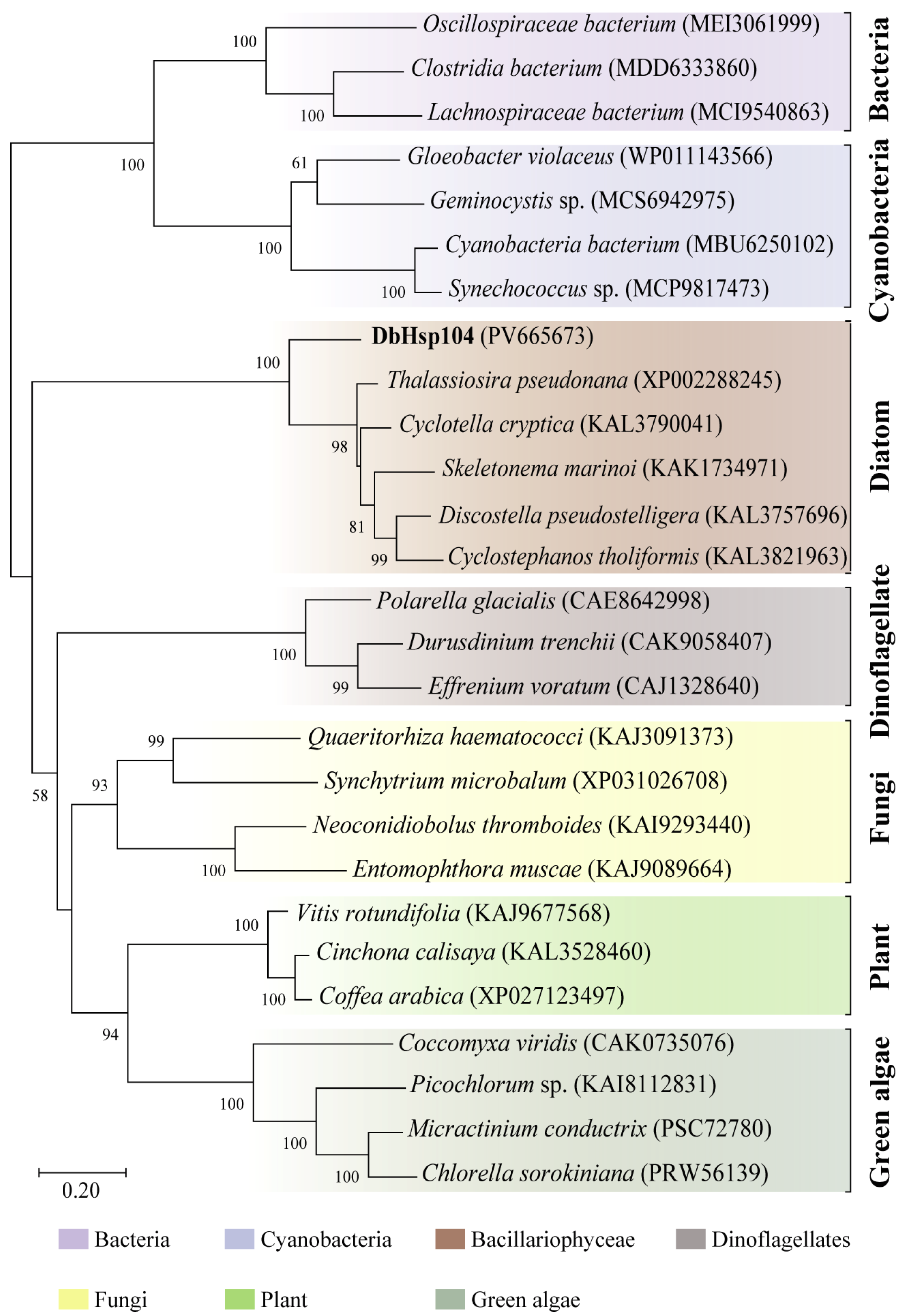
3.3. Phylogenetic Relevance and Motif Characteristic of DbHsp104
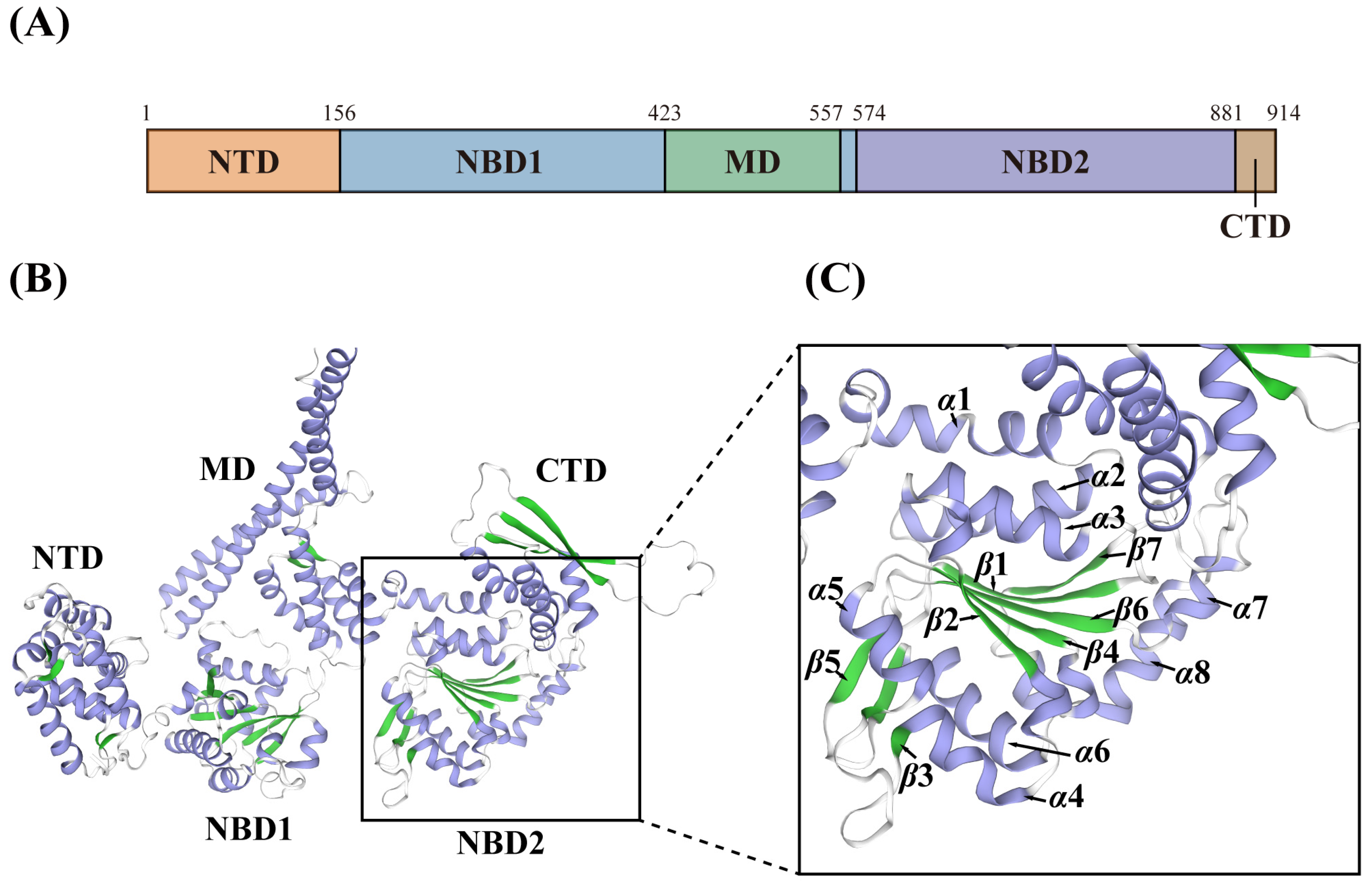
3.4. Structural Features of DbHsp104
3.5. Transcriptional Changes in Dbhsp104 Under Temperature Stress
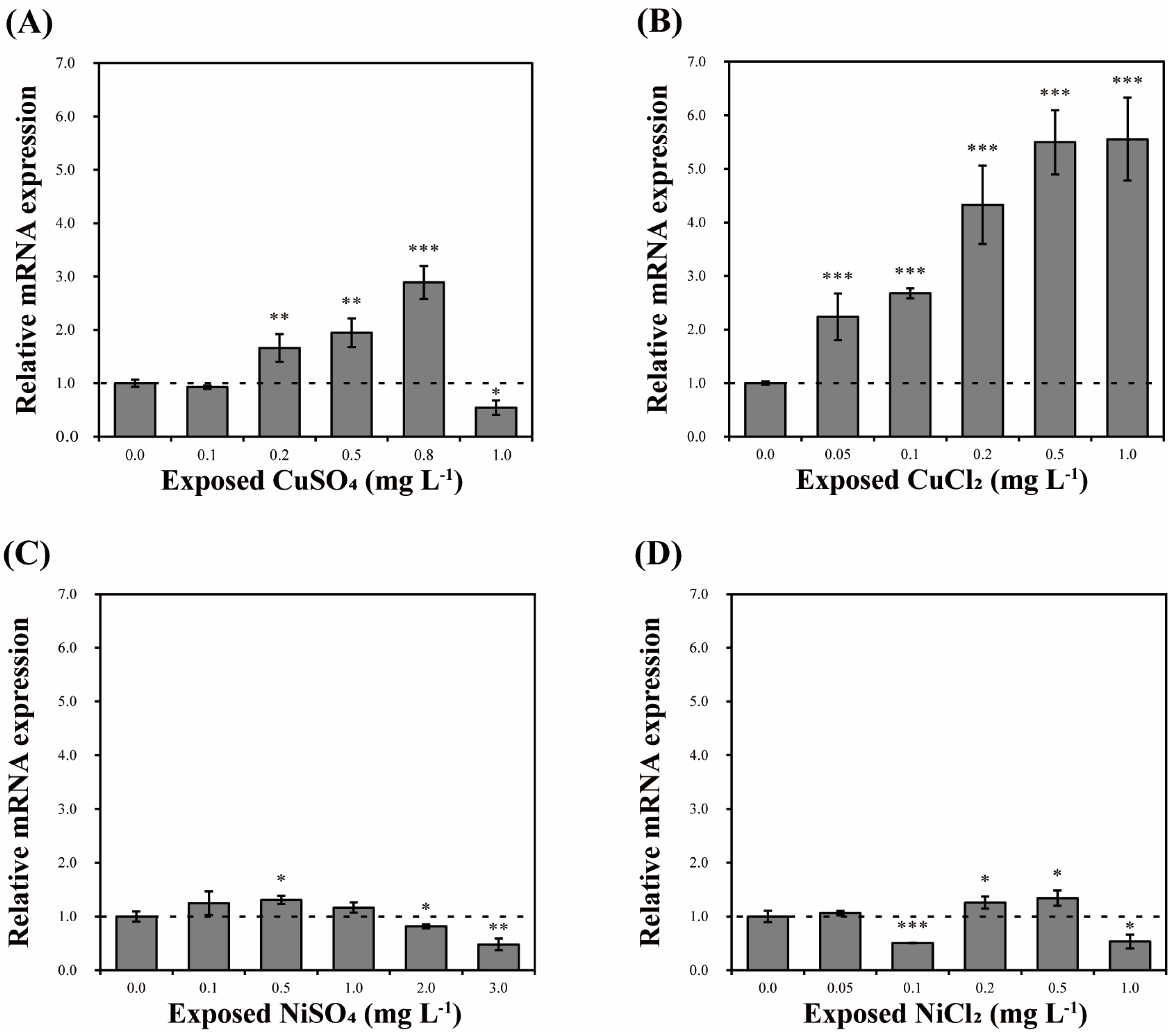
3.6. Effect of Pollutants on DbHsp104 Expression
4. Discussion
4.1. Molecular Features of Hsp and Hsp104
4.2. Structural Features of Eukaryotic DbHsp104
4.3. Effect of Thermal Stress on DbHsp104 Gene Expression
4.4. Effects of Acute Toxicity on Hsp104 Transcription
4.5. Minor Effects of EDCs on Hsp104 System
4.6. DbHsp104 as a Potential Biomarker
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HSP | heat shock protein |
| CuSO4 | copper sulfate |
| CuCl2 | copper chloride |
| NiSO4 | nickel sulfate |
| BPA | bisphenol A |
| PCB | polychlorinated biphenyl |
| EDS | endosulfan |
| EDCs | endocrine-disrupting chemicals |
| P-loop NTPase | P-loop containing nucleoside triphosphate hydrolase |
| NTD | N-terminal domain |
| NBD | nucleotide-binding domains |
| CTD | C-terminal domain |
| CLP | R Clp repeat domain |
| ML | Maximum likelihood |
| ANOVA | One-way analysis of variance |
References
- Hildebrand, M. Diatoms, biomineralization processes, and genomics. Chem. Rev. 2008, 108, 4855–4874. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V. The life of diatoms in the world’s oceans. Nature 2009, 459, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Brzezinski, M.A.; Dickson, M.L.; Nelson, D.M.; Sambrotto, R. Ratios of Si, C, and N uptake by microplankton in the Southern Ocean. Deep Sea Res. Part II Top. Stud. Oceanogr. 2003, 50, 619–633. [Google Scholar] [CrossRef]
- Sommers, M.D.; Naselli-Flores, L.; Padisák, J. Ecosystem services provided by freshwater and marine diatoms: Silica cell wall and high reproduction rate as key traits. Hydrobiologia 2022, 849, 123–140. [Google Scholar] [CrossRef]
- Fritz, C.; Masouras, A.; Pinheiro, P. Diatoms as bioindicators for health assessments of ephemeral freshwater ecosystems: A comprehensive review. Sci. Total Environ. 2024, 904, 166766. [Google Scholar] [CrossRef]
- Stauber, J.L.; Davies, C.M. Use and limitations of microbial bioassays for assessing copper bioavailability in the aquatic environment. Environ. Rev. 2000, 8, 255–301. [Google Scholar] [CrossRef]
- Gerringa, L.J.A.; Rijstenbil, J.W.; Poortvliet, T.C.W.; Van Drie, J.; Schot, M.C. Speciation of copper and responses of the marine diatom Ditylum brightwellii upon increasing copper concentrations. Aquat. Toxicol. 1995, 31, 77–90. [Google Scholar] [CrossRef]
- Costa, A.P.T.; Schneck, F. Diatoms as indicators in running waters: Trends of studies on biological assessment and monitoring. Environ. Monit. Assess. 2022, 194, 695. [Google Scholar] [CrossRef]
- Blanco, S. What do diatom indices indicate? Modeling the specific pollution sensitivity index. Environ. Sci. Pollut. Res. 2024, 31, 29449–29459. [Google Scholar] [CrossRef]
- Moreno-Garrido, I.; Lubián, L.M.; Soares, A.M. Influence of cellular density on determination of EC50 in microalgal growth inhibition tests. Ecotoxicol. Environ. Saf. 2000, 47, 112–116. [Google Scholar] [CrossRef]
- Carvalho, R.N.; Bopp, S.K.; Lettieri, T. Transcriptomics responses in marine diatom Thalassiosira pseudonana exposed to the polycyclic aromatic hydrocarbon benzo[a]pyrene. PLoS ONE 2011, 6, e26985. [Google Scholar] [CrossRef]
