Genome-Wide Screening for MYB Transcription Factors Involved in Flavonoid Glycoside Biosynthesis in Carthamus tinctorius L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of MYBs in Safflower Genome
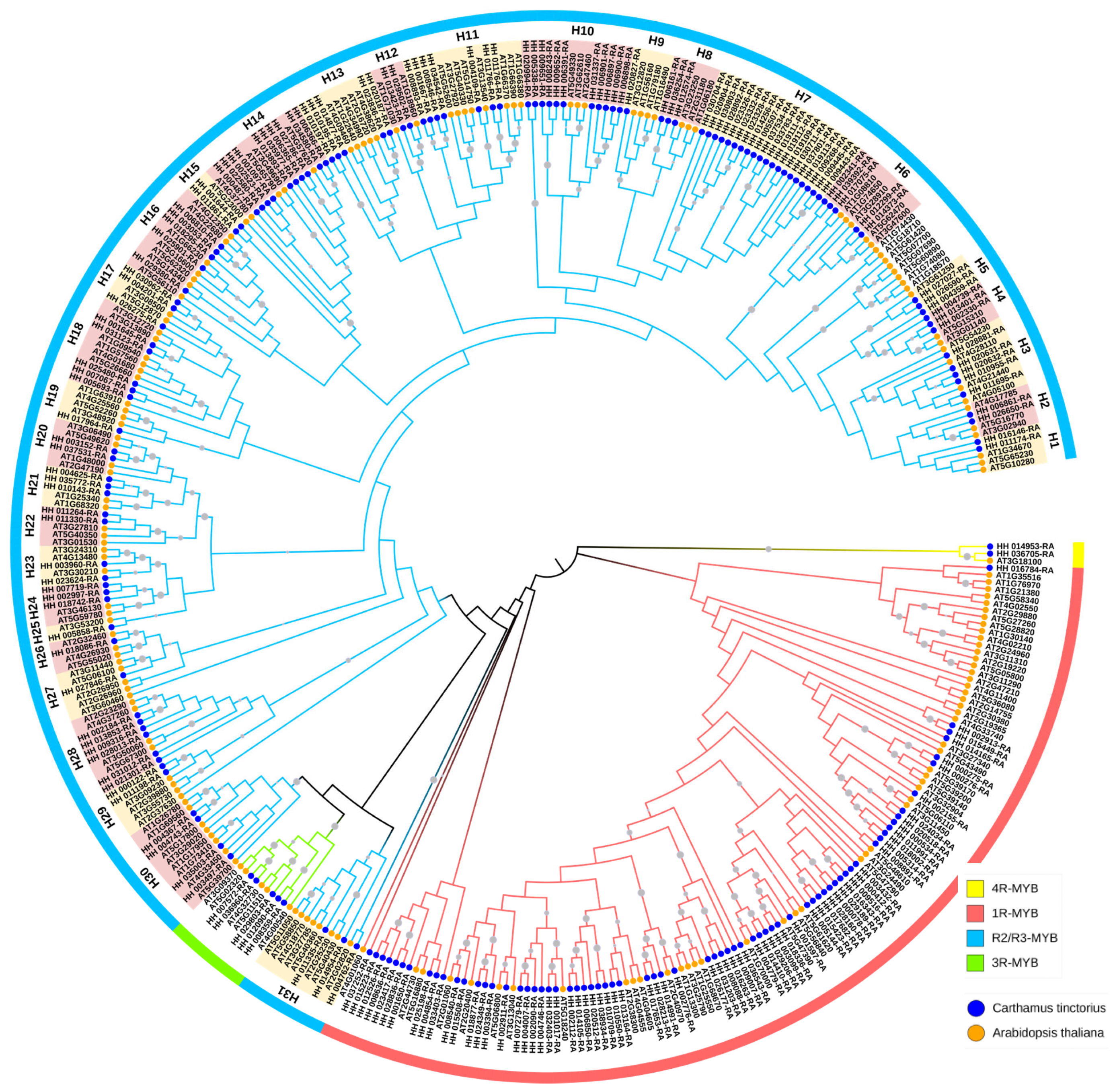
2.2. Evolutionary Classification and Phylogenetic Analysis of CtMYB Gene Family
2.3. Conserved Motif Analysis, Gene Structures, and Chromosomal Mapping of MYBs
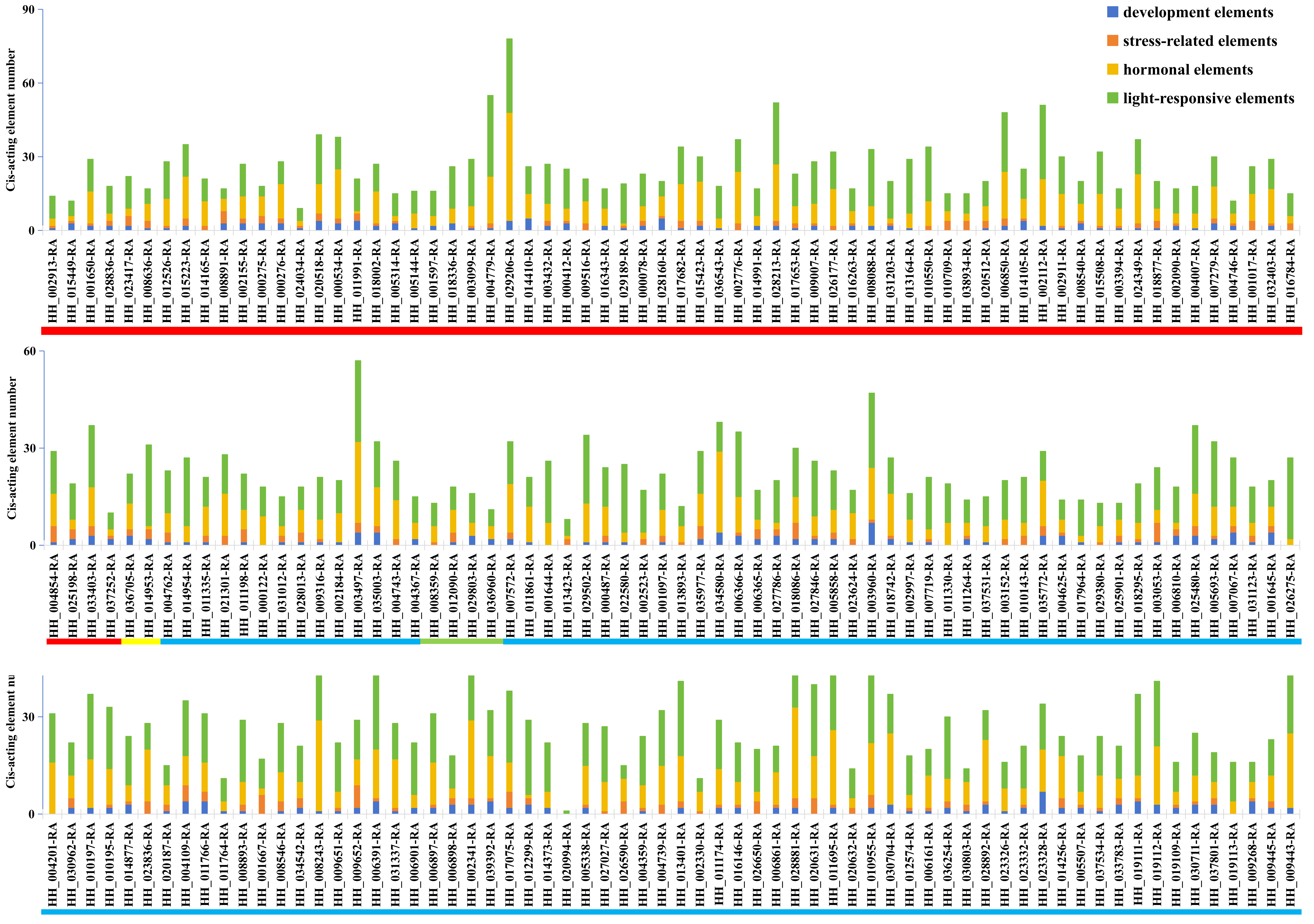
2.4. Cis-Acting Elements Analysis in the Promoter Region of MYB
2.5. Transcriptome Analysis of Safflower
2.6. Screening of MYB Transcription Factors Regulating Flavonoid Glycosides via Co-Expression Analysis
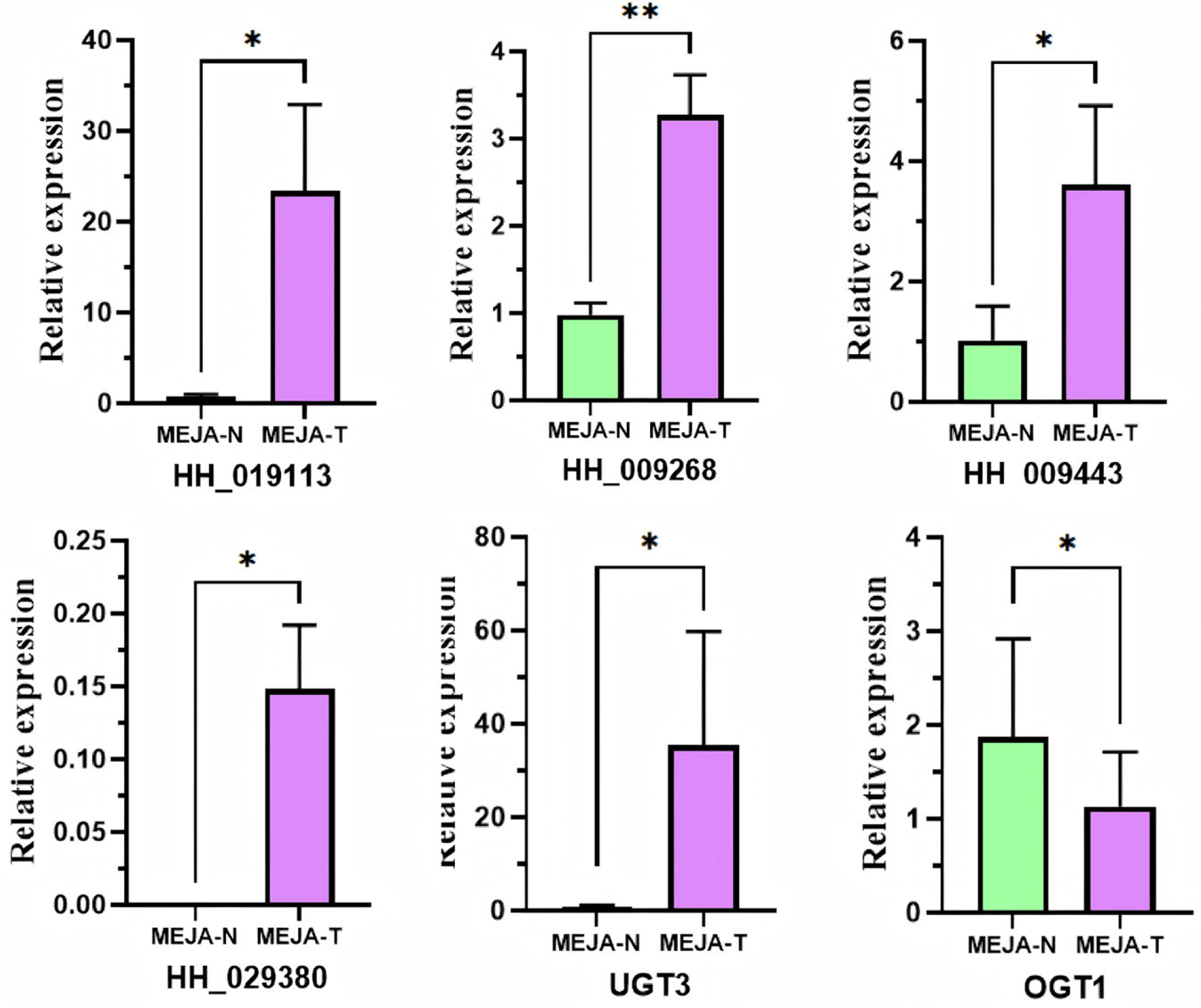
2.7. MeJA Treatment and qPCR Verification
3. Results
3.1. Identification of MYBs in the Safflower Genome
3.2. Evolutionary Classification and Phylogenetic Analysis of Ct MYB Gene Family
3.3. Conserved Motif Analysis, Gene Structures, and Chromosomal Mapping of MYBs
3.4. Cis-Acting Elements Analysis in the Promoter Region of MYBs
3.5. Expression Profiling of MYB Genes of Safflower Under Different Development Stages, Light Intensities and Tissues
3.6. Screening of MYB Transcription Factors Regulating Flavonoid Glycosides via Co-Expression Analysis
3.7. MYB Gene Expression Was Enhanced by MeJA Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| C.tinctorius | Carthamus tinctorius L |
| A. thaliana | Arabidopsis thaliana |
| O. sativa | Oryza sativa |
| HYSA | Hydroxysafflor yellow A |
| UGT | UDP-glycosyltransferase |
| OGT | O-linked N-acetylglucosamine (GlcNAc) transferase |
| MeJA | Methyl jasmonate |
| bHLH | Basic helix–loop–helix |
| CHS | Chalcone synthase |
| F3H | Flavanone 3-hydroxylase |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| CDS | Coding sequences |
| UTR | Untranslatted regions |
| qPCR | Quantitative real-time PCR |
| LTR | Long terminal repeat |
