Identification of Key Genes Associated with Endoplasmic Reticulum Stress in Calcium Oxalate Kidney Stones
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. ScRNA-Seq Analysis
2.3. Identification of Key Genes
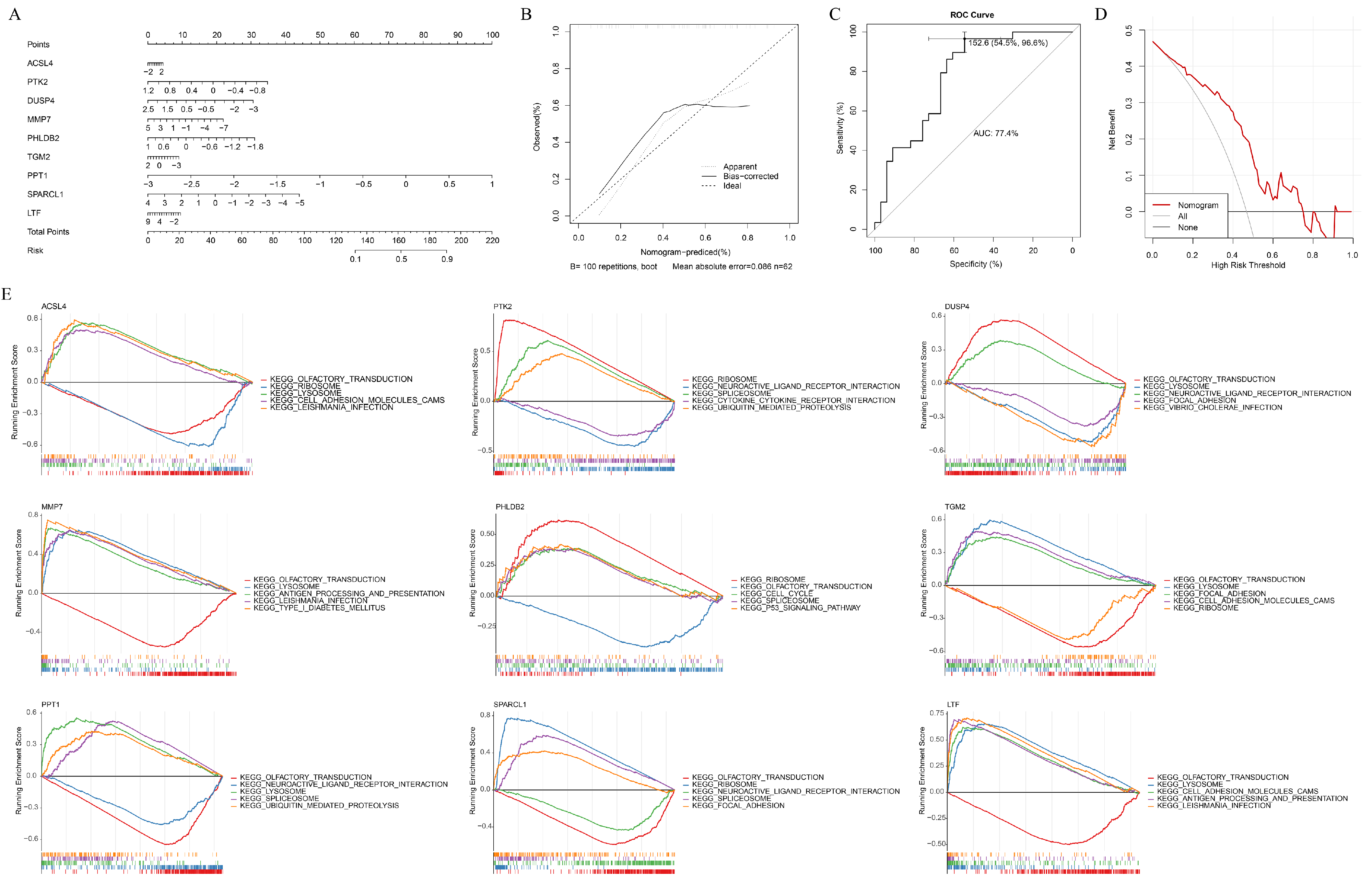
2.4. Construction of Nomogram
2.5. Gene Set Enrichment Analysis (GSEA)
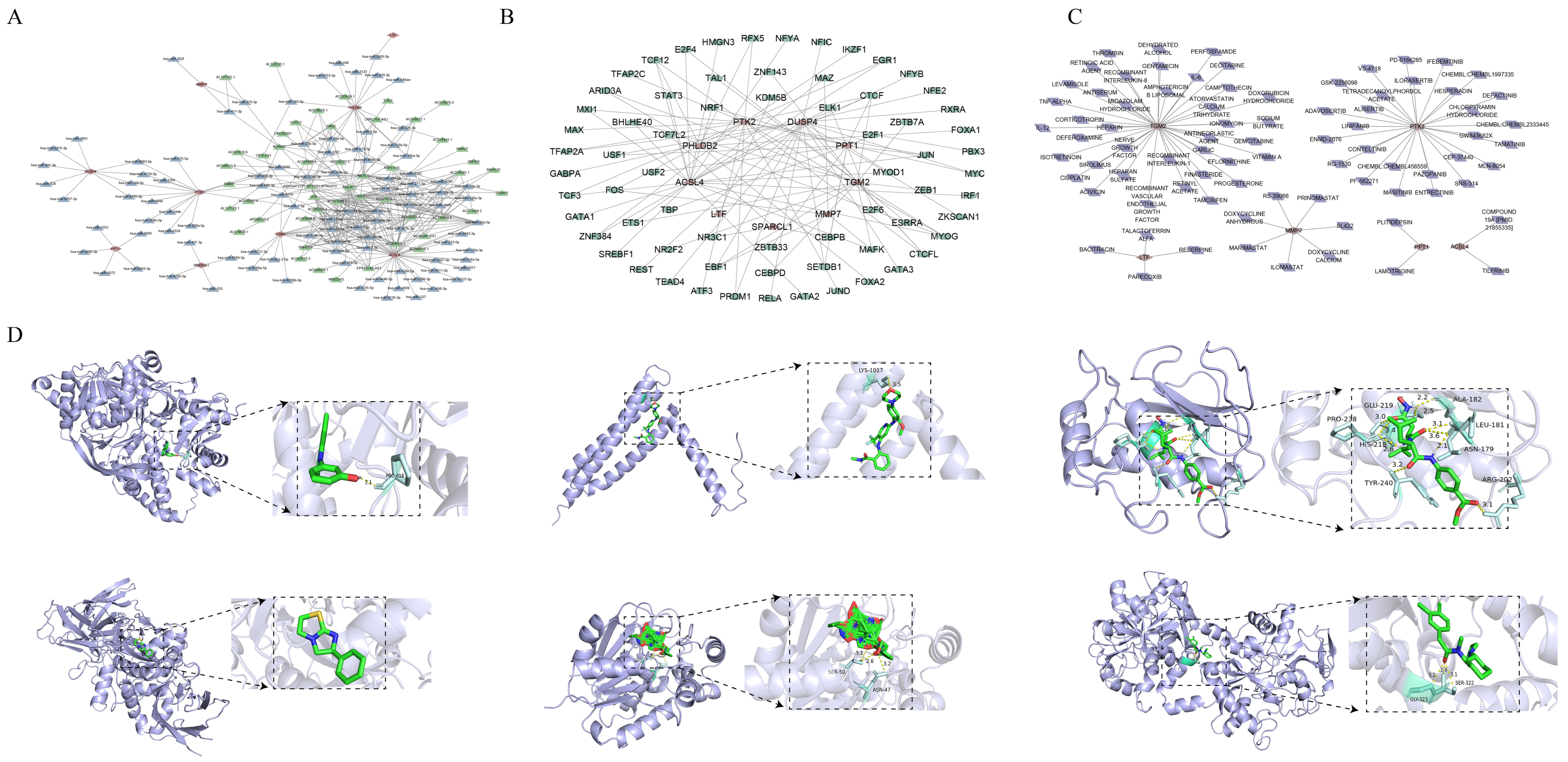
2.6. Drug Prediction and Molecular Docking
2.7. Regulatory Network and Drug Prediction of Key Genes
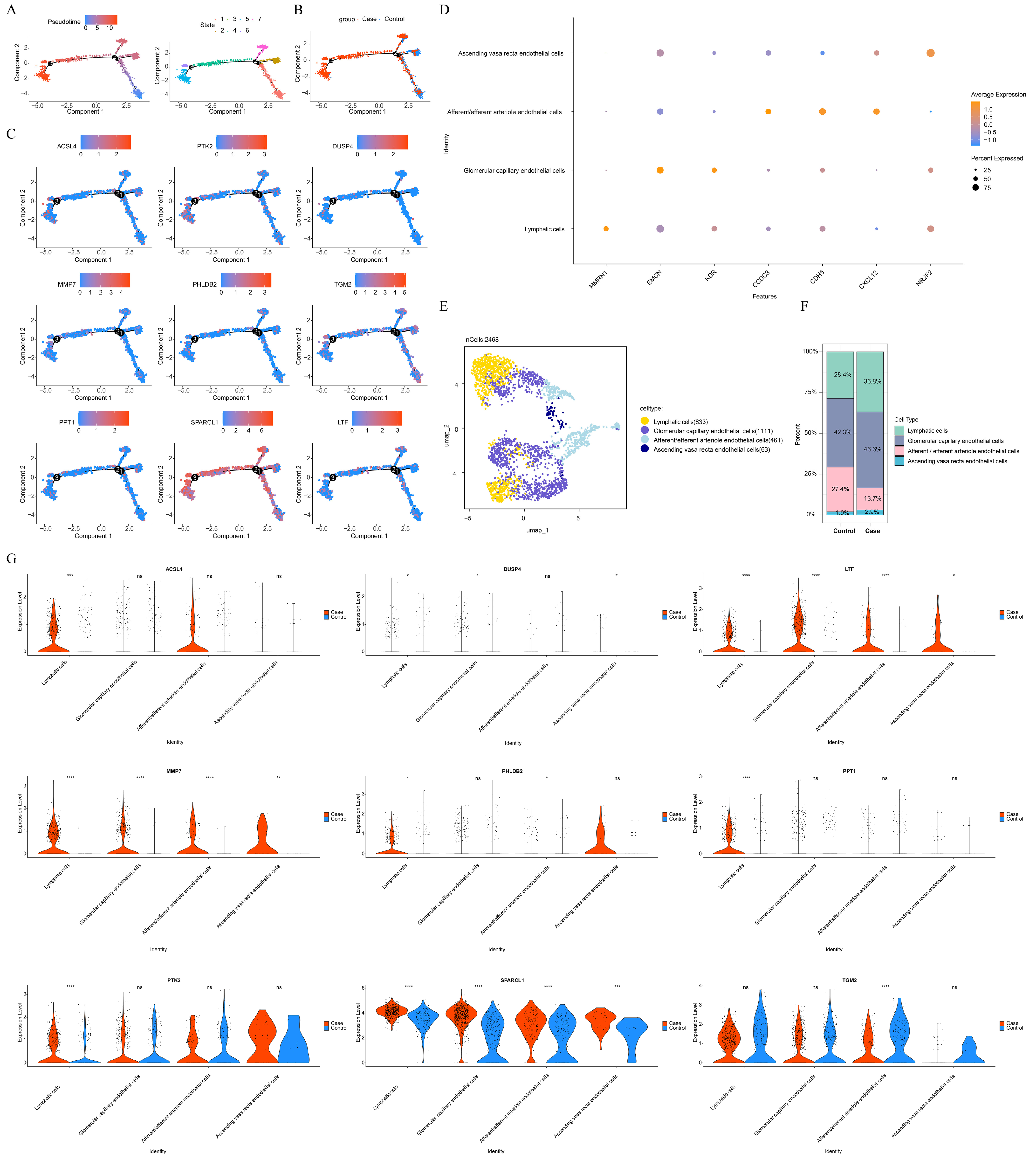
2.8. Pseudotime Analysis and Secondary Clustering
2.9. Cell Culture
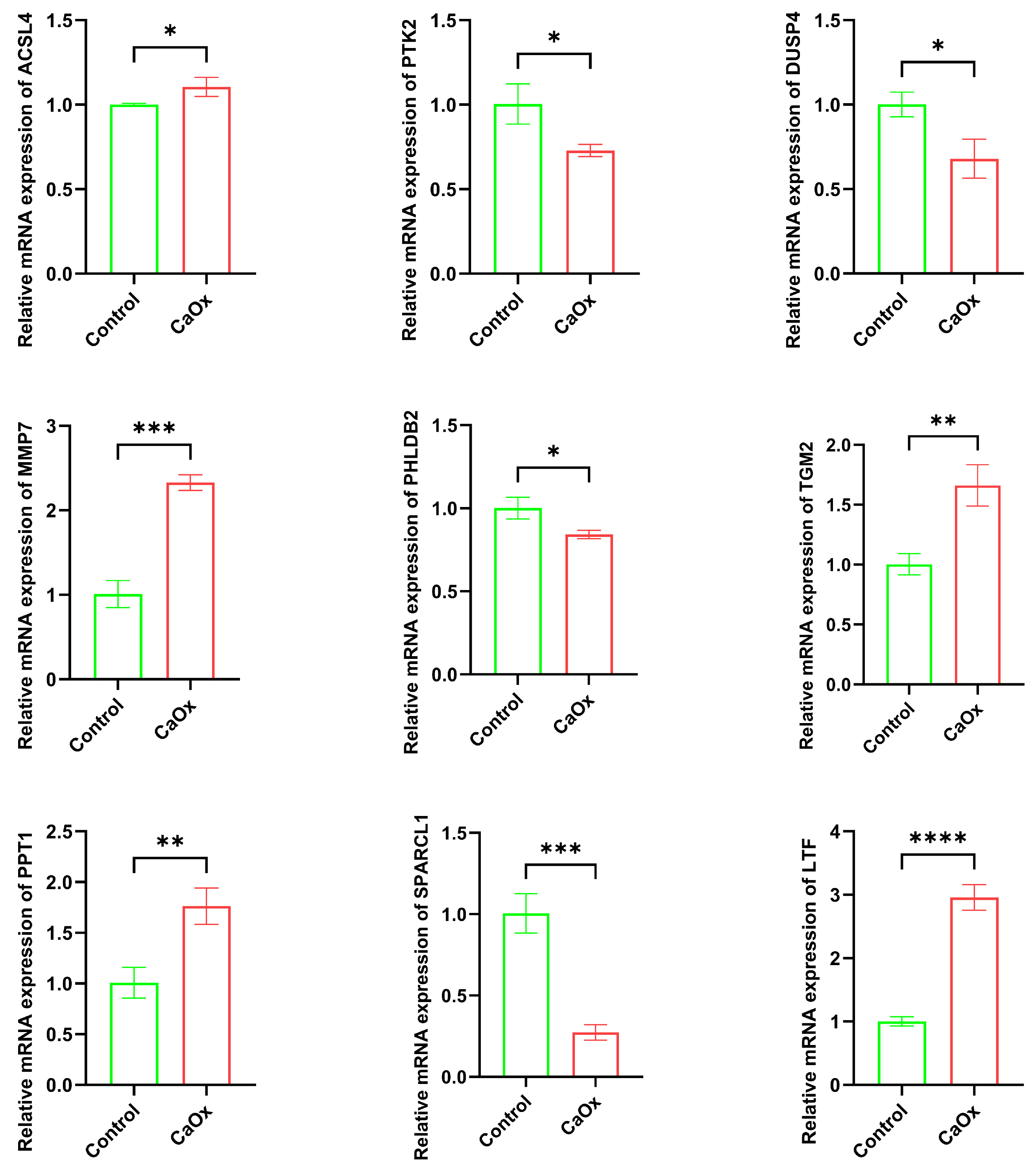
2.10. Quantitative RT-PCR
2.11. Western Blot
2.12. Statistical Analysis
3. Results
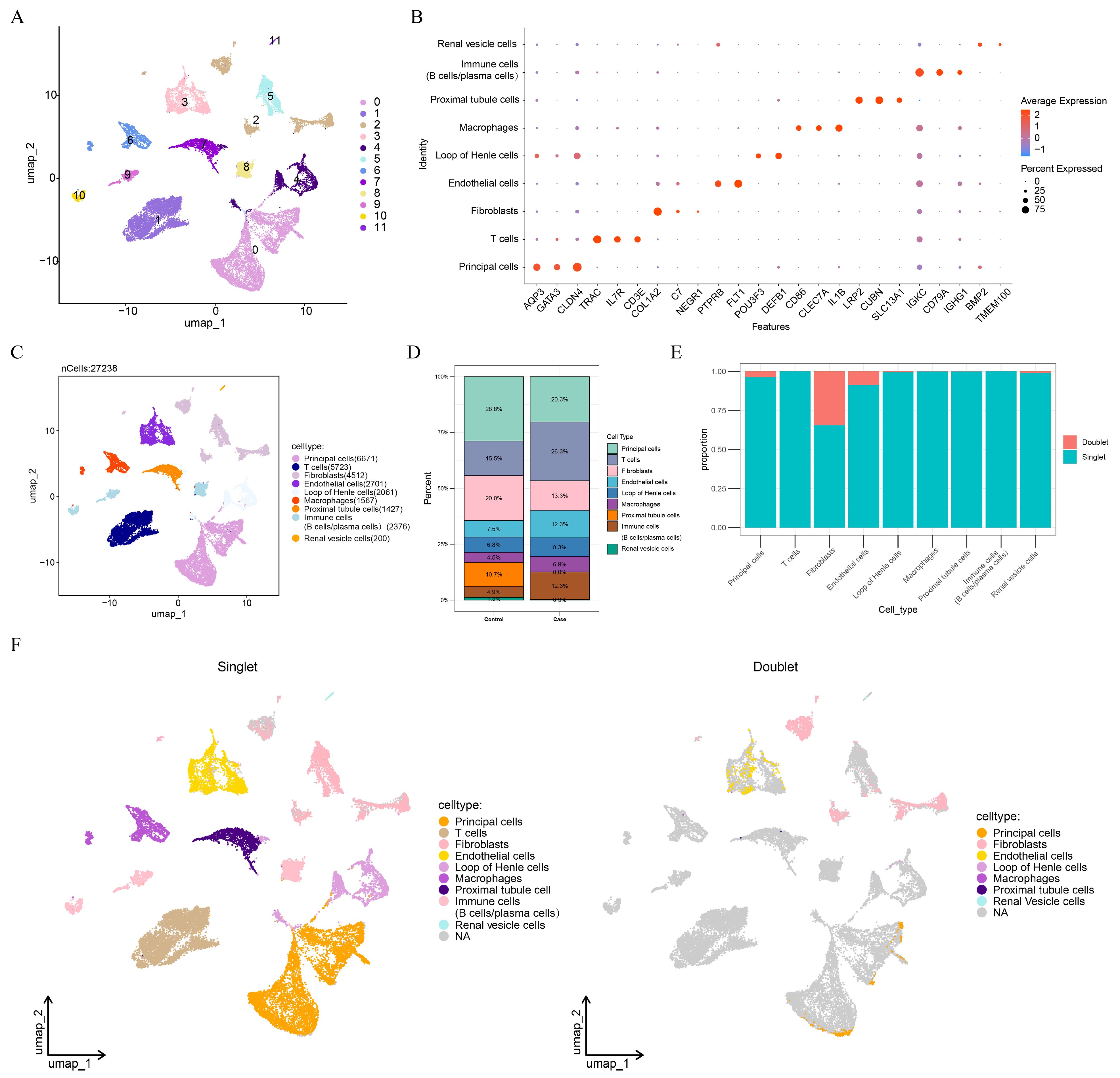
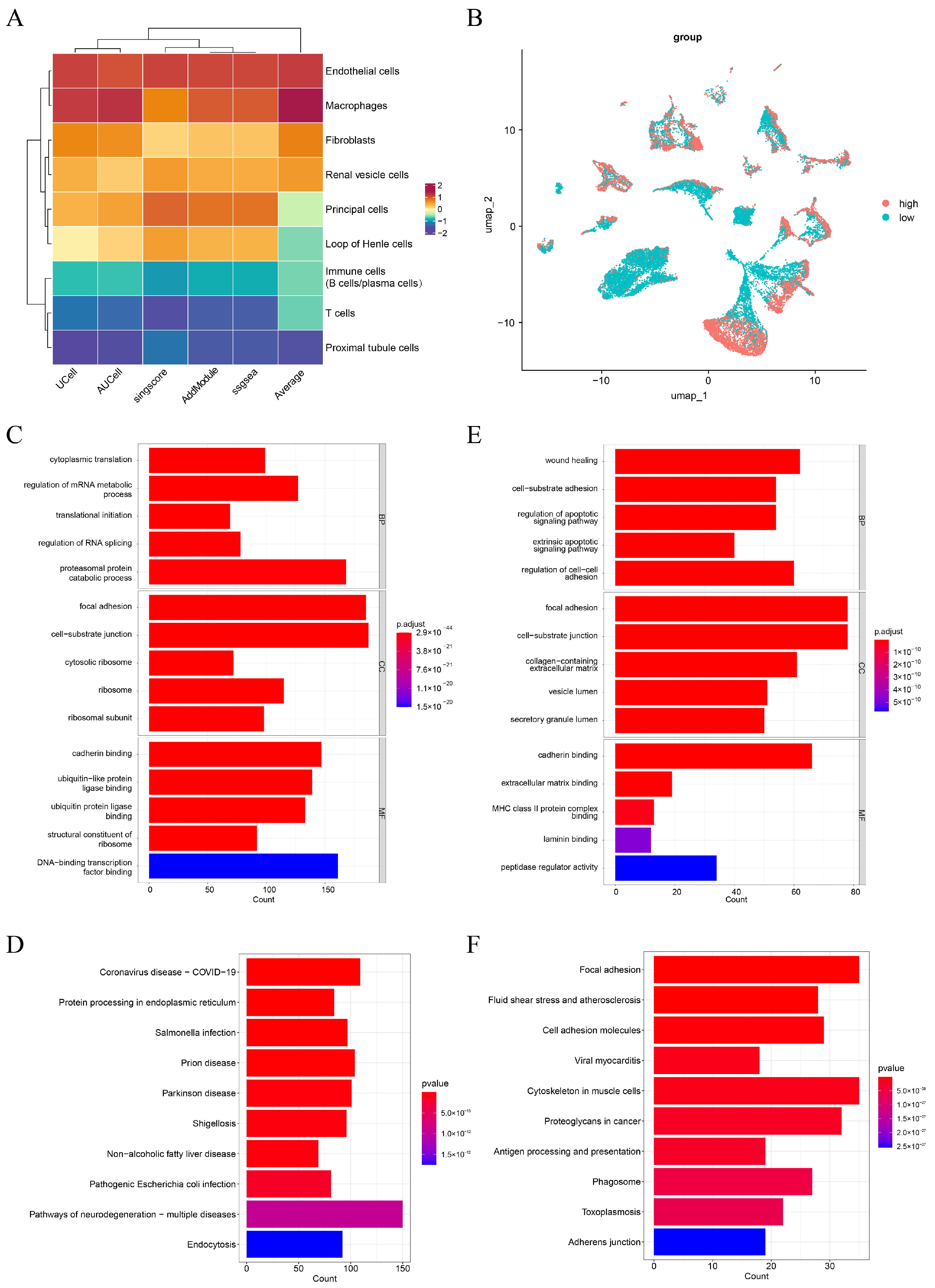
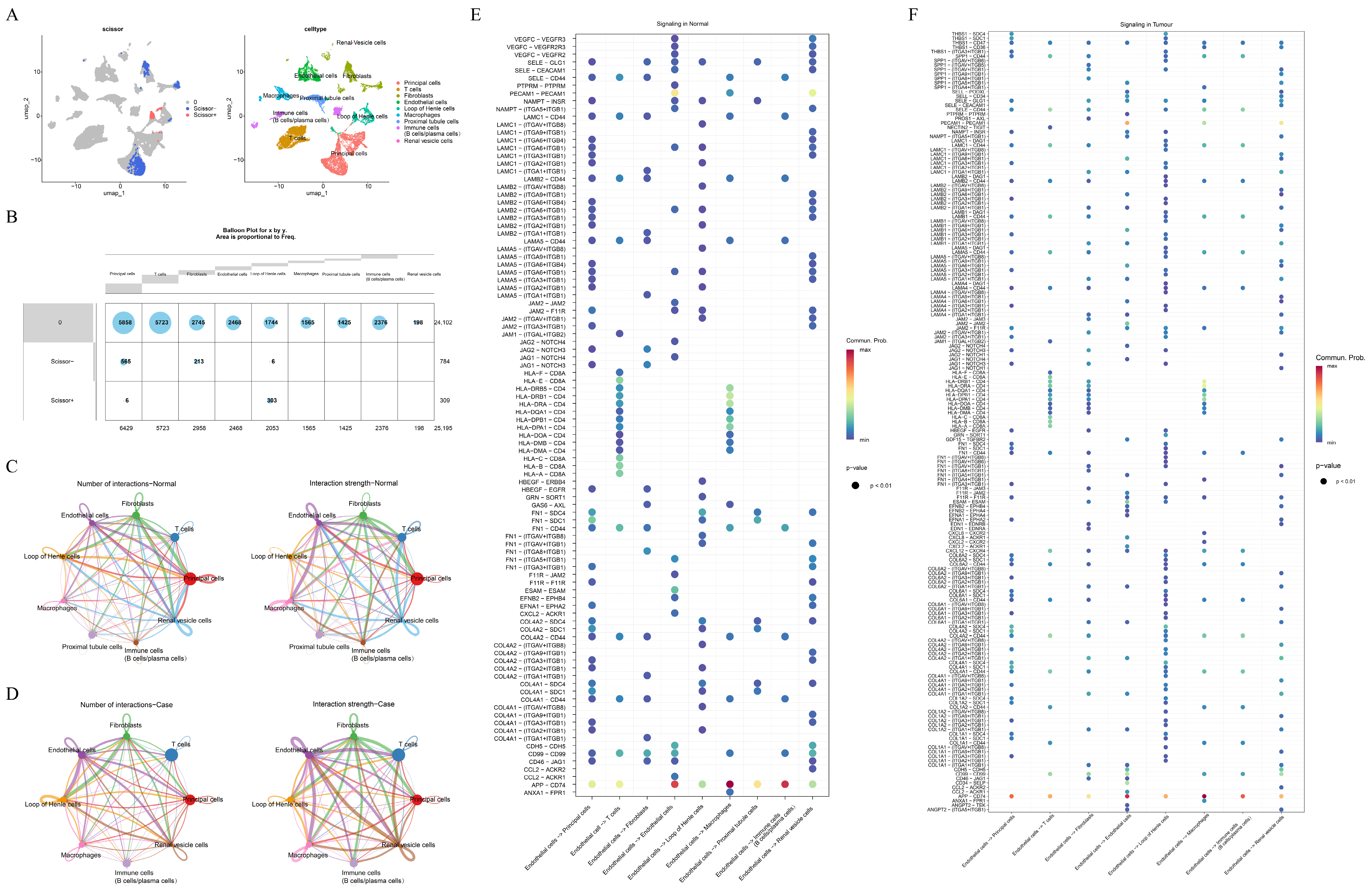
3.1. Endothelial Cells Were Selected as Key Cells in Caox Kidney Stones
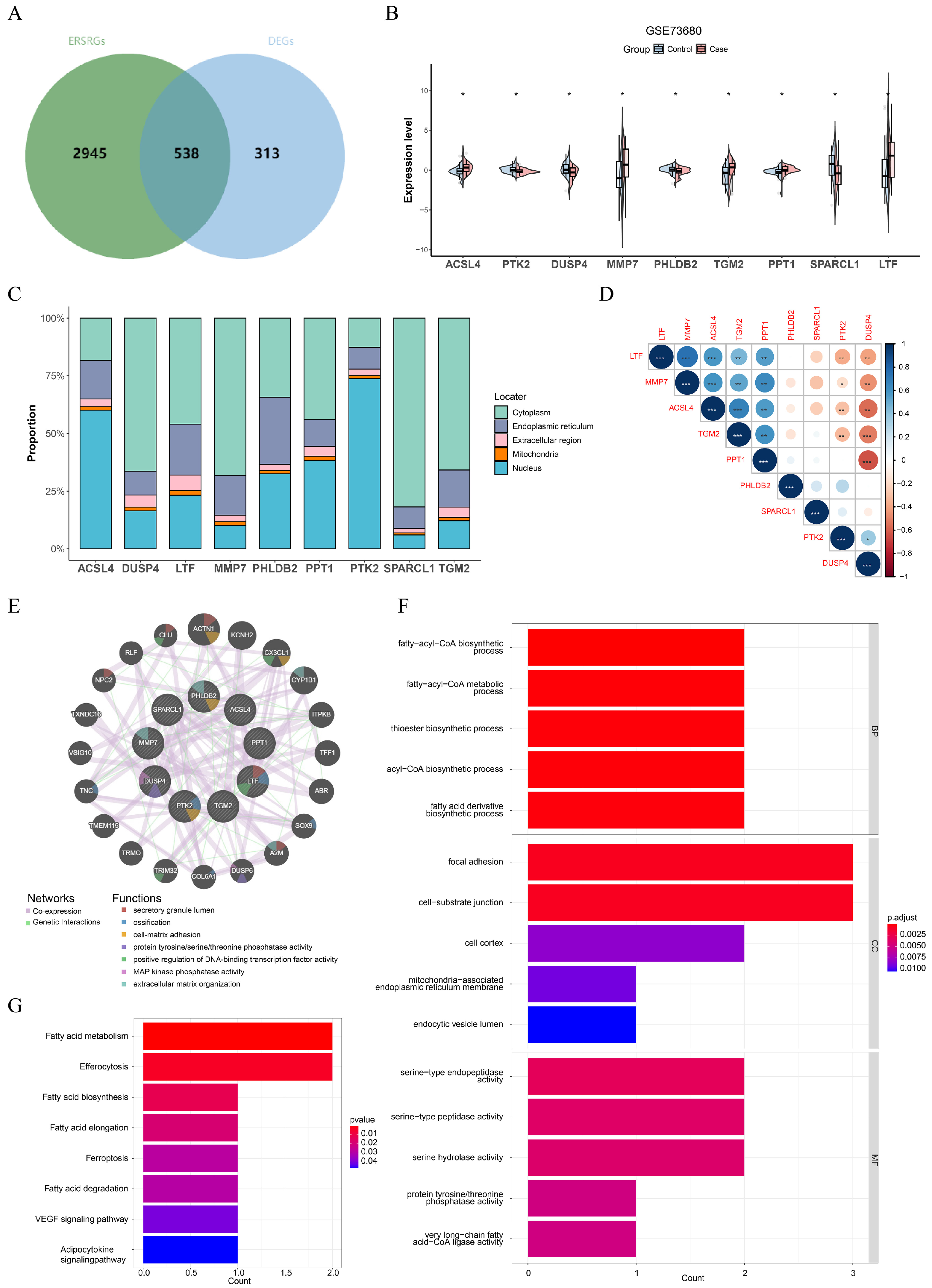
3.2. ACSL4, PTK2, DUSP4, MMP7, PHLDB2, TGM2, PPT1, SPARCL1, and LTF Were Identified as Key Genes in CaOx Kidney Stones
3.3. Key Genes Accurately Predicted the Risk of Caox Kidney Stones
3.4. Regulatory Networks and Potential Therapeutic Drugs of Key Genes
3.5. The Differential Expression of Key Genes in Key Cell Subtypes
3.6. Key Gene mRNA Expression (qRT-PCR)
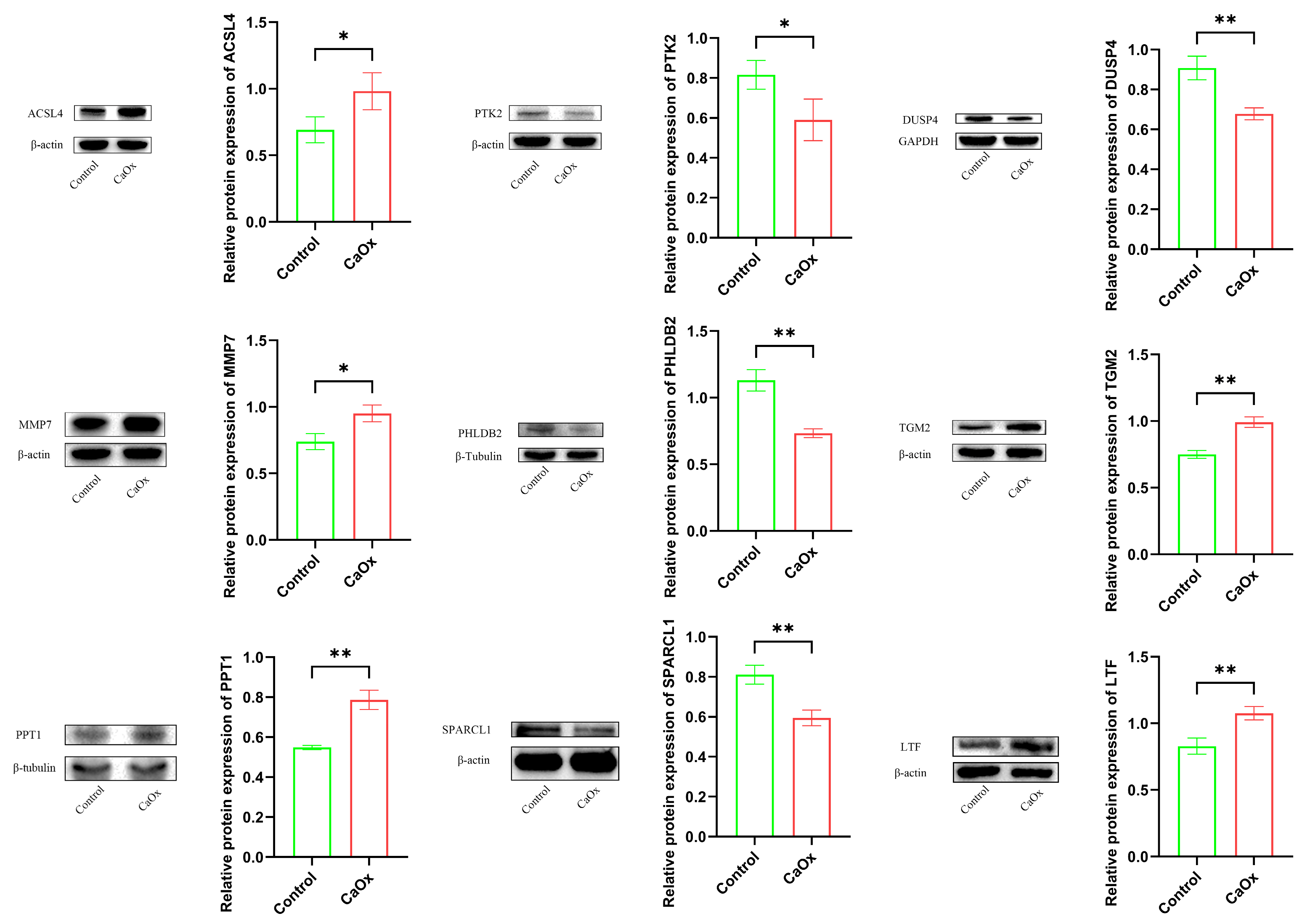
3.7. Key Protein Expression Analysis (Western Blotting)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Popova, E.; Tkachev, S.; Shapoval, A.; Karpenko, A.; Lee, Y.; Chislov, P.; Ershov, B.; Golub, D.; Galechyan, G.; Bogoedov, D.; et al. Kidney Stones as Minerals: How Methods from Geology Could Inform Urolithiasis Treatment. J. Clin. Med. 2025, 14, 997. [Google Scholar] [CrossRef]
- Gianvincenzo, P.D.; Leyes, M.F.; Boonkam, K.; Puentes, A.F.; Reyes, S.G.; Nardi, A.N.; Olivieri, A.; Pummarin, S.; Kamonsutthipaijit, N.; Amenitsch, H.; et al. Supramolecular citrate poly allylamine hydrochloride nanoparticles for citrate delivery and calcium oxalate nanocrystal dissolution. J. Colloid Interface Sci. 2024, 669, 667–678. [Google Scholar] [CrossRef]
- Thongprayoon, C.; Krambeck, A.E.; Rule, A.D. Determining the true burden of kidney stone disease. Nat. Rev. Nephrol. 2020, 16, 736–746. [Google Scholar] [CrossRef]
- Khan, A. Prevalence, pathophysiological mechanisms and factors affecting urolithiasis. Int. Urol. Nephrol. 2018, 50, 799–806. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Amier, Y.; Yuan, D.; Xun, Y.; Yu, X. Advancements in Nanomedicine for the Diagnosis and Treatment of Kidney Stones. Int. J. Nanomed. 2025, 20, 1401–1423. [Google Scholar] [CrossRef]