- Hirst, H.; Jüttner, I.; Ormerod, S.J. Comparing the responses of diatoms and macro-invertebrates to metals in upland streams of Wales and Cornwall. Freshw. Biol. 2002, 47, 1752–1765. [Google Scholar] [CrossRef]
- Blanco, S.; Bécares, E. Are biotic indices sensitive to river toxicants? A comparison of metrics based on diatoms and macro-invertebrates. Chemosphere 2010, 79, 18–25. [Google Scholar] [CrossRef]
- Morin, S.; Cordonier, A.; Lavoie, I.; Arini, A.; Blanco, S.; Duong, T.T.; Sabater, S. Consistency in diatom response to metal-contaminated environments. In Emerging and Priority Pollutants in Rivers: Bringing Science into River Management Plans; Springer: Berlin/Heidelberg, Germany, 2012; pp. 117–146. [Google Scholar]
- Kim Tiam, S.; Lavoie, I.; Doose, C.; Hamilton, P.B.; Fortin, C. Morphological, physiological and molecular responses of Nitzschia palea under cadmium stress. Ecotoxicology 2018, 27, 675–688. [Google Scholar] [CrossRef]
- Rijstenbil, J.W.; Derksen, J.W.M.; Gerringa, L.J.A.; Poortvliet, T.C.W.; Sandee, A.; Van den Berg, M.; Wijnholds, J.A. Oxidative stress induced by copper: Defense and damage in the marine planktonic diatom Ditylum brightwellii, grown in continuous cultures with high and low zinc levels. Mar. Biol. 1994, 119, 583–590. [Google Scholar] [CrossRef]
- Horvatić, J.; Peršić, V. The effect of Ni2+, Co2+, Zn2+, Cd2+ and Hg2+ on the growth rate of marine diatom Phaeodactylum tricornutum Bohlin: Microplate growth inhibition test. Bull. Environ. Contam. Toxicol. 2007, 79, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Liping, W.; Zheng, B. Toxic effects of fluoranthene and copper on marine diatom Phaeodactylum tricornutum. J. Environ. Sci. 2008, 20, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Janknegt, P.J.; Van de Poll, W.H.; Visser, R.J.; Rijstenbil, J.W.; Buma, A.G. Oxidative stress responses in the marine Antarctic diatom Chaetoceros brevis (Bacillariophyceae) during photoacclimation. J. Phycol. 2008, 44, 957–966. [Google Scholar] [CrossRef]
- Dulle, J.E.; Stein, K.C.; True, H.L. Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS ONE 2014, 9, e87521. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Padmini, E. Physiological adaptations of stressed fish to polluted environments: Role of heat shock proteins. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2010; Volume 206, pp. 1–27. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sharma, A.; Mishra, M.; Mishra, R.K.; Chowdhuri, D.K. Heat shock proteins in toxicology: How close and how far? Life Sci. 2010, 86, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, Y.; Taulien, J.; Borkovich, K.A.; Lindquist, S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992, 11, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Kim, R.S.; Lee, S.B.; Lee, S.; Tsai, F.T. Deciphering the mechanism and function of Hsp100 unfoldases from protein structure. Biochem. Soc. Trans. 2022, 50, 1725–1736. [Google Scholar] [CrossRef]
- Lee, J.; Sung, N.; Mercado, J.M.; Hryc, C.F.; Chang, C.; Lee, S.; Tsai, F.T. Overlapping and specific functions of the Hsp104 N domain define its role in protein disaggregation. Sci. Rep. 2017, 7, 11184. [Google Scholar] [CrossRef]
- Schirmer, E.C.; Glover, J.R.; Singer, M.A.; Lindquist, S. Hsp104/ClpB proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 1996, 21, 289–296. [Google Scholar] [CrossRef]