| MBS | MYB binding site |
References
- Zhang, L.-L.; Tian, K.; Tang, Z.-H.; Chen, X.-J.; Bian, Z.-X.; Wang, Y.-T.; Lu, J.-J. Phytochemistry and Pharmacology of Carthamus tinctorius L. Am. J. Chin. Med. 2016, 44, 197–226. [Google Scholar] [CrossRef]
- Xue, X.; Deng, Y.; Wang, J.; Zhou, M.; Liao, L.; Wang, C.; Peng, C.; Li, Y. Hydroxysafflor yellow A, a natural compound from Carthamus tinctorius L. with good effect of alleviating atherosclerosis. Phytomedicine 2021, 91, 153694. [Google Scholar] [CrossRef]
- Bai, X.; Wang, W.-X.; Fu, R.-J.; Yue, S.-J.; Gao, H.; Chen, Y.-Y.; Tang, Y.-P. Therapeutic Potential of Hydroxysafflor Yellow A on Cardio-Cerebrovascular Diseases. Front. Pharmacol. 2020, 11, 01265. [Google Scholar] [CrossRef]
- Ren, C.; Xi, Z.; Xian, B.; Chen, C.; Huang, X.; Jiang, H.; Chen, J.; Peng, C.; Pei, J. Identification and Characterization of CtUGT3 as the Key Player of Astragalin Biosynthesis in Carthamus tinctorius L. J. Agric. Food Chem. 2023, 71, 16221–16232. [Google Scholar] [CrossRef]
- Xian, B.; Zhou, Y.; Hu, Y.; Peng, Y.; Song, X.; Xi, Z.; Li, Y.; Yan, J.; Ren, C.; Pei, J.; et al. Genome-wide screen and multi-omics analysis reveal OGT1 participate in the biosynthesis of safflower flavonoid glycosides. Hortic. Res. 2024, 11, uhae261. [Google Scholar] [CrossRef]
- Xi, Z.; Li, Y.; Liu, S.; Wang, D.; Guo, J.; Xian, B.; Rao, K.; Chen, C.; Peng, Y.; Zhou, Y.; et al. Functional analysis and molecular characterization of UGT95A2, a specialized glycosyltransferase for flavonoid 3′-O-glycosylation in Carthamus tinctorius L. Plant J. 2025, 122, e70213. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Y.; Ye, Z.; Zuo, H.; Li, P.; Zhao, X.; Chen, Z.; Chen, C.; Zhao, J. Roles of trichomes in tea plant resistance against multiple abiotic and biotic stresses. Beverage Plant Res. 2022, 2, 14. [Google Scholar] [CrossRef]
