Mapping-by-Sequencing via eBSRmap (Easy Bulk Segregate RNA Mapping) in a B73 EMS Mutant Population
Abstract
1. Introduction
2. Materials and Methods
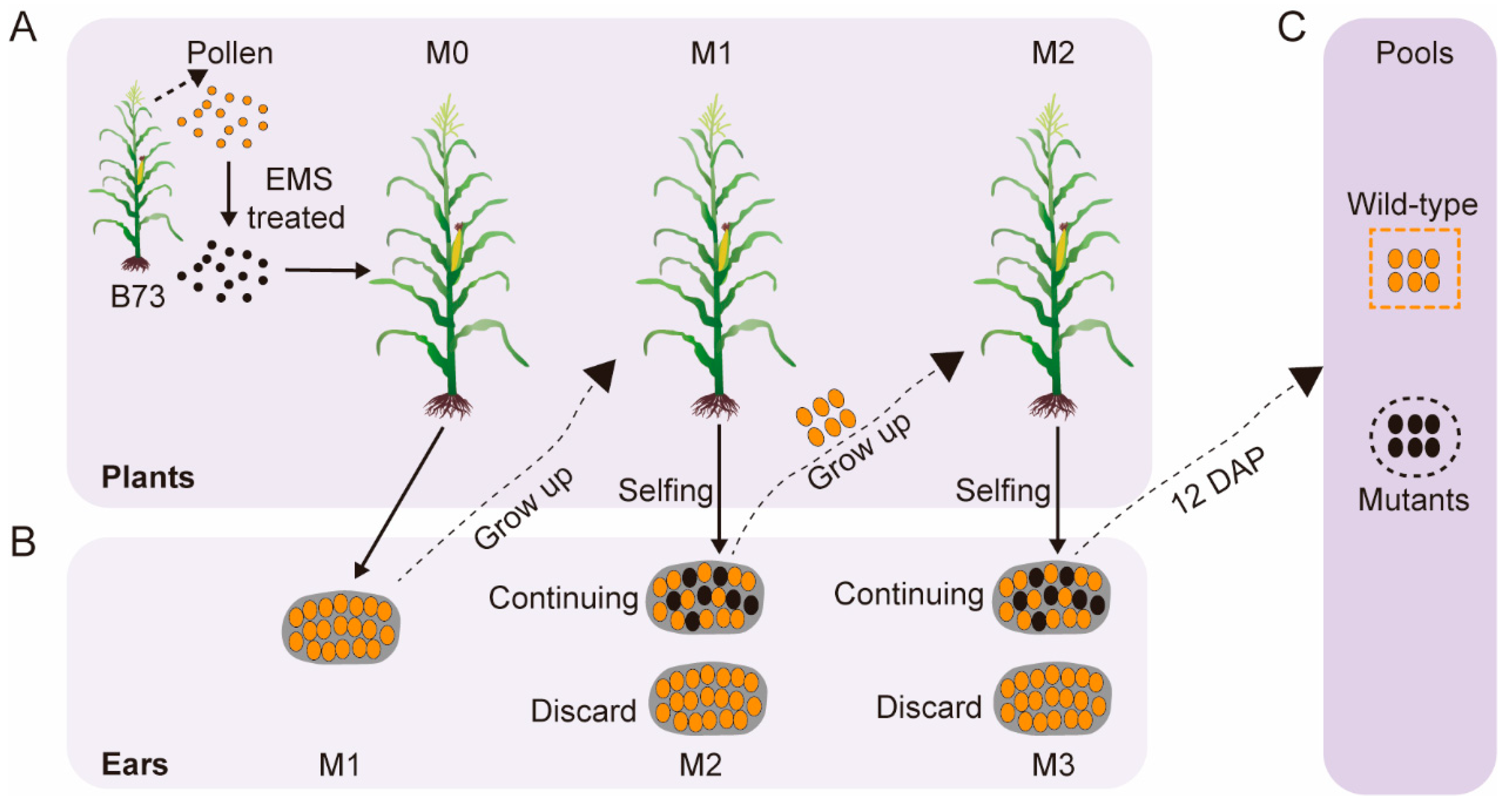
2.1. Plant Materials
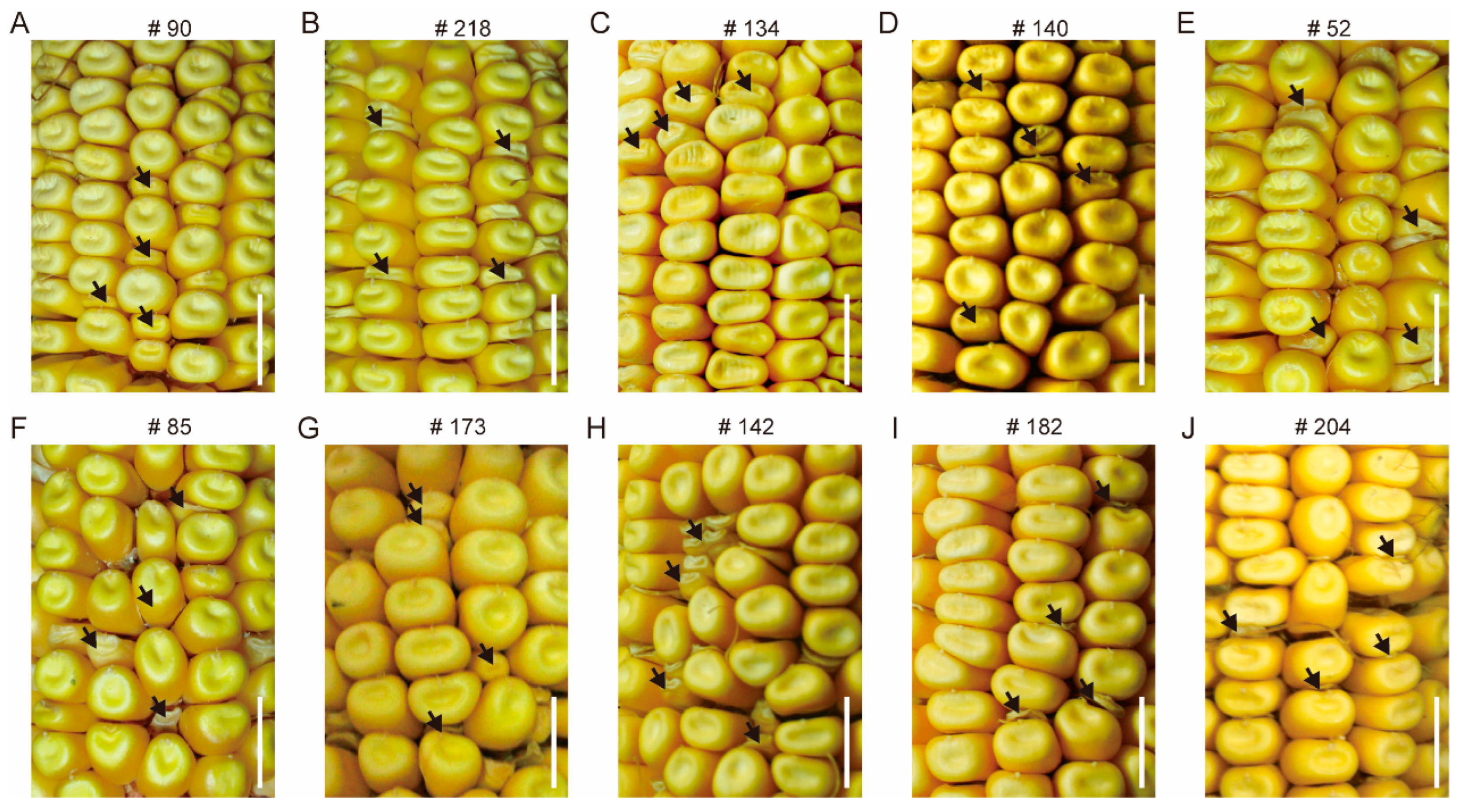
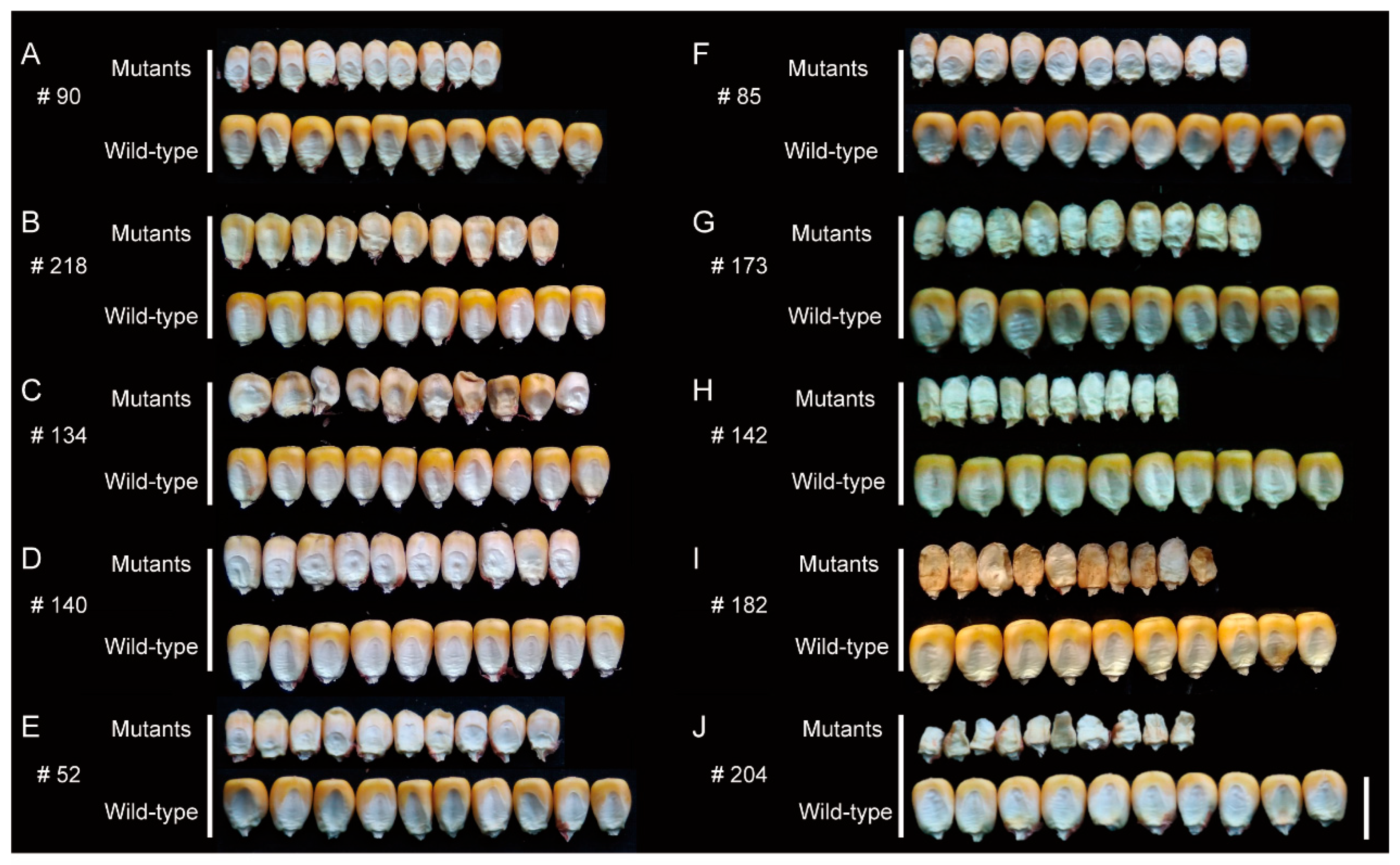
2.2. Classification of Kernel Phenotype
2.3. cDNA Library Preparation and Data Analysis
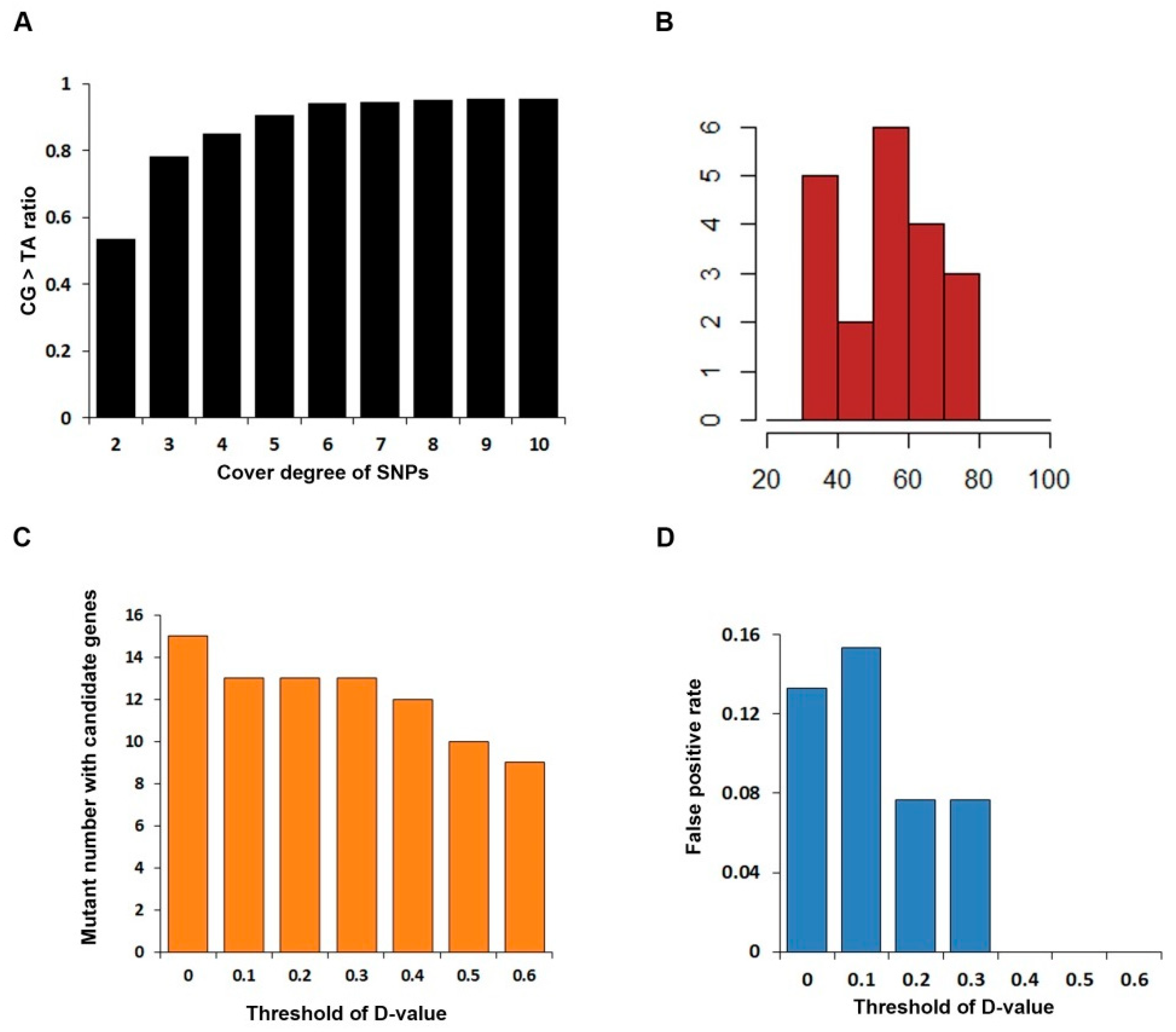
2.4. Statistical Simulation of D-Value Threshold
2.5. Analysis of SNP Linkage
2.6. Validation of Co-Segregation
2.7. Allelic Test
2.8. Validation by CRISPR/Cas9 Technologies
2.9. Accession Numbers
2.10. Primers in This Study
| Mutant ID | Primer ID | Primer Sequence | Amplification Interval |
|---|---|---|---|
| #105 | CY16_105CGF | 5′-TGTGGAATGAGATGGAAAGTGC-3′ | chr1: 276804254–276804810 |
| CY16_105CGR | 5′-CCGCAGATGCCTCTTTTTC-3′ | ||
| #119 | CY16_119CGF | 5′-TGATTGCCCTTTACTCTATTGGGTA-3′ | chr3: 134956480–134957182 |
| CY16_119CGR | 5′-GTAACGCAACTTATCAAGAGCATCA-3′ | ||
| #180 | CY16_180CGF | 5′-CCATTGGACTCCCCTTTTGCT-3′ | chr8: 14483769–14484442 |
| CY16_180CGR | 5′-AACAGGGCAAGTATTTCATGG-3′ | ||
| #205 | CY16_205CGF | 5′-TATTGGGAGGAAAAATGATGCG-3′ | chr5: 41112547–41112992 |
| CY16_205CGR | 5′-GACAATACTTGAGTGCCTACTGAAT-3′ | ||
| #206 | CY16_206CGF | 5′-TGATAGGAGGACAGAGGGAAAG-3′ | chr2: 50703053–50703671 |