- Sweeny, E.A.; Shorter, J. Prion proteostasis: Hsp104 meets its supporting cast. Prion 2008, 2, 135–140. [Google Scholar] [CrossRef]
- DeSantis, M.E.; Shorter, J. The elusive middle domain of Hsp104 and ClpB: Location and function. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 29–39. [Google Scholar] [CrossRef]
- Shorter, J.; Southworth, D.R. Spiraling in control: Structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb. Perspect. Biol. 2019, 11, a034033. [Google Scholar] [CrossRef]
- Barnett, M.E.; Zolkiewska, A.; Zolkiewski, M. Structure and activity of ClpB from Escherichia coli: Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 2000, 275, 37565–37571. [Google Scholar] [CrossRef]
- Cashikar, A.G.; Schirmer, E.C.; Hattendorf, D.A.; Glover, J.R.; Ramakrishnan, M.S.; Ware, D.M.; Lindquist, S.L. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 2002, 9, 751–760. [Google Scholar] [CrossRef]
- Doyle, S.M.; Wickner, S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem. Sci. 2009, 34, 40–48. [Google Scholar] [CrossRef]
- Mackay, R.G.; Helsen, C.W.; Tkach, J.M.; Glover, J.R. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry 2008, 47, 1918–1927. [Google Scholar] [CrossRef]
- Banwait, J.K.; Lucius, A.L. Quantitative insights into processivity of an Hsp100 protein disaggregase on folded protein substrates. Biophys. J. 2024, 124, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Rioflorido, I.; Hong, S.W.; Larkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J. 2007, 49, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Queraltó, C.; Álvarez, R.; Ortega, C.; Díaz-Yáñez, F.; Paredes-Sabja, D.; Gil, F. Role and regulation of Clp proteases: A target against gram-positive bacteria. Bacteria 2023, 2, 21–36. [Google Scholar] [CrossRef]
- Canterford, G.S.; Canterford, D.R. Toxicity of heavy metals to the marine diatom Ditylum brightwellii (West) Grunow: Correlation between toxicity and metal speciation. J. Mar. Biol. Assoc. UK 1980, 60, 227–242. [Google Scholar] [CrossRef]
- Ryther, J.H.; Guillard, R.R.L. Studies of marine planktonic diatoms: II. Use of Cyclotella nana Hustedt for assays of vitamin B12 in sea water. Can. J. Microbiol. 1962, 8, 437–445. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Masindi, V.; Muedi, K.L. Environmental contamination by heavy metals. Heavy Met. 2018, 10, 115–133. [Google Scholar]
- Lee, M.A.; Guo, R.; Ki, J.S. Different transcriptional responses of heat shock protein 20 in the marine diatom Ditylum brightwellii exposed to metals and endocrine-disrupting chemicals. Environ. Toxicol. 2014, 29, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Ki, J.S. Evaluation and validation of internal control genes for studying gene expression in the dinoflagellate Prorocentrum minimum using real-time PCR. Eur. J. Protistol. 2012, 48, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Sanders, B.M. Stress proteins in aquatic organisms: An environmental perspective. Crit. Rev. Toxicol. 1993, 23, 49–75. [Google Scholar] [CrossRef]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef]
- Moreira-de-Sousa, C.; de Souza, R.B.; Fontanetti, C.S. HSP70 as a biomarker: An excellent tool in environmental contamination analysis—A review. Water Air Soil Pollut. 2018, 229, 264. [Google Scholar] [CrossRef]
- Kirstein, J.; Molière, N.; Dougan, D.A.; Turgay, K. Adapting the machine: Adaptor proteins for Hsp104/ClpB and AAA+ proteases. Nat. Rev. Microbiol. 2009, 7, 589–599. [Google Scholar] [CrossRef]