- Lin, X.; Ding, C.; Xiao, W.; Wang, J.; Lin, Z.; Sun, X.; Li, S.; Pan, Z.; Zeng, R.; Song, Y. A molecular switch OsWRKY10-OsVQ8 orchestrates rice diterpenoid phytoalexin biosynthesis for broad-spectrum disease resistance. New Phytol. 2025, 246, 2243–2262. [Google Scholar] [CrossRef]
- Virag, E.; Hegedus, G.; Nagy, A.; Pallos, J.P.; Kutasy, B. Temporal Shifts in Hormone Signaling Networks Orchestrate Soybean Floral Development Under Field Conditions: An RNA-Seq Study. Int. J. Mol. Sci. 2025, 26, 6455. [Google Scholar] [CrossRef]
- Hong, Y.; Lv, Y.; Zhang, J.; Ahmad, N.; Li, X.; Yao, N.; Liu, X.; Li, H. The safflower MBW complex regulates HYSA accumulation through degradation by the E3 ligase CtBB1. J. Integr. Plant Biol. 2023, 65, 1277–1296. [Google Scholar] [CrossRef]
- Tan, Z.; Lu, D.; Li, L.; Su, X.; Sun, Y.; Wang, L.; Yu, Y.; Wan, X.; Xu, L.; Li, C.; et al. Comprehensive analysis of safflower R2R3-MYBs reveals the regulation mechanism of CtMYB76 on flavonol biosynthesis. Ind. Crops Prod. 2025, 227, 120795. [Google Scholar] [CrossRef]
- Wang, R.; Ren, C.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.; Wang, J.; Pei, J.; Chen, J. Integrated Metabolomics and Transcriptome Analysis of Flavonoid Biosynthesis in Safflower (Carthamus tinctorius L.) With Different Colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar] [CrossRef]
- Chen, J.; Guo, S.; Hu, X.; Wang, R.; Jia, D.; Li, Q.; Yin, X.; Liao, X.; Hu, Z.; Wang, P.; et al. Whole-genome and genome-wide association studies improve key agricultural traits of safflower for industrial and medicinal use. Hortic. Res. 2023, 10, uhad197. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43(W1), W39–W49. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15. [Google Scholar] [CrossRef]
