Conservation and Tandem Duplication of tRNA Genes in Plant Species
Abstract
1. Introduction
2. Materials and Methods
2.1. tRNA-Coding Gene Identification
2.2. Sequence Alignment and Kn/Ks Estimation
2.3. GC Content Calculation
2.4. Phylogenetic Analysis of tRNA Genes
2.5. Identification of Tandem Duplication Event in tRNA Genes
3. Results
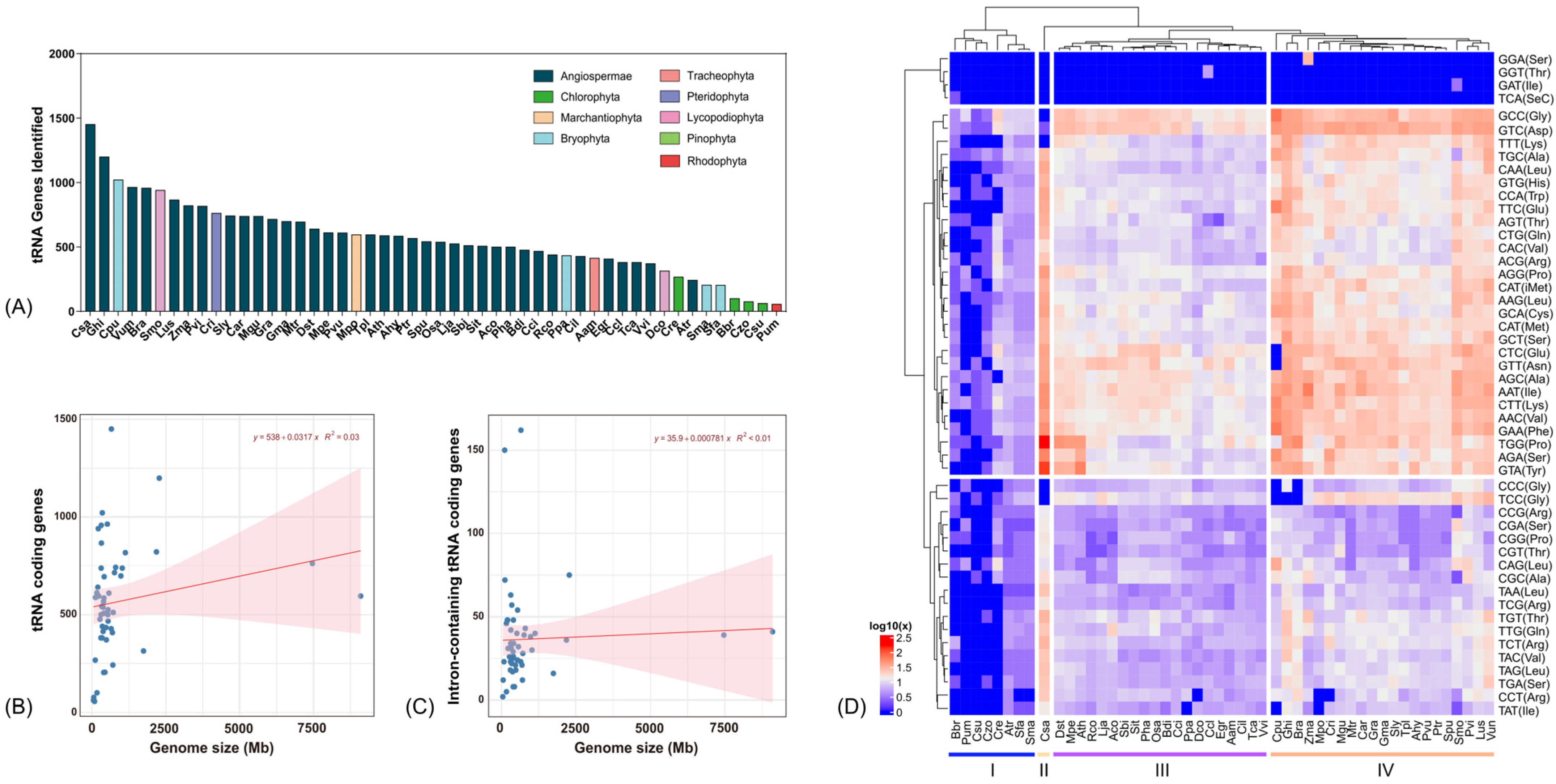
3.1. tRNA-Coding Genes in 50 Plant Species
3.2. Intron-Containing tRNAs in Plants
3.3. The GC Content of tRNA-Coding Genes
3.4. Phylogenetic Analysis of tRNA-Coding Genes
3.5. Identification of Tandem Duplication Event During tRNA Gene Expansion
3.6. Conservation of Proline tRNA Isotypes Among Different Plant Species
4. Discussion
4.1. Conservation of tRNA Genes Among Plant Species
4.2. Tandem Duplication Is a Universal Driving Force for the tRNA Gene Evolution
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reynolds, N.M.; Vargas-Rodriguez, O.; Söll, D.; Crnković, A. The central role of tRNA in genetic code expansion. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 3001–3008. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N. Studies on nucleic acids of living fossils. J. Biochem. 1971, 69, 761–770. [Google Scholar] [CrossRef]
- Agrawal, S.; Karcher, D.; Ruf, S.; Bock, R. The functions of chloroplast glutamyl-tRNA in translation and tetrapyrrole biosynthesis. Plant Physiol. 2020, 183, 263–276. [Google Scholar] [CrossRef]
- Liu, B.; Cao, J.; Wang, X.; Guo, C.; Liu, Y.; Wang, T. Deciphering the tRNA-derived small RNAs: Origin, development, and future. Cell Death Dis. 2021, 13, 24. [Google Scholar] [CrossRef]