- Sanchez, Y.; Lindquist, S.L. HSP104 required for induced thermotolerance. Science 1990, 248, 1112–1115. [Google Scholar] [CrossRef]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Parsell, D.A.; Kowal, A.S.; Singer, M.A.; Lindquist, S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 1994, 372, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Lum, R.; Tkach, J.M.; Vierling, E.; Glover, J.R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004, 279, 29139–29146. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.D.; Ho, S.; Glover, J.R. Saccharomyces cerevisiae Hsp104 enhances the chaperone capacity of human cells and inhibits heat stress-induced proapoptotic signaling. Biochemistry 2004, 43, 8107–8115. [Google Scholar] [CrossRef] [PubMed]
- Kaimal, J.M.; Kandasamy, G.; Gasser, F.; Andréasson, C. Coordinated Hsp110 and Hsp104 activities power protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2017, 37, e00027-17. [Google Scholar] [CrossRef]
- Oguchi, Y.; Kummer, E.; Seyffer, F.; Berynskyy, M.; Anstett, B.; Zahn, R.; Bukau, B. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 2012, 19, 1338–1346. [Google Scholar] [CrossRef]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef]
- Parsell, D.A.; Kowal, A.S.; Lindquist, S. Saccharomyces cerevisiae Hsp104 protein: Purification and characterization of ATP-induced structural changes. J. Biol. Chem. 1994, 269, 4480–4487. [Google Scholar] [CrossRef]
- Parsell, D.A.; Sanchez, Y.; Stitzel, J.D.; Lindquist, S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 1991, 353, 270–273. [Google Scholar] [CrossRef]
- Sweeny, E.A.; Jackrel, M.E.; Go, M.S.; Sochor, M.A.; Razzo, B.M.; DeSantis, M.E.; Shorter, J. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 2015, 57, 836–849. [Google Scholar] [CrossRef]
- Gates, S.N.; Yokom, A.L.; Lin, J.; Jackrel, M.E.; Rizo, A.N.; Kendsersky, N.M.; Southworth, D.R. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 2017, 357, 273–279. [Google Scholar] [CrossRef]
- Lee, S.; Sowa, M.E.; Watanabe, Y.H.; Sigler, P.B.; Chiu, W.; Yoshida, M.; Tsai, F.T. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell 2003, 115, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Bösl, B.; Grimminger, V.; Walter, S. The molecular chaperone Hsp104—A molecular machine for protein disaggregation. J. Struct. Biol. 2006, 156, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.J.; Clarke, A.K. The Escherichia coli heat shock protein ClpB restores acquired thermotolerance to a cyanobacterial clpB deletion mutant. Cell Stress Chaperones 2000, 5, 255. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Bröms, J.E.; Kumar, R.; Sjöstedt, A. The role of ClpB in bacterial stress responses and virulence. Front. Mol. Biosci. 2021, 8, 668910. [Google Scholar] [CrossRef]
- Eriksson, M.J.; Schelin, J.; Miskiewicz, E.; Clarke, A.K. Novel form of ClpB/HSP100 protein in the cyanobacterium Synechococcus. J. Bacteriol. 2001, 183, 7392–7396. [Google Scholar] [CrossRef]
- Kreis, E.; Niemeyer, J.; Merz, M.; Scheuring, D.; Schroda, M. CLPB3 is required for the removal of chloroplast protein aggregates and thermotolerance in Chlamydomonas. J. Exp. Bot. 2023, 74, 3714–3728. [Google Scholar] [CrossRef]