| CY16_206CGR | 5′-CAAAGTGAAGTCAGAACAGCA-3′ | ||
| #225 | CY16_225CGF | 5′-CGGCTCCACGGTAAAAATCAT-3′ | chr5: 77316940–77317587 |
| CY16_225CGR | 5′-CCCTTCTCTGTTCCCCATTCT-3′ | ||
| #226 | CY16_226CGF | 5′-TGATAGGAGGACAGAGGGAAAG-3′ | chr2: 50703053–50703410 |
| CY16_226CGR | 5′-TGAACTCTCACTGACTGCCGTAG-3′ | ||
| #254 | CY16_254CGF | 5′-TTCACAGCAAGCCCAAGACCG-3′ | chr7: 2078837–2079339 |
| CY16_254CGR | 5′-AAAAAAGGGCAAGGGTCAGAT-3′ | ||
| #2 | CY16_2CGF | 5′-CATAATCCGAATCAAGACAACCC-3′ | chr5: 74947055–74947634 |
| CY16_2CGR | 5′-ACTGTATTCTCCTTGGGTCATCTCC-3′ | ||
| #32 | CY16_32CGF | 5′-TTGAGTTACTTCAGTTGATGCC-3′ | chr6: 142178366–142178996 |
| CY16_32CGR | 5′-GGACTATGCTCTTCATTGTGTTTG-3′ | ||
| #43 | CY16_43CGF | 5′-ACTGCTCTGTTTGATTTTAGTGCTG-3′ | chr5: 3927508–3928212 |
| CY16_43CGR | 5′-CCAATGACATATCCGAGAGTTT-3′ | ||
| #82 | CY16_82CGF | 5′-TTCAACAACTTCTTAGCCGCCTTAC-3′ | chr5: 77316835–77317586 |
| CY16_82CGR | 5′-CCCTTCTCTGTTCCCCATTCT-3′ |
3. Results
3.1. Construction of B73 EMS Mutant Population and Bulked Sergeant Library
3.2. Discover Candidate Gene by SNP Calling
3.3. Identification of SNP Markers Tightly Linked to the Mutant Gene
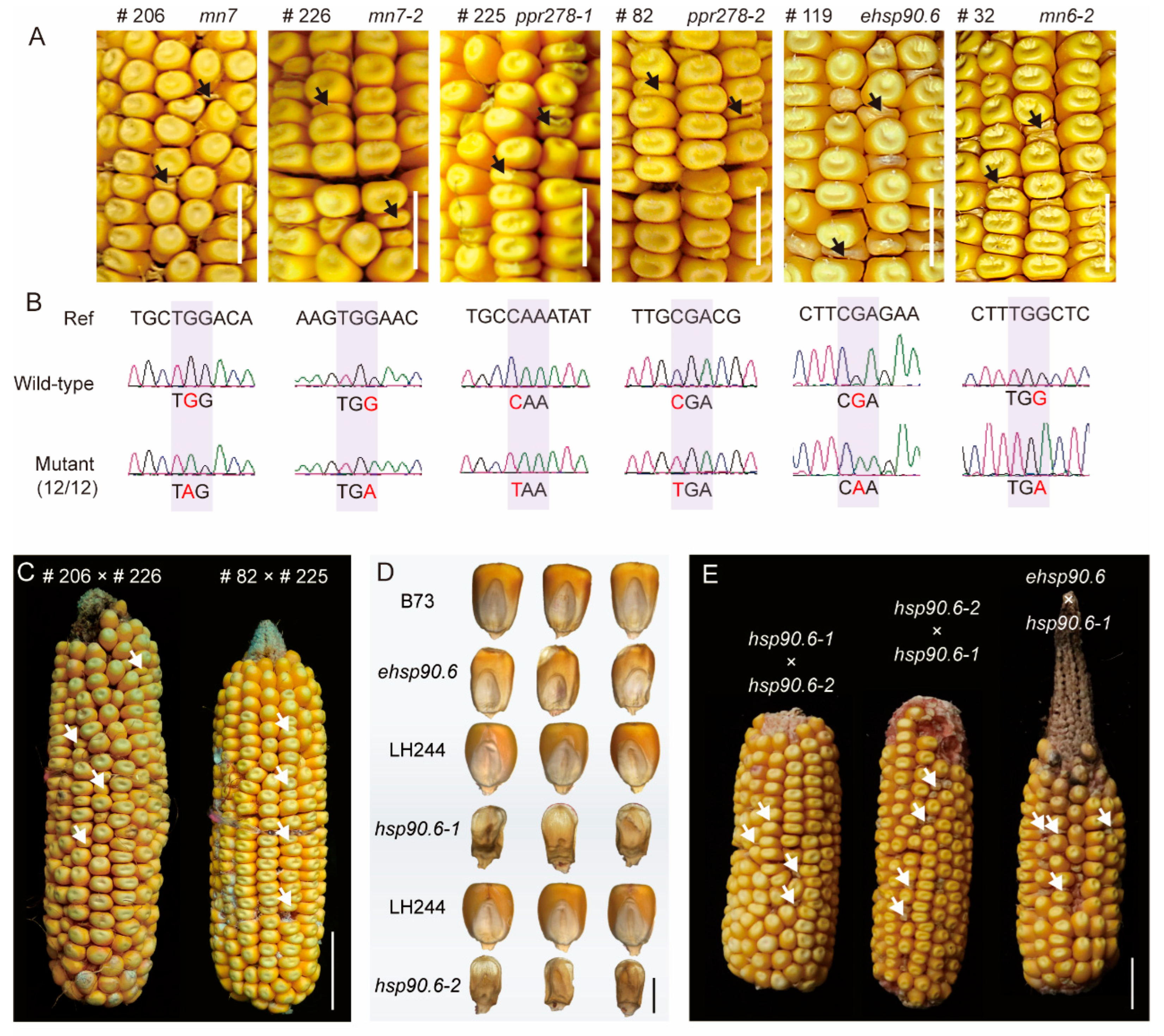
3.4. Confirmation of Candidate Genes
4. Discussion
4.1. Advantages of eBSRmap
4.2. Potential Problems and Strategies for Improvement
5. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L.; et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073, Erratum in Science 1989, 245, 1437. https://doi.org/10.1126/science.245.4925.1437.d. [Google Scholar] [CrossRef]
- Arondel, V.; Lemieux, B.; Hwang, I.; Gibson, S.; Goodman, H.M.; Somerville, C.R. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 1992, 258, 1353–1355. [Google Scholar] [CrossRef] [PubMed]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar] [PubMed]
- Taramino, G.; Tingey, S. Simple sequence repeats for germplasm analysis and mapping in maize. Genome 1996, 39, 277–287. [Google Scholar] [CrossRef]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef]
- Taramino, G.; Sauer, M.; Stauffer, J.L., Jr.; Multani, D.; Niu, X.; Sakai, H.; Hochholdinger, F. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 2007, 50, 649–659. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Wang, G.; Wang, F.; Zhang, X.; Zhong, M.; Zhang, J.; Lin, D.; Tang, Y.; Xu, Z.; et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 2012, 24, 3447–3462. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Feng, F.; Qi, W.; Lv, Y.; Yan, S.; Xu, L.; Yang, W.; Yuan, Y.; Chen, Y.; Zhao, H.; Song, R. OPAQUE11 Is a Central Hub of the Regulatory Network for Maize Endosperm Development and Nutrient Metabolism. Plant Cell 2018, 30, 375–396. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Qi, W.; Song, R. Maize Dek15 Encodes the Cohesin-Loading Complex Subunit SCC4 and Is Essential for Chromosome Segregation and Kernel Development. Plant Cell 2019, 31, 465–485. [Google Scholar] [CrossRef]
- Qi, W.; Lu, L.; Huang, S.; Song, R. Maize Dek44 Encodes Mitochondrial Ribosomal Protein L9 and Is Required for Seed Development. Plant Physiol. 2019, 180, 2106–2119. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Feng, F.; Qi, W.; Xu, L.; Yao, D.; Wang, Q.; Song, R. Dek35 Encodes a PPR Protein that Affects cis-Splicing of Mitochondrial nad4 Intron 1 and Seed Development in Maize. Mol. Plant 2017, 10, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Myers, A.M.; James, M.G.; Lin, Q.; Yi, G.; Stinard, P.S.; Hennen-Bierwagen, T.A.; Becraft, P.W. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 2011, 23, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ma, L.; Wang, X.; Zhao, Z.; Wang, W.; Fan, Y.; Liu, K.; Jiang, T.; Xiong, Z.; Song, Q.; et al. Genome-Wide Association Study Identifies a Rice Panicle Blast Resistance Gene, Pb2, Encoding NLR Protein. Int. J. Mol. Sci. 2022, 23, 5668. [Google Scholar] [CrossRef]