- Yan, X.; Xia, Y.; Li, B.; Ye, Z.; Li, L.; Yuan, T.; Song, B.; Yu, W.; Rao, T.; Ning, J.; et al. The SOX4/EZH2/SLC7A11 signaling axis mediates ferroptosis in calcium oxalate crystal deposition-induced kidney injury. J. Transl. Med. 2024, 22, 9. [Google Scholar] [CrossRef]
- Khan, S.R. Reactive oxygen species, inflammation and calcium oxalate nephrolithiasis. Transl. Androl. Urol. 2014, 3, 256–276. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, J.; Wang, S.; Zhao, Y.; Wang, H.; Wang, Y.; Ma, Y.; Yang, J. Endoplasmic Reticulum Stress: Triggers Microenvironmental Regulation and Drives Tumor Evolution. Cancer Med. 2025, 14, e70684. [Google Scholar] [CrossRef]
- Wiese, W.; Galita, G.; Siwecka, N.; Rozpędek-Kamińska, W.; Slupianek, A.; Majsterek, I. Endoplasmic Reticulum Stress in Acute Myeloid Leukemia: Pathogenesis, Prognostic Implications, and Therapeutic Strategies. Int. J. Mol. Sci. 2025, 26, 3092. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, C.; Ji, L.; Zhang, H. Endoplasmic reticulum stress in acute pancreatitis: Exploring the molecular mechanisms and therapeutic targets. Cell Stress Chaperones 2025, 30, 119–129. [Google Scholar] [CrossRef]
- Sun, Y.; Kang, J.; Guan, X.; Xu, H.; Wang, X.; Deng, Y. Regulation of endoplasmic reticulum stress on the damage and apoptosis of renal tubular epithelial cells induced by calcium oxalate crystals. Urolithiasis 2021, 49, 291–299. [Google Scholar] [CrossRef]
- Sun, Y.; Kang, J.; Tao, Z.; Wang, X.; Liu, Q.; Li, D.; Guan, X.; Xu, H.; Liu, Y.; Deng, Y. Effect of endoplasmic reticulum stress-mediated excessive autophagy on apoptosis and formation of kidney stones. Life Sci. 2020, 244, 117232. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Hamamoto, S.; Okada, A.; Unno, R.; Kamisawa, H.; Naiki, T.; Ando, R.; Mizuno, K.; Kawai, N.; Tozawa, K.; et al. Genome-Wide Gene Expression Profiling of Randall’s Plaques in Calcium Oxalate Stone Formers. J. Am. Soc. Nephrol. 2017, 28, 333–347. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e324. [Google Scholar] [CrossRef]