- Santos, T.F.; Pereira, H.; Schüler, L.; Maia, I.B.; Jacinto, R.; Bombo, G.; Varela, J. Enhancement of heat tolerance by salt stress in Tetraselmis striata CTP4: Impacts on HSP gene expression, pigments, and proximal composition. J. Appl. Phycol. 2025, 37, 287–301. [Google Scholar] [CrossRef]
- Sielaff, B.; Tsai, F.T. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 2010, 402, 30–37. [Google Scholar] [CrossRef]
- Oakley, C.A.; Durand, E.; Wilkinson, S.P.; Peng, L.; Weis, V.M.; Grossman, A.R.; Davy, S.K. Thermal shock induces host proteostasis disruption and endoplasmic reticulum stress in the model symbiotic cnidarian Aiptasia. J. Proteome Res. 2017, 16, 2121–2134. [Google Scholar] [CrossRef]
- Xing, C.; Li, J.; Yuan, H.; Yang, J. Physiological and transcription level responses of microalgae Auxenochlorella protothecoides to cold and heat induced oxidative stress. Environ. Res. 2022, 211, 113023. [Google Scholar] [CrossRef]
- Tian, X.; Lin, X.; Xie, Q.; Liu, J.; Luo, L. Effects of temperature and light on microalgal growth and nutrient removal in turtle aquaculture wastewater. Biology 2024, 13, 901. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Ebenezer, V.; Ki, J.S. Transcriptional responses of heat shock protein 70 (Hsp70) to thermal, bisphenol A, and copper stresses in the dinoflagellate Prorocentrum minimum. Chemosphere 2012, 89, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.T.; Yi, A.X.; Ip, J.C.; Mak, S.S.; Leung, K.M. Photosynthetic and transcriptional responses of the marine diatom Thalassiosira pseudonana to the combined effect of temperature stress and copper exposure. Mar. Pollut. Bull. 2017, 124, 938–945. [Google Scholar] [CrossRef]
- Sathasivam, R.; Ki, J.S. Heat shock protein genes in the green alga Tetraselmis suecica and their role against redox and non-redox active metals. Eur. J. Protistol. 2019, 69, 37–51. [Google Scholar] [CrossRef]
- Abassi, S.; Wang, H.; Ki, J.S. Molecular cloning of heat shock protein 70 and HOP from the freshwater green alga Closterium ehrenbergii and their responses to stress. Cell Stress Chaperones 2020, 25, 1117–1123. [Google Scholar] [CrossRef]
- Halladay, J.T.; Craig, E.A. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol. Cell. Biol. 1995, 15, 4890–4897. [Google Scholar] [CrossRef]
- Seppä, L.; Hänninen, A.L.; Makarow, M. Upregulation of the Hsp104 chaperone at physiological temperature during recovery from thermal insult. Mol. Microbiol. 2004, 52, 217–225. [Google Scholar] [CrossRef]
- Yamamoto, N.; Maeda, Y.; Ikeda, A.; Sakurai, H. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot. Cell 2008, 7, 783–790. [Google Scholar] [CrossRef]
- Kitagawa, M.; Wada, C.; Yoshioka, S.; Yura, T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32). J. Bacteriol. 1991, 173, 4247–4253. [Google Scholar] [CrossRef]
- Ingmer, H.; Vogensen, F.K.; Hammer, K.; Kilstrup, M. Disruption and analysis of the clpB, clpC, and clpE genes in Lactococcus lactis: ClpE, a new Clp family in gram-positive bacteria. J. Bacteriol. 1999, 181, 2075–2083. [Google Scholar] [CrossRef]