- Wang, C.; He, W.; Li, K.; Yu, Y.; Zhang, X.; Yang, S.; Wang, Y.; Yu, L.; Huang, W.; Yu, H.; et al. Genetic Diversity Analysis and GWAS of Plant Height and Ear Height in Maize Inbred Lines from South-East China. Plants 2025, 14, 481. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, F.; Tian, L.; Ding, Y.; Qi, J.; Zhang, H.; Mu, X.; Ma, Z.; Xia, L.; Tang, B. Molecular mapping of quantitative trait loci for 3 husk traits using genotyping by sequencing in maize (Zea mays L.). G3 2022, 12, jkac198. [Google Scholar] [CrossRef]
- Trampe, B.; Dos Santos, I.G.; Frei, U.K.; Ren, J.; Chen, S.; Lübberstedt, T. QTL mapping of spontaneous haploid genome doubling using genotyping-by-sequencing in maize (Zea mays L.). Theor. Appl. Genet. 2020, 133, 2131–2140. [Google Scholar] [CrossRef]
- Tran, Q.H.; Bui, N.H.; Kappel, C.; Dau, N.T.N.; Nguyen, L.T.; Tran, T.T.; Khanh, T.D.; Trung, K.H.; Lenhard, M.; Vi, S.L. Mapping-by-Sequencing via MutMap Identifies a Mutation in ZmCLE7 Underlying Fasciation in a Newly Developed EMS Mutant Population in an Elite Tropical Maize Inbred. Genes 2020, 11, 281. [Google Scholar] [CrossRef]
- Zheng, M.; Yang, T.; Liu, X.; Lü, G.; Zhang, P.; Jiang, B.; Zhou, S.; Lu, Y.; Lan, H.; Zhang, S.; et al. qRf8-1, a Novel QTL for the Fertility Restoration of Maize CMS-C Identified by QTL-seq. G3 2020, 10, 2457–2464. [Google Scholar] [CrossRef]
- Ni, J.; You, C.; Chen, Z.; Tang, D.; Wu, H.; Deng, W.; Wang, X.; Yang, J.; Bao, R.; Liu, Z.; et al. Deploying QTL-seq rapid identification and separation of the major QTLs of tassel branch number for fine-mapping in advanced maize populations. Mol. Breed. 2023, 43, 88. [Google Scholar] [CrossRef]