- Chen, G.; Qi, H.; Jiang, L.; Sun, S.; Zhang, J.; Yu, J.; Liu, F.; Zhang, Y.; Du, S. Integrating single-cell RNA-Seq and machine learning to dissect tryptophan metabolism in ulcerative colitis. J. Transl. Med. 2024, 22, 1121. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Lv, M.; Luo, Y.; Ke, K.; Zhang, B.; Wang, M.; Zhang, K.; Li, H. Mechanistic studies of Ca2+-induced classical pyroptosis pathway promoting renal adhesion on calcium oxalate kidney stone formation. Sci. Rep. 2025, 15, 6669. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yang, R.; Xie, L.; Wang, R.; Li, Y.; Lu, Y.; Liu, B.; Dai, S.; Zheng, S.; Dong, K.; Dong, R. Integration of single-nuclei and spatial transcriptomics to decipher tumor phenotype predictive of relapse-free survival in Wilms tumor. Front. Immunol. 2025, 16, 1539897. [Google Scholar] [CrossRef]
- Sun, D.; Guan, X.; Moran, A.E.; Wu, L.Y.; Qian, D.Z.; Schedin, P.; Dai, M.S.; Danilov, A.V.; Alumkal, J.J.; Adey, A.C.; et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2022, 40, 527–538. [Google Scholar] [CrossRef]
- Cao, L.; Lu, X.; Wang, X.; Wu, H.; Miao, X. From single-cell to spatial transcriptomics: Decoding the glioma stem cell niche and its clinical implications. Front. Immunol. 2024, 15, 1475235. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, J.; Pi, J.; Hu, D.; Xu, J.; Zhao, Y.; Wang, Y. Identification and Validation of Genes Related to Macrophage Polarization and Cell Death Modes Under Mycobacterium tuberculosis Infection. J. Inflamm. Res. 2024, 17, 1397–1411. [Google Scholar] [CrossRef]
- Tang, Q.; Nie, F.; Kang, J.; Chen, W. mRNALocater: Enhance the prediction accuracy of eukaryotic mRNA subcellular localization by using model fusion strategy. Mol. Ther. 2021, 29, 2617–2623. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Bao, E.; Yang, Y.; Jiang, B.; Wang, B.; Liu, Y.; Yang, L.; Xia, L.; Zhu, P. An exploratory study evaluated the 30 most commonly reported medications in the United States food and drug administration’s adverse event reporting system that are associated with the occurrence of kidney stones. Front. Pharmacol. 2024, 15, 1377679. [Google Scholar] [CrossRef]
- Alexander, R.T.; McArthur, E.; Jandoc, R.; Welk, B.; Hayward, J.S.; Jain, A.K.; Braam, B.; Flockerzi, V.; Garg, A.X.; Quinn, R.R. Antihypertensive medications and the risk of kidney stones in older adults: A retrospective cohort study. Hypertens. Res. 2017, 40, 837–842. [Google Scholar] [CrossRef]
- Ali, A.I.; Abdelfadel, A.; Rohiem, M.F.; Hassan, A. Semirigid ureteroscopy and tamsulosin therapy as dilatation methods before flexible ureteroscopy: Evaluation and benefits. World J. Urol. 2024, 42, 75. [Google Scholar] [CrossRef]