- de Oliveira, N.E.M.; Abranches, J.; Gaca, A.O.; Laport, M.S.; Damaso, C.R.; Bastos, M.D.C.D.F.; Giambiagi-deMarval, M. clpB, a class III heat-shock gene regulated by CtsR, is involved in thermotolerance and virulence of Enterococcus faecalis. Microbiology 2011, 157, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Miot, M.; Reidy, M.; Doyle, S.M.; Hoskins, J.R.; Johnston, D.M.; Genest, O.; Wickner, S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA 2011, 108, 6915–6920. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lee, M.A.; Ki, J.S. Different transcriptional responses of heat shock protein 70/90 in the marine diatom Ditylum brightwellii exposed to metal compounds and endocrine-disrupting chemicals. Chemosphere 2013, 92, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hu, Z.; Shang, L.; Chai, Z.; Tang, Y.Z. Transcriptional responses of the heat shock protein 20 (Hsp20) and 40 (Hsp40) genes to temperature stress and alteration of life cycle stages in the harmful alga Scrippsiella trochoidea (Dinophyceae). Biology 2020, 9, 408. [Google Scholar] [CrossRef]
- Blaby, I.K.; Blaby-Haas, C.E.; Pérez-Pérez, M.E.; Schmollinger, S.; Fitz-Gibbon, S.; Lemaire, S.D.; Merchant, S.S. Genome-wide analysis on Chlamydomonas reinhardtii reveals the impact of hydrogen peroxide on protein stress responses and overlap with other stress transcriptomes. Plant J. 2015, 84, 974–988. [Google Scholar] [CrossRef]
- Yu, A.; Li, P.; Tang, T.; Wang, J.; Chen, Y.; Liu, L. Roles of Hsp70s in stress responses of microorganisms, plants, and animals. Biomed. Res. Int. 2015, 2015, 510319. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Ilahi, I. Environmental chemistry and ecotoxicology of hazardous heavy metals: Environmental persistence, toxicity, and bioaccumulation. J. Chem. 2019, 2019, 6730305. [Google Scholar] [CrossRef]
- Fan, J.J.; Wang, S.; Tang, J.P.; Zhao, J.L.; Wang, L.; Wang, J.X.; Yang, Y. Bioaccumulation of endocrine disrupting compounds in fish with different feeding habits along the largest subtropical river, China. Environ. Pollut. 2019, 247, 999–1008. [Google Scholar] [CrossRef]
- Hong, Y.J.; Liao, W.; Yan, Z.F.; Bai, Y.C.; Feng, C.L.; Xu, Z.X.; Xu, D.Y. Progress in the research of the toxicity effect mechanisms of heavy metals on freshwater organisms and their water quality criteria in China. J. Chem. 2020, 2020, 9010348. [Google Scholar] [CrossRef]
- Santhosh, K.; Kamala, K.; Ramasamy, P.; Musthafa, M.S.; Almujri, S.S.; Asdaq, S.M.B.; Sivaperumal, P. Unveiling the silent threat: Heavy metal toxicity devastating impact on aquatic organisms and DNA damage. Mar. Pollut. Bull. 2024, 200, 116139. [Google Scholar] [CrossRef]
- Devi, A.; De Silva, Y.S.K.; Tyagi, L.; Aaryashree. The individual and combined effects of microplastics and heavy metals on marine organisms. Microplastics 2025, 4, 38. [Google Scholar] [CrossRef]
- Yruela, I. Copper in plants: Acquisition, transport and interactions. Funct. Plant Biol. 2009, 36, 409–430. [Google Scholar] [CrossRef]
- Kumar, K.S.; Dahms, H.-U.; Lee, J.S.; Kim, H.C.; Lee, W.C.; Shin, K.H. Algal photosynthetic responses to toxic metals and herbicides assessed by chlorophyll a fluorescence. Ecotoxicol. Environ. Saf. 2014, 104, 51–71. [Google Scholar] [CrossRef]
- Chen, G.; Li, J.; Han, H.; Du, R.; Wang, X. Physiological and molecular mechanisms of plant responses to copper stress. Int. J. Mol. Sci. 2022, 23, 12950. [Google Scholar] [CrossRef]
- Wang, H.; Sathasivam, R.; Ki, J.S. Physiological effects of copper on the freshwater alga Closterium ehrenbergii Meneghini (Conjugatophyceae) and its potential use in toxicity assessments. Algae 2017, 32, 131–137. [Google Scholar] [CrossRef]