- Li, S.; Ahmed, W.; Jiang, T.; Yang, D.; Yang, L.; Hu, X.; Zhao, M.; Peng, X.; Yang, Y.; Zhang, W.; et al. Amino acid metabolism pathways as key regulators of nitrogen distribution in tobacco: Insights from transcriptome and WGCNA analyses. BMC Plant Biol. 2025, 25, 393. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Wang, R.; Xian, B.; Ren, C.; Liu, Q.; Wu, Q.; Pei, J. Integrated metabolomics and transcriptome analysis on flavonoid biosynthesis in safflower (Carthamus tinctorius L.) under MeJA treatment. BMC Plant Biol. 2020, 20, 353. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef]
- Ao, B.; Han, Y.; Wang, S.; Wu, F.; Zhang, J. Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress. Int. J. Mol. Sci. 2022, 23, 7243. [Google Scholar] [CrossRef]
- Chen, J.; Tang, X.; Ren, C.; Chen, X.; He, W.; Zhang, S.; Wu, Q.; Pei, J. Safflower MYB Transcription Factor Gene Cloning and Expression Analysis. Acta Pharm. Sin. 2018, 53, 141–146. [Google Scholar] [CrossRef]
- Jin, F.; Ding, L.; Luo, J.; Nie, S.; Fang, Z. Research Progress on MYB Transcription Factors in Rice. J. Plant Genet. Resour. 2023, 24, 917–926. [Google Scholar] [CrossRef]
- Du, H.; Feng, B.-R.; Yang, S.-S.; Huang, Y.-B.; Tang, Y.-X. The R2R3-MYB Transcription Factor Gene Family in Maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef]
- Wei, H. Evolutionary Mechanisms and Functional Studies of MYB Genes in Soybean. Ph.D. Thesis, Graduate School of Chinese Academy of Sciences (Northeast Institute of Geography and Agroecology), Changchun, China, 2015. [Google Scholar]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef]
- Li, Z.; Peng, R.; Tian, Y.; Han, H.; Xu, J.; Yao, Q. Genome-Wide Identification and Analysis of the MYB Transcription Factor Superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef]