- Dong, C.; He, Z.; Liao, W.; Jiang, Q.; Song, C.; Song, Q.; Su, X.; Xiong, Y.; Wang, Y.; Meng, L.; et al. CHAC1 Mediates Endoplasmic Reticulum Stress-Dependent Ferroptosis in Calcium Oxalate Kidney Stone Formation. Adv. Sci. 2025, 12, e2403992. [Google Scholar] [CrossRef]
- Dong, F.; Jiang, S.; Tang, C.; Wang, X.; Ren, X.; Wei, Q.; Tian, J.; Hu, W.; Guo, J.; Fu, X.; et al. Trimethylamine N-oxide promotes hyperoxaluria-induced calcium oxalate deposition and kidney injury by activating autophagy. Free Radic. Biol. Med. 2022, 179, 288–300. [Google Scholar] [CrossRef]
- Wang, B.; Tan, Z.; She, W.; Wang, X.; Guan, X.; Tao, Z.; Guo, F.; Xu, H.; Deng, Y. Characterizing Chemokine Signaling Pathways and Hub Genes in Calcium Oxalate-Induced Kidney Stone Formation: Insights from Rodent Models. Biochem. Genet. 2025. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, Y.; Kang, J.; He, Z.; Liu, Q.; Wu, J.; Li, D.; Wang, X.; Tao, Z.; Guan, X.; et al. Role of ROS-Induced NLRP3 Inflammasome Activation in the Formation of Calcium Oxalate Nephrolithiasis. Front. Immunol. 2022, 13, 818625. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.E.; Brunt, V.E.; Shiu, Y.T.; Bunsawat, K. Endothelial dysfunction in chronic kidney disease: A clinical perspective. Am. J. Physiol. Heart Circ. Physiol. 2025, 329, H135–H153. [Google Scholar] [CrossRef]
- Wang, J.H.; Lin, Y.L.; Hsu, B.G. Endothelial dysfunction in chronic kidney disease: Mechanisms, biomarkers, diagnostics, and therapeutic strategies. Tzu Chi Med. J. 2025, 37, 125–134. [Google Scholar] [CrossRef]
- Lee, H.J.; Jeong, S.J.; Park, M.N.; Linnes, M.; Han, H.J.; Kim, J.H.; Lieske, J.C.; Kim, S.H. Gallotannin suppresses calcium oxalate crystal binding and oxalate-induced oxidative stress in renal epithelial cells. Biol. Pharm. Bull. 2012, 35, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Huang, F.; Gao, M.; Liu, M.; Zhang, Y.; Tang, L.; Wu, J.; Yu, H.; He, C.; Chen, J.; et al. Osteogenic-Like Microenvironment of Renal Interstitium Induced by Osteomodulin Contributes to Randall’s Plaque Formation. Adv. Sci. 2024, 11, e2405875. [Google Scholar] [CrossRef]
- Sun, Y.; Li, B.; Song, B.; Xia, Y.; Zhou, X.; Lin, F.; Rao, T.; Cheng, F. CREB1/CRTC2 regulated tubular epithelial-derived exosomal miR-93-3p promotes kidney injury induced by calcium oxalate via activating M1 polarization and macrophage extracellular trap formation. J. Nanobiotechnol. 2025, 23, 204. [Google Scholar] [CrossRef]