- Abassi, S.; Wang, H.; Ponmani, T.; Ki, J.S. Small heat shock protein genes of the green algae Closterium ehrenbergii: Cloning and differential expression under heat and heavy metal stresses. Environ. Toxicol. 2019, 34, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ki, J.S. Molecular identification, differential expression and protective roles of iron/manganese superoxide dismutases in the green algae Closterium ehrenbergii against metal stress. Eur. J. Protistol. 2020, 74, 125689. [Google Scholar] [CrossRef] [PubMed]
- Ebenezer, V.; Lim, W.A.; Ki, J.S. Effects of the algicides CuSO4 and NaOCl on various physiological parameters in the harmful dinoflagellate Cochlodinium polykrikoides. J. Appl. Phycol. 2014, 26, 2357–2365. [Google Scholar] [CrossRef]
- Guo, R.; Wang, H.; Suh, Y.S.; Ki, J.S. Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate Cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genom. 2016, 17, 29. [Google Scholar] [CrossRef]
- Guo, R.; Ki, J.S. Differential transcription of heat shock protein 90 (HSP90) in the dinoflagellate Prorocentrum minimum by copper and endocrine-disrupting chemicals. Ecotoxicology 2012, 21, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Wang, H.; Abassi, S.; Ki, J.S. The herbicide alachlor severely affects photosystem function and photosynthetic gene expression in the marine dinoflagellate Prorocentrum minimum. J. Environ. Sci. Health Part B 2020, 55, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ki, J.S. Identification of a metacaspase gene in the bloom-forming dinoflagellate Prorocentrum minimum and its putative function involved in programmed cell death. Curr. Microbiol. 2021, 78, 3577–3585. [Google Scholar] [CrossRef] [PubMed]
- Bui, Q.T.N.; Kim, H.S.; Ki, J.S. Two chloroplast superoxide dismutase genes (MnSOD and CuZnSOD) in the freshwater dinoflagellate Palatinus apiculatus and their high-level gene expression in response to algicidal agents. J. Appl. Phycol. 2025, 14, 1–13. [Google Scholar] [CrossRef]
- Florence, T.M.; Stauber, J.L. Toxicity of copper complexes to the marine diatom Nitzschia closterium. Aquat. Toxicol. 1986, 8, 11–26. [Google Scholar] [CrossRef]
- Kim, T.; Byeon, H.; An, Y.; Rayamajhi, V.; Lee, J.; Lee, J.D.; Jung, S.M. Acute toxicity of antifouling agents CuSO4, ZnPT, and CuPT on marine diatoms Skeletonema costatum and Navicula sp. Mar. Environ. Res. 2025, 207, 107084. [Google Scholar] [CrossRef]
- Laporte, D.; Rodríguez, F.; González, A.; Zúñiga, A.; Castro-Nallar, E.; Sáez, C.A.; Moenne, A. Copper-induced concomitant increases in photosynthesis, respiration, and C, N and S assimilation revealed by transcriptomic analyses in Ulva compressa (Chlorophyta). BMC Plant Biol. 2020, 20, 25. [Google Scholar] [CrossRef]
- Luengluetham, P.; Chotikarn, P.; Nopparat, J.; Buapet, P. Ecotoxicological assessment of copper and zinc in a common aquatic plant Ceratophyllum demersum: Physiological effects and biomarker responses. Aquat. Bot. 2023, 188, 103678. [Google Scholar] [CrossRef]
- Santiago, A.M.; Gonçalves, D.L.; Morano, K.A. Mechanisms of sensing and response to proteotoxic stress. Exp. Cell Res. 2020, 395, 112240. [Google Scholar] [CrossRef]
- Colunga Biancatelli, R.M.; Solopov, P.A.; Day, T.; Gregory, B.; Osei-Nkansah, M.; Dimitropoulou, C.; Catravas, J.D. HSP70 is a critical regulator of HSP90 inhibitor’s effectiveness in preventing HCl-induced chronic lung injury and pulmonary fibrosis. Int. J. Mol. Sci. 2024, 25, 1920. [Google Scholar] [CrossRef]
- Azizullah, A.; Khan, S.; Gao, G.; Gao, K. The interplay between bisphenol A and algae—A review. J. King Saud Univ. Sci. 2022, 34, 102050. [Google Scholar] [CrossRef]