- Xu, M.; Fu, J.; Ni, Y.; Zhang, C. Genome—Wide analysis of the MYB gene family in pumpkin. Peerj 2024, 12, e17304. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, S.; Zuo, D.; Wang, Q.; Lv, L.; Zhang, Y.; Cheng, H.; Yu, J.Z.; Song, G. Identification of MYB gene family and functional analysis of GhMYB4 in cotton (Gossypium spp.). Mol. Genet. Genom. 2023, 298, 755–766. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Liu, X.; Yao, N. Genome-wide analysis and transcriptional reprogrammings of MYB superfamily revealed positive insights into abiotic stress responses and anthocyanin accumulation in Carthamus tinctorius. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef]
- Sukumaran, S.; Lethin, J.; Liu, X.; Pelc, J.; Zeng, P.; Hassan, S.; Aronsson, H. Genome-Wide Analysis of MYB Transcription Factors in the Wheat Genome and Their Roles in Salt Stress Response. Cells 2023, 12, 1431. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Li, H.; Yang, J.; Ma, R.; An, X.; Pan, F.; Zhang, S.; Fu, Y. Genome-wide identification and expression analysis of MYB gene family in Cajanus cajan and CcMYB107 improves plant drought tolerance. Physiol. Plant. 2023, 175, e13954. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, J.; Xu, H.; Wang, S.; Xu, J.; Wang, Y.; Dai, X.; Yu, H.; Xiong, K.; Chen, Q.; et al. Molecular insights into ThMYB14-mediated flavonoid accumulation in Tetrastigma hemsleyanum Diels et Gilg in response to water stress. Ind. Crops Prod. 2025, 226, 120708. [Google Scholar] [CrossRef]
- Albani, D.; Hammond-Kosack, M.C.; Smith, C.; Conlan, S.; Colot, V.; Holdsworth, M.; Bevan, M.W. The wheat transcriptional activator SPA: A seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 1997, 9, 171–184. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Ding, J.; Han, J.; Shi, L. GlMPC activated by GCN4 regulates secondary metabolism under nitrogen limitation conditions in Ganoderma lucidum. Mbio 2023, 14, e0135623. [Google Scholar] [CrossRef]
- Yang, H.; Chen, C.; Han, L.; Zhang, X.; Yue, M.; Ma, Q. Genome-Wide Identification and Expression Analysis of the MYB Transcription Factor Family in Salvia nemorosa. Genes 2024, 15, 110. [Google Scholar] [CrossRef]