- Sun, K.; Shi, Y.; Yan, C.; Wang, S.; Han, L.; Li, F.; Xu, X.; Wang, Y.; Sun, J.; Kang, Z.; et al. Glycolysis-Derived Lactate Induces ACSL4 Expression and Lactylation to Activate Ferroptosis during Intervertebral Disc Degeneration. Adv. Sci. 2025, 12, e2416149. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, X.; Jing, D.; Lin, Y.; Gao, R.; Zhao, Q.; Da, H.; Ren, Y.; Cao, Q.; Liu, N.; et al. ACSL4 knockdown inhibits colorectal cancer progression through stimulating anti-tumor immunity. Neoplasia 2025, 67, 101194. [Google Scholar] [CrossRef]
- Li, L.; Ye, Z.; Xia, Y.; Li, B.; Chen, L.; Yan, X.; Yuan, T.; Song, B.; Yu, W.; Rao, T.; et al. YAP/ACSL4 Pathway-Mediated Ferroptosis Promotes Renal Fibrosis in the Presence of Kidney Stones. Biomedicines 2023, 11, 2692. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Zhou, K.; Huang, M.; Sun, Y.; Kang, J.; Su, Q.; Zhao, Y.; Liu, Q.; Li, C. Ferroptosis in calcium oxalate kidney stone formation and the possible regulatory mechanism of ANKRD1. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119452. [Google Scholar] [CrossRef]
- Hirohama, D.; Abedini, A.; Moon, S.; Surapaneni, A.; Dillon, S.T.; Vassalotti, A.; Liu, H.; Doke, T.; Martinez, V.; Md Dom, Z.; et al. Unbiased Human Kidney Tissue Proteomics Identifies Matrix Metalloproteinase 7 as a Kidney Disease Biomarker. J. Am. Soc. Nephrol. 2023, 34, 1279–1291. [Google Scholar] [CrossRef]
- Avello, A.; Guerrero-Mauvecin, J.; Sanz, A.B. Urine MMP7 as a kidney injury biomarker. Clin. Kidney J. 2024, 17, sfad233. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Liang, H.; Liu, X.; Liu, H.; Zhuang, Z.; Hou, J. Depletion of PHLDB2 Suppresses Epithelial-Mesenchymal Transition and Enhances Anti-Tumor Immunity in Head and Neck Squamous Cell Carcinoma. Biomolecules 2024, 14, 232. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Zheng, Q.; Lu, Z.; Chen, Y.; Shen, D.; Xue, D.; Jiang, M.; Ding, L.; Zhang, J.; et al. Oncometabolite L-2-hydroxyglurate directly induces vasculogenic mimicry through PHLDB2 in renal cell carcinoma. Int. J. Cancer 2021, 148, 1743–1755. [Google Scholar] [CrossRef]
- Supianto, M.; Yoo, D.K.; Hwang, H.; Oh, H.B.; Jhung, S.H.; Lee, H.J. Linker-Preserved Iron Metal-Organic Framework-Based Lateral Flow Assay for Sensitive Transglutaminase 2 Detection in Urine Through Machine Learning-Assisted Colorimetric Analysis. ACS Sens. 2024, 9, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.A.; Sayyed, N.D.; Lee, Y.J.; Jeon, H.Y.; Kim, E.B.; Hong, S.H.; Cho, S.; Kim, M.; Ha, K.S. Midazolam Ameliorates Hyperglycemia-Induced Glomerular Endothelial Dysfunction by Inhibiting Transglutaminase 2 in Diabetes. Int. J. Mol. Sci. 2022, 23, 753. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wang, W.G.; Cao, J.; Hu, X.C. SPARCL1 suppresses cell migration and invasion in renal cell carcinoma. Mol. Med. Rep. 2017, 16, 7784–7790. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Q.; Lv, S.; Qin, L. Prognostic value of serum lipids in newly diagnosed acute promyelocytic leukemia. Front. Oncol. 2025, 15, 1522239. [Google Scholar] [CrossRef]
- Westphal, S.; Stoppe, C.; Gruenewald, M.; Bein, B.; Renner, J.; Cremer, J.; Coburn, M.; Schaelte, G.; Boening, A.; Niemann, B.; et al. Genome-wide association study of myocardial infarction, atrial fibrillation, acute stroke, acute kidney injury and delirium after cardiac surgery—A sub-analysis of the RIPHeart-Study. BMC Cardiovasc. Disord. 2019, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, N.; Zhang, B.; Zhuang, P.; Tan, B.; Cai, C.; He, N.; Nie, H.; Xiang, S.; Chen, C. Identification of aging-related biomarkers and immune infiltration analysis in renal stones by integrated bioinformatics analysis. Sci. Rep. 2025, 15, 21650. [Google Scholar] [CrossRef]
- Zhu, Z.; Zuo, S.; Zhu, Z.; Wang, C.; Du, Y.; Chen, F. THSWD upregulates the LTF/AMPK/mTOR/Becn1 axis and promotes lysosomal autophagy in hepatocellular carcinoma cells by regulating gut flora and metabolic reprogramming. Int. Immunopharmacol. 2025, 148, 114091. [Google Scholar] [CrossRef]
- Tian, T.; Han, H.; Huang, J.; Ma, J.; Ran, R. DBI as a Novel Immunotherapeutic Candidate in Colorectal Cancer: Dissecting Genetic Risk and the Immune Landscape via GWAS, eQTL, and pQTL. Biomedicines 2025, 13, 1115. [Google Scholar] [CrossRef]
- Chiu, I.J.; Hsu, Y.H.; Chang, J.S.; Yang, J.C.; Chiu, H.W.; Lin, Y.F. Lactotransferrin Downregulation Drives the Metastatic Progression in Clear Cell Renal Cell Carcinoma. Cancers 2020, 12, 847. [Google Scholar] [CrossRef]
- Zhu, G.; Jin, L.; Guo, Y.; Sun, L.; Li, S.; Zhou, F. Establishment and application of a nomogram diagram for predicting calcium oxalate stones in patients with urinary tract stones. Urolithiasis 2024, 52, 40. [Google Scholar] [CrossRef]
- Maßberg, D.; Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef]
- Shepard, B.D.; Pluznick, J.L. How does your kidney smell? Emerging roles for olfactory receptors in renal function. Pediatr. Nephrol. 2016, 31, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wei, H.; Huang, G.; Jin, L. CCL7 and olfactory transduction pathway activation play an important role in the formation of CaOx and CaP kidney stones. Front. Genet. 2023, 14, 1267545. [Google Scholar] [CrossRef] [PubMed]