- Wang, H.; Guo, R.; Ki, J.S. 6.0 K microarray reveals differential transcriptomic responses in the dinoflagellate Prorocentrum minimum exposed to polychlorinated biphenyl (PCB). Chemosphere 2018, 195, 398–409. [Google Scholar] [CrossRef]
- Menone, M.L.; Pesce, S.F.; Díaz, M.P.; Moreno, V.J.; Wunderlin, D.A. Endosulfan induces oxidative stress and changes on detoxication enzymes in the aquatic macrophyte Myriophyllum quitense. Phytochemistry 2008, 69, 1150–1157. [Google Scholar] [CrossRef]
- Guo, R.; Youn, S.H.; Ki, J.S. Heat shock protein 70 and 90 genes in the harmful dinoflagellate Cochlodinium polykrikoides: Genomic structures and transcriptional responses to environmental stresses. Int. J. Genom. 2015, 2015, 484626. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rohilla, M.S.; Tiwari, P.K. Developmental and hyperthermia-induced expression of the heat shock proteins HSP60 and HSP70 in tissues of the housefly Musca domestica: An in vitro study. Genet. Mol. Biol. 2007, 30, 159–168. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Cui, Y.; Chen, N. Removal of copper ions from wastewater: A review. Int. J. Environ. Res. Public Health 2023, 20, 3885. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, I.; Nazir, A.; Saxena, D.K.; Chowdhuri, D.K. Heat shock response: Hsp70 in environmental monitoring. J. Biochem. Mol. Toxicol. 2003, 17, 249–254. [Google Scholar] [CrossRef]
- Mitra, T.; Mahanty, A.; Ganguly, S.; Purohit, G.K.; Mohanty, S.; Parida, P.K.; Mohanty, B.P. Expression patterns of heat shock protein genes in Rita rita from natural riverine habitat as biomarker response against environmental pollution. Chemosphere 2018, 211, 535–546. [Google Scholar] [CrossRef]


| Chemicals | Treatments (mg L−1) | |||||
|---|---|---|---|---|---|---|
| CuSO4 | 0.0 | 0.1 | 0.2 | 0.5 | 0.8 | 1.0 |
| CuCl2 | 0.0 | 0.05 | 0.1 | 0.2 | 0.5 | 1.0 |
| NiSO4 | 0.0 | 0.1 | 0.5 | 1.0 | 2.0 | 3.0 |
| NiCl2 | 0.0 | 0.05 | 0.1 | 0.2 | 0.5 | 1.0 |
| BPA | 0.0 | 0.05 | 0.25 | 0.5 | 1.0 | 2.0 |
| PCB | 0.0 | 0.001 | 0.005 | 0.01 | 0.05 | 0.1 |
| EDS | 0.0 | 0.0001 | 0.0002 | 0.0005 | 0.001 | 0.002 |
| Chemicals | EC50 (mg L−1) | Reference |
|---|---|---|
| CuSO4 | 0.406 | [45] |
| CuCl2 | 1.455 | |
| NiSO4 | 3.468 | |
| NiCl2 | 0.720 | |
| BPA | 0.039 | |
| PCB | 0.002 | |
| EDS | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-S.; Lee, J.-W.; Ki, J.-S. Heat Shock Protein 104 (Hsp104) in the Marine Diatom Ditylum brightwellii: Identification and Transcriptional Responses to Environmental Stress. Genes 2025, 16, 1408. https://doi.org/10.3390/genes16121408
Kim H-S, Lee J-W, Ki J-S. Heat Shock Protein 104 (Hsp104) in the Marine Diatom Ditylum brightwellii: Identification and Transcriptional Responses to Environmental Stress. Genes. 2025; 16(12):1408. https://doi.org/10.3390/genes16121408
Chicago/Turabian StyleKim, Han-Sol, Jong-Won Lee, and Jang-Seu Ki. 2025. "Heat Shock Protein 104 (Hsp104) in the Marine Diatom Ditylum brightwellii: Identification and Transcriptional Responses to Environmental Stress" Genes 16, no. 12: 1408. https://doi.org/10.3390/genes16121408
APA StyleKim, H.-S., Lee, J.-W., & Ki, J.-S. (2025). Heat Shock Protein 104 (Hsp104) in the Marine Diatom Ditylum brightwellii: Identification and Transcriptional Responses to Environmental Stress. Genes, 16(12), 1408. https://doi.org/10.3390/genes16121408