- Wang, H.; Yan, X.; Du, Q.; Yan, P.; Xi, J.; Meng, X.; Li, X.; Liu, H.; Liu, G.; Fu, Z.; et al. Maize Dek407 Encodes the Nitrate Transporter 1.5 and Is Required for Kernel Development. Int. J. Mol. Sci. 2023, 24, 17471. [Google Scholar] [CrossRef]
- Yang, T.; Guo, L.; Ji, C.; Wang, H.; Wang, J.; Zheng, X.; Xiao, Q.; Wu, Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 2021, 33, 104–128. [Google Scholar] [CrossRef]
- Neuffer, M.G.; Sheridan, W.F. Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 1980, 95, 929–944. [Google Scholar] [CrossRef]
- Dolfini, S.; Consonni, G.; Viotti, C.; Dal Prà, M.; Saltini, G.; Giulini, A.; Pilu, R.; Malgioglio, A.; Gavazzi, G. A mutational approach to the study of seed development in maize. J. Exp. Bot. 2007, 58, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, W.F.; Neuffer, M.G. Defective Kernel Mutants of Maize II. Morphological and Embryo Culture Studies. Genetics 1980, 95, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, B.; Ma, B.; Wang, Y.; Wang, H.; Sun, X.; Tan, B.C. Maize Dek51 encodes a DEAD-box RNA helicase essential for pre-rRNA processing and seed development. Cell Rep. 2024, 43, 114673. [Google Scholar] [CrossRef]