- Motahharynia, A.; Moein, S.; Kiyanpour, F.; Moradzadeh, K.; Yaqubi, M.; Gheisari, Y. Olfactory receptors contribute to progression of kidney fibrosis. NPJ Syst. Biol. Appl. 2022, 8, 8. [Google Scholar] [CrossRef]
| Gene Name | Forward Sequence (5′ → 3′) | Reverse Sequence (5′ → 3′) |
|---|---|---|
| ACSL4 | GTGAAAGAATACCTGGACTGG | AGAGAGTGTAAGCGGAGAAG |
| PTK2 | CAGTATTGACAGGGAGGATG | AGGCGGTTTCTTTGGTGGAG |
| DUSP4 | GCGTCAGTCCAATAGGTCAG | CAGAAACTTCCCATCACCAG |
| MMP7 | AGCTCATGGGGACTCCTACC | GTCCAGCGTTCATCCTCATC |
| PHLDB2 | CTGAATATCAACGGAACATCG | TCTCTCTGAGCCTGCTGAAC |
| TGM2 | GAAGGAGGAGACAGGGATGG | CAGCGGTGTTGTTGGTGATG |
| PPT1 | GTATCGCAACCACAGCATCT | TCCGAATCTACAGGGTCCAC |
| SPARCL1 | ACGGTAGCACCTGACAACAC | ATGGTGGGAATCGTCTTCTGT |
| LTF | GAACCGTACTTCAGCTACTCTG | CTCATACTCGTCCCTTTCAGC |
| β-actin | CCTGGCACCCAGCACAAT | GGGCCGGACTCGTCATAC |
| ID | FastaID | mRNALocater | Cytoplasm | Endoplasmic Reticulum | Extracellular Region | Mitochondria | Nucleus | Protein Localization Annotation (From Uniprot) |
|---|---|---|---|---|---|---|---|---|
| ACSL4 | NC_000023.11:c109733257-109641335 | Nucleus | 0.1836 | 0.167 | 0.0336 | 0.0152 | 0.6006 | Mitochondrion outer membrane, Microsome membrane, Endoplasmic reticulum membrane, Cell membrane |
| DUSP4 | NC_000008.11:c29350684-29333064 | Cytoplasm | 0.6639 | 0.1029 | 0.0533 | 0.0151 | 0.1648 | Nucleus |
| LTF | NC_000003.12:c46485234-46435645 | Cytoplasm | 0.46 | 0.2209 | 0.0668 | 0.0204 | 0.2318 | Isoform 1, Secreted, Cytoplasmic granule, Note: Secreted into most exocrine fluids by various endothelial cells. Stored in the secondary gran-ules of neutro-phils. Isoform DeltaLf, Cyto-plasm, Nucleus, Note: Mainly lo-calized in the cy-toplasm. |
| MMP7 | NC_000011.10:c102530747-102520508 | Cytoplasm | 0.6825 | 0.1725 | 0.0277 | 0.0164 | 0.1009 | Secreted, extracel-lular space, extra-cellular matrix |
| PHLDB2 | NC_000003.12:111732496-111976517 | Cytoplasm | 0.3434 | 0.2906 | 0.0277 | 0.0134 | 0.3249 | Cytoplasm, Cytoplasm, cell cortex, Membrane; Peripheral membrane protein |
| PPT1 | NC_000001.11:c40097252-40071461 | Cytoplasm | 0.4397 | 0.1168 | 0.0428 | 0.0177 | 0.383 | Cell projection, podosome, Note: Translocates to the plasma membrane at high levels of PtdIns-(3,4,5)-P3. At low levels of PtdIns-(3,4,5)-P3 is translocated to vesicular compartments. Localized to the myotube podosome cortex that surrounds the core Lysosome, Secreted, Golgi apparatus, Endoplasmic retic-ulum |