- Yaginuma, S.; Shiraishi, T.; Igarashi, K. Developmental transition of the flavonoid contents in safflower leaves during stress-loaded cultivation. Biosci. Biotechnol. Biochem. 2003, 67, 1691–1698. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, X.; Lin, N.; Yu, S.; Fernie, A.R.; Zhao, J. CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator–repressor network. Hortic. Res. 2021, 8, 110. [Google Scholar] [CrossRef]
- Jing, T.; Du, W.; Qian, X.; Wang, K.; Luo, L.; Zhang, X.; Deng, Y.; Li, B.; Gao, T.; Zhang, M.; et al. UGT89AC1-mediated quercetin glucosylation is induced upon herbivore damage and enhances Camellia sinensis resistance to insect feeding. Plant Cell Environ. 2024, 47, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Xian, B.; Peng, L.; Wu, X.; Zhang, J.; Li, Y.; Hu, Y.; Chen, J. Genome-Wide Screening for MYB Transcription Factors Involved in Flavonoid Glycoside Biosynthesis in Carthamus tinctorius L. Genes 2025, 16, 1376. https://doi.org/10.3390/genes16111376
Yu X, Xian B, Peng L, Wu X, Zhang J, Li Y, Hu Y, Chen J. Genome-Wide Screening for MYB Transcription Factors Involved in Flavonoid Glycoside Biosynthesis in Carthamus tinctorius L. Genes. 2025; 16(11):1376. https://doi.org/10.3390/genes16111376
Chicago/Turabian StyleYu, Xiaohan, Bin Xian, Lijun Peng, Xunjian Wu, Juncheng Zhang, Yuanyuan Li, Yueying Hu, and Jiang Chen. 2025. "Genome-Wide Screening for MYB Transcription Factors Involved in Flavonoid Glycoside Biosynthesis in Carthamus tinctorius L." Genes 16, no. 11: 1376. https://doi.org/10.3390/genes16111376
APA StyleYu, X., Xian, B., Peng, L., Wu, X., Zhang, J., Li, Y., Hu, Y., & Chen, J. (2025). Genome-Wide Screening for MYB Transcription Factors Involved in Flavonoid Glycoside Biosynthesis in Carthamus tinctorius L. Genes, 16(11), 1376. https://doi.org/10.3390/genes16111376