- Gutiérrez-Marcos, J.F.; Dal Prà, M.; Giulini, A.; Costa, L.M.; Gavazzi, G.; Cordelier, S.; Sellam, O.; Tatout, C.; Paul, W.; Perez, P.; et al. Empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 2007, 19, 196–210. [Google Scholar] [CrossRef]
- Yi, F.; Gu, W.; Li, J.; Chen, J.; Hu, L.; Cui, Y.; Zhao, H.; Guo, Y.; Lai, J.; Song, W. Miniature Seed6, encoding an endoplasmic reticulum signal peptidase, is critical in seed development. Plant Physiol. 2021, 185, 985–1001. [Google Scholar] [CrossRef]
- Yang, J.; Cui, Y.; Zhang, X.; Yang, Z.; Lai, J.; Song, W.; Liang, J.; Li, X. Maize PPR278 Functions in Mitochondrial RNA Splicing and Editing. Int. J. Mol. Sci. 2022, 23, 3035. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Z.; Fei, X.; Zhang, M.; Cui, Y.; Zhang, X.; Tan, K.; E, L.; Zhao, H.; Lai, J.; et al. HEAT SHOCK PROTEIN 90.6 interacts with carbon and nitrogen metabolism components during seed development. Plant Physiol. 2023, 191, 2316–2333. [Google Scholar] [CrossRef]
- Li, J.; Gu, W.; Yang, Z.; Chen, J.; Yi, F.; Li, T.; Li, J.; Zhou, Y.; Guo, Y.; Song, W.; et al. ZmELP1, an Elongator complex subunit, is required for the maintenance of histone acetylation and RNA Pol II phosphorylation in maize kernels. Plant Biotechnol. J. 2024, 22, 1251–1268. [Google Scholar] [CrossRef]