| PTK2 | NC_000008.11:c141002079-140657900 | Nucleus | 0.1269 | 0.0943 | 0.0287 | 0.0123 | 0.7378 | Cell junction, focal adhesion, Cell membrane by simi-larity; Peripheral membrane protein, Cytoplasm, peri-nuclear region, Cy-toplasm, cell cortex, Cytoplasm, cyto-skeleton, Cyto-plasm, cytoskeleton, microtubule organ-izing center, cen-trosome, Nucleus, Cytoplasm, cyto-skeleton, cilium ba-sal body, Cyto-plasm, Note: Con-stituent of focal ad-hesions. Detected at microtubules. |
| SPARCL1 | NC_000004.12:c87529376-87473335 | Cytoplasm | 0.8185 | 0.0933 | 0.0195 | 0.0092 | 0.0595 | Secreted, extracellular space, extracellular matrix |
| TGM2 | NC_000020.11:c38168475-38127385 | Cytoplasm | 0.6584 | 0.1614 | 0.0438 | 0.0153 | 0.121 | Cytoplasm, cytosol, Nucleus, Chromosome, Secreted, extracellular space, extracellular matrix, Cell membrane, Mitochondrion, Note: Mainly localizes to the cytosol, present at much lower level in the nucleus and chromatin, Also secreted via a non-classical secretion pathway to the extracellular matrix, Isoform 2, Cytoplasm, perinuclear region |
| Gene Symbol | UniProt | Chemical Name | Kcal/mol |
|---|---|---|---|
| ACSL4 | O60488 | TILFRINIB | −8.3 |
| PTK2 | Q05397 | VS-4718 | −7 |
| MMP7 | P09237 | RS 39066 | −7.9 |
| TGM2 | P21980 | LEVAMISOLE | −5.9 |
| PPT1 | P50897 | PLITIDEPSIN | −12.8 |
| LTF | P02788 | TALACTOFERRIN ALFA | −7.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Z.; She, W.; Wang, B.; Wang, X.; Guan, X.; Tao, Z.; Deng, Y. Identification of Key Genes Associated with Endoplasmic Reticulum Stress in Calcium Oxalate Kidney Stones. Genes 2025, 16, 1338. https://doi.org/10.3390/genes16111338
Tan Z, She W, Wang B, Wang X, Guan X, Tao Z, Deng Y. Identification of Key Genes Associated with Endoplasmic Reticulum Stress in Calcium Oxalate Kidney Stones. Genes. 2025; 16(11):1338. https://doi.org/10.3390/genes16111338
Chicago/Turabian StyleTan, Zhenkun, Wusheng She, Boqiang Wang, Xiang Wang, Xiaofeng Guan, Zhiwei Tao, and Yaoliang Deng. 2025. "Identification of Key Genes Associated with Endoplasmic Reticulum Stress in Calcium Oxalate Kidney Stones" Genes 16, no. 11: 1338. https://doi.org/10.3390/genes16111338
APA StyleTan, Z., She, W., Wang, B., Wang, X., Guan, X., Tao, Z., & Deng, Y. (2025). Identification of Key Genes Associated with Endoplasmic Reticulum Stress in Calcium Oxalate Kidney Stones. Genes, 16(11), 1338. https://doi.org/10.3390/genes16111338