- Wilusz, J.E. Controlling translation via modulation of tRNA levels. Wiley Interdiscip. Rev. RNA 2015, 6, 453–470. [Google Scholar] [CrossRef]
- Zhang, W.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H. tRNAs as regulators in gene expression. Sci. China Ser. C Life Sci. 2009, 52, 245–252. [Google Scholar] [CrossRef]
- Chery, M.; Drouard, L. Plant tRNA functions beyond their major role in translation. J. Exp. Bot. 2023, 74, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Santana, C.; Attolini, C.S.O.; Kirsten, T.; Engelhardt, J.; Prohaska, S.J.; Steigele, S.; Stadler, P.F. Genomic organization of eukaryotic tRNAs. BMC Genom. 2010, 11, 270. [Google Scholar] [CrossRef]
- Michaud, M.; Cognat, V.; Duchêne, A.M.; Maréchal-Drouard, L. A global picture of tRNA genes in plant genomes. Plant J. 2011, 66, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.; Ibba, M. Bacterial transfer RNAs. FEMS Microbiol. Rev. 2015, 39, 280–300. [Google Scholar] [CrossRef]
- Monloy, K.C.; Planta, J. tRNA gene content, structure, and organization in the flowering plant lineage. Front. Plant Sci. 2024, 15, 1486612. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. The Origins of Genome Architecture; Sinauer Associates: Sunderland, MA, USA, 2007. [Google Scholar]
- Tang, H. Genome assembly, rearrangement, and repeats. Chem. Rev. 2007, 107, 3391–3406. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Ayan, G.B.; Park, H.J.; Gallie, J. The birth of a bacterial tRNA gene by large-scale, tandem duplication events. Elife 2020, 9, e57947. [Google Scholar] [CrossRef]