- Everaert, C.; Verwilt, J.; Verniers, K.; Vandamme, N.; Marcos Rubio, A.; Vandesompele, J.; Mestdagh, P. Blocking Abundant RNA Transcripts by High-Affinity Oligonucleotides during Transcriptome Library Preparation. Biol. Proced. Online 2023, 25, 7. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, O.; Kato, S.; Poulain, S.; Plessy, C. Targeted reduction of highly abundant transcripts using pseudo-random primers. Biotechniques 2016, 60, 169–174. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Yang, J.; Zhao, H.; Zhao, H.; Zhang, X. Mapping-by-Sequencing via eBSRmap (Easy Bulk Segregate RNA Mapping) in a B73 EMS Mutant Population. Genes 2025, 16, 1337. https://doi.org/10.3390/genes16111337
Cui Y, Yang J, Zhao H, Zhao H, Zhang X. Mapping-by-Sequencing via eBSRmap (Easy Bulk Segregate RNA Mapping) in a B73 EMS Mutant Population. Genes. 2025; 16(11):1337. https://doi.org/10.3390/genes16111337
Chicago/Turabian StyleCui, Yang, Jing Yang, Haiming Zhao, Hainan Zhao, and Xiangbo Zhang. 2025. "Mapping-by-Sequencing via eBSRmap (Easy Bulk Segregate RNA Mapping) in a B73 EMS Mutant Population" Genes 16, no. 11: 1337. https://doi.org/10.3390/genes16111337
APA StyleCui, Y., Yang, J., Zhao, H., Zhao, H., & Zhang, X. (2025). Mapping-by-Sequencing via eBSRmap (Easy Bulk Segregate RNA Mapping) in a B73 EMS Mutant Population. Genes, 16(11), 1337. https://doi.org/10.3390/genes16111337