- Theologis, A.; Ecker, J.R.; Palm, C.J.; Federspiel, N.A.; Kaul, S.; White, O.; Alonso, J.; Altafi, H.; Araujo, R.; Bowman, C.L.; et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 2020, 408, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; Lindemann, B.F.; Tietze, M.; Winkler-Oswatitsch, R.; Dress, A.; Von Haeseler, A. How old is the genetic code? Statistical geometry of tRNA provides an answer. Science 1989, 244, 673–679. [Google Scholar] [CrossRef]
- Kim, Y.; Opron, K.; Burton, Z.F. A tRNA-and anticodon-centric view of the evolution of aminoacyl-tRNA synthetases, tRNAomes, and the genetic code. Life 2019, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Burton, Z.F. Evolution of life on Earth: tRNA, aminoacyl-tRNA synthetases and the genetic code. Life 2020, 10, 21. [Google Scholar] [CrossRef]
- Masta, S.E.; Boore, J.L. Parallel evolution of truncated transfer RNA genes in Arachnid mitochondrial genomes. Mol. Biol. Evol. 2008, 25, 949–959. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Cognat, V.; Pawlak, G.; Pflieger, D.; Drouard, L. PlantRNA 2.0: An updated database dedicated to tRNAs of photosynthetic eukaryotes. Plant J. 2022, 112, 1112–1119. [Google Scholar] [CrossRef]
- Komatsu, R.; Sawada, R.; Umehara, T.; Tamura, K. Proline might have been the first amino acid in the primitive genetic code. J. Mol. Evol. 2014, 78, 310–312. [Google Scholar] [CrossRef]
- Morgado, S.M.; Vicente, A.C.P. Exploring tRNA gene cluster in archaea. Mem. Inst. Oswaldo Cruz 2019, 114, e180348. [Google Scholar] [CrossRef] [PubMed]


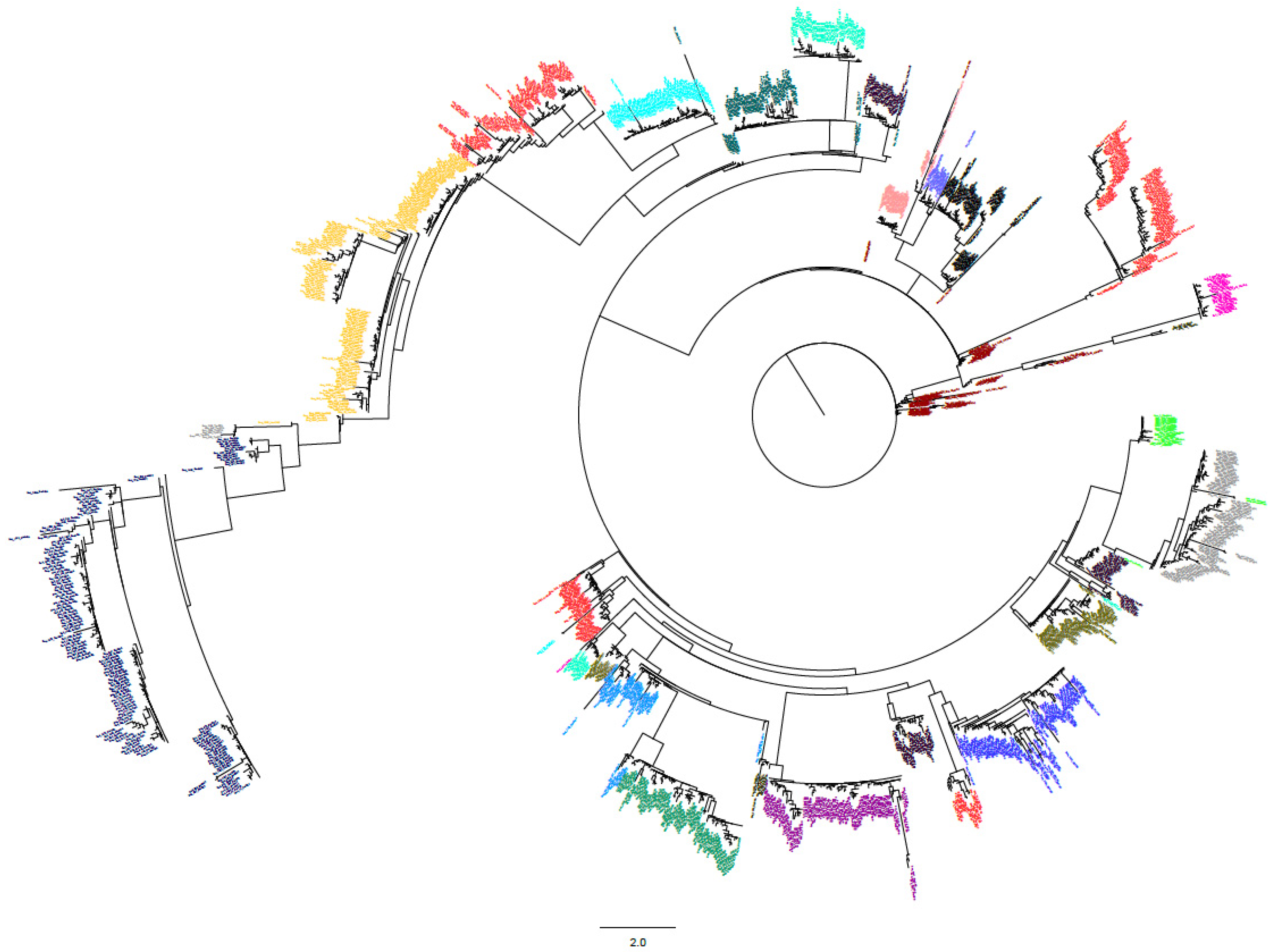

| Clusters | Enriched tRNAs |
|---|---|
| 0 | GCA(Cys); CGA(Ser); CAA(Leu); CCT(Arg); CAT(Met); GCC(Gly); GTT(Asn) |
| 1 | AGT(Thr); CTC(Glu); CTG(Gln); CCC(Gly); CCA(Trp); TTT(Lys) |
| 2 | TCC(Gly); CTT(Lys); AAC(Val); CAC(Val); AAT(Ile); GTC(Asp); CAG(Leu); TAA(Leu); TAT(Ile) |
| 3 | TCT(Arg); CGT(Thr); GCT(Ser); AGG(Pro); TGG(Pro) |
| 4 | AGA(Ser); TTC(Glu) |
| 5 | GAA(Phe); AGC(Ala); GTG(His); CGC(Ala); TAG(Leu) |
| 6 | CCG(Arg); TCG(Arg); TTG(Gln); AAG(Leu); TGT(Thr) |
| 7 | TGC(Ala); ACG(Arg); TGA(Ser); TAC(Val) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Xiong, Y.; Chen, Y.; Jadoon, S.; Yu, H.; Liu, Z.; Liu, K.; Qiu, L.; Wang, J. Conservation and Tandem Duplication of tRNA Genes in Plant Species. Genes 2025, 16, 1307. https://doi.org/10.3390/genes16111307
Zhang F, Xiong Y, Chen Y, Jadoon S, Yu H, Liu Z, Liu K, Qiu L, Wang J. Conservation and Tandem Duplication of tRNA Genes in Plant Species. Genes. 2025; 16(11):1307. https://doi.org/10.3390/genes16111307
Chicago/Turabian StyleZhang, Fan, Yajun Xiong, Yijie Chen, Sawaira Jadoon, Huan Yu, Zhiyu Liu, Kanglin Liu, Lijuan Qiu, and Jun Wang. 2025. "Conservation and Tandem Duplication of tRNA Genes in Plant Species" Genes 16, no. 11: 1307. https://doi.org/10.3390/genes16111307
APA StyleZhang, F., Xiong, Y., Chen, Y., Jadoon, S., Yu, H., Liu, Z., Liu, K., Qiu, L., & Wang, J. (2025). Conservation and Tandem Duplication of tRNA Genes in Plant Species. Genes, 16(11), 1307. https://doi.org/10.3390/genes16111307






