Integrated GWAS and Transcriptome Analysis to Identify Genes Underlying Plant Height and Ear Height Plasticity in Maize Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Field Assessment
2.2. Genotype Identification
2.3. Quantitative Evaluation of Phenotypic Plasticity
2.4. Genotype-Phenotype Association Analysis
2.5. Transcriptome Data Analysis
2.6. Candidate Gene Prediction
3. Results
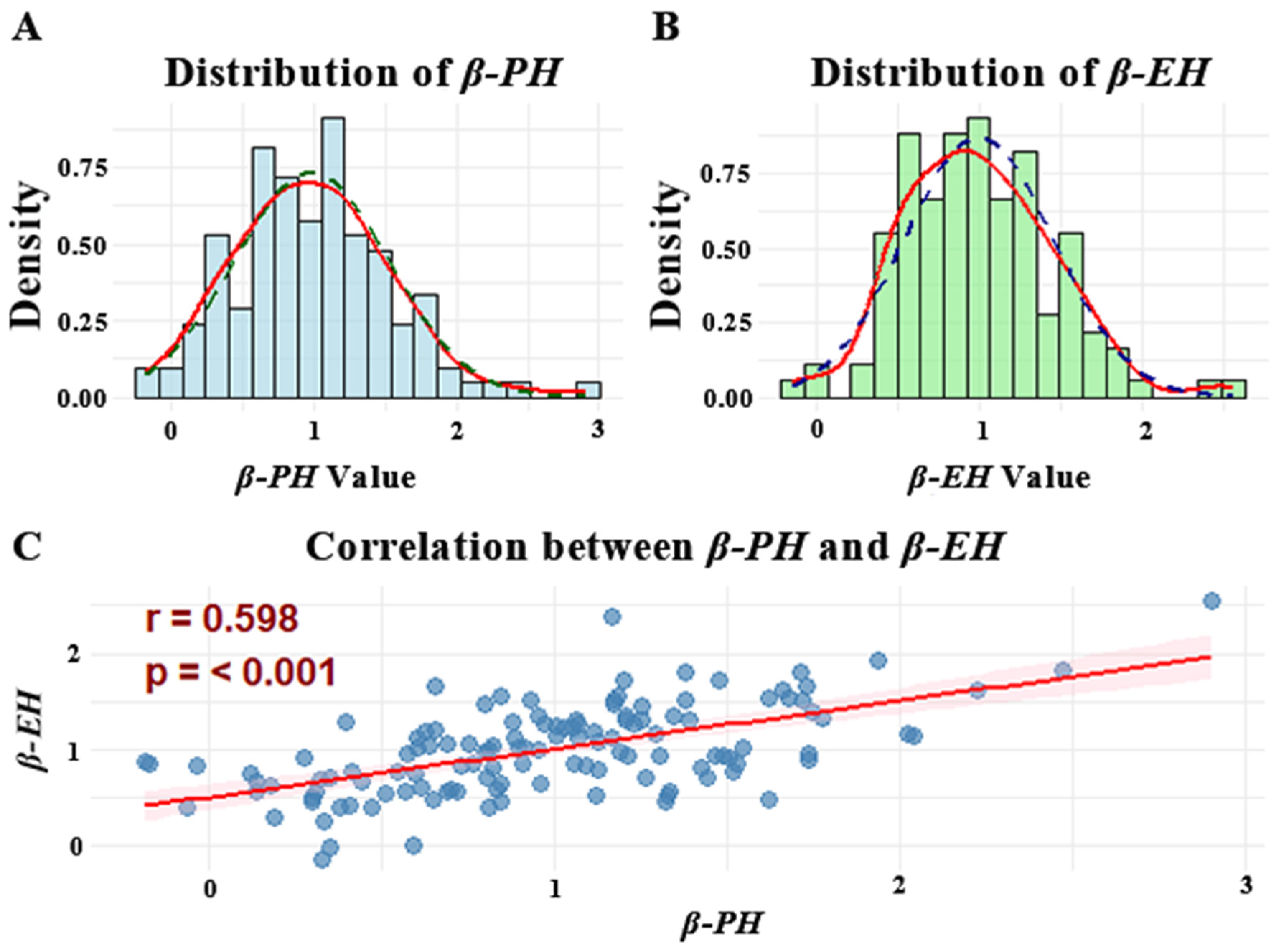
3.1. Plasticity Analysis of PH and EH
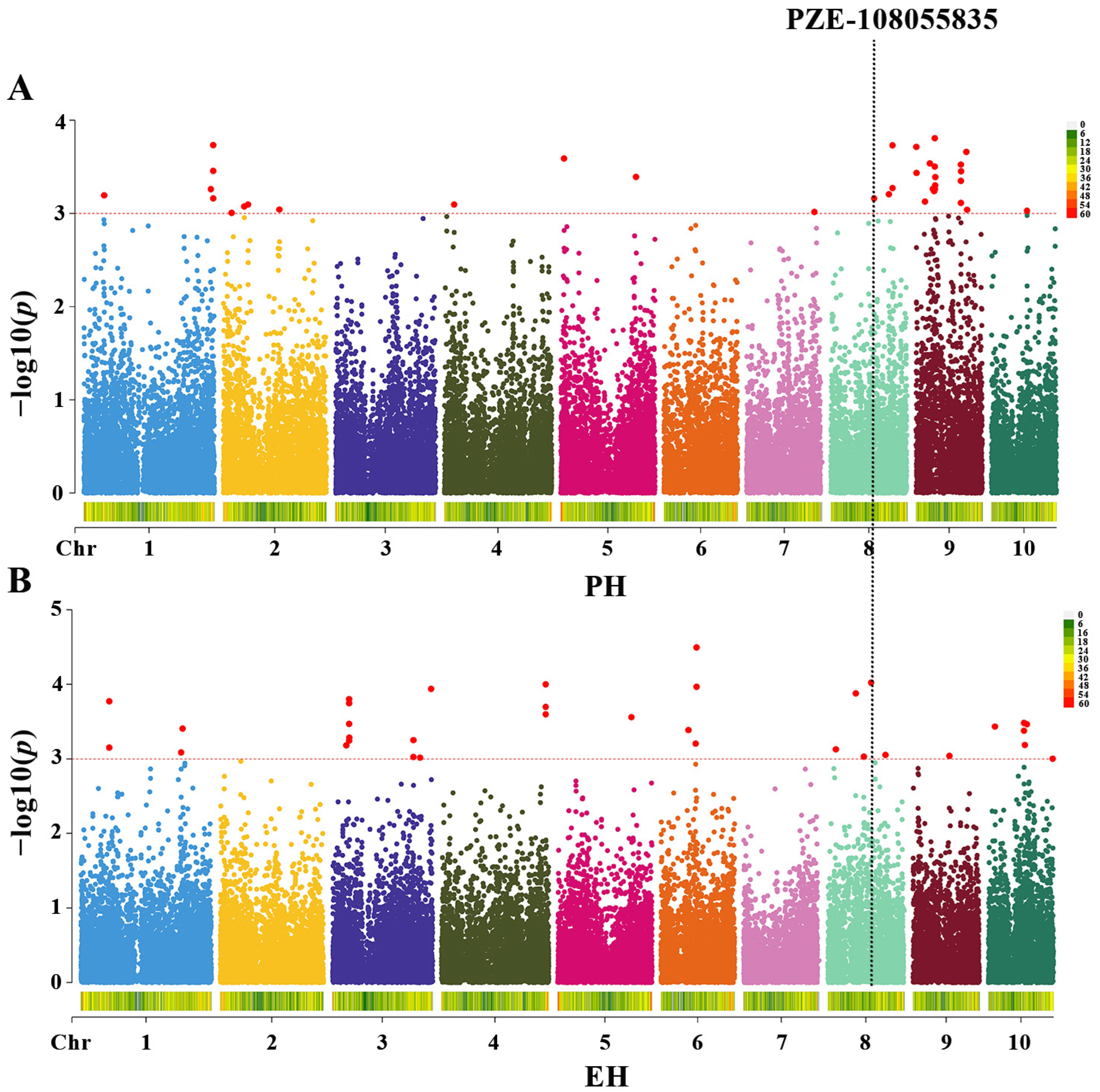
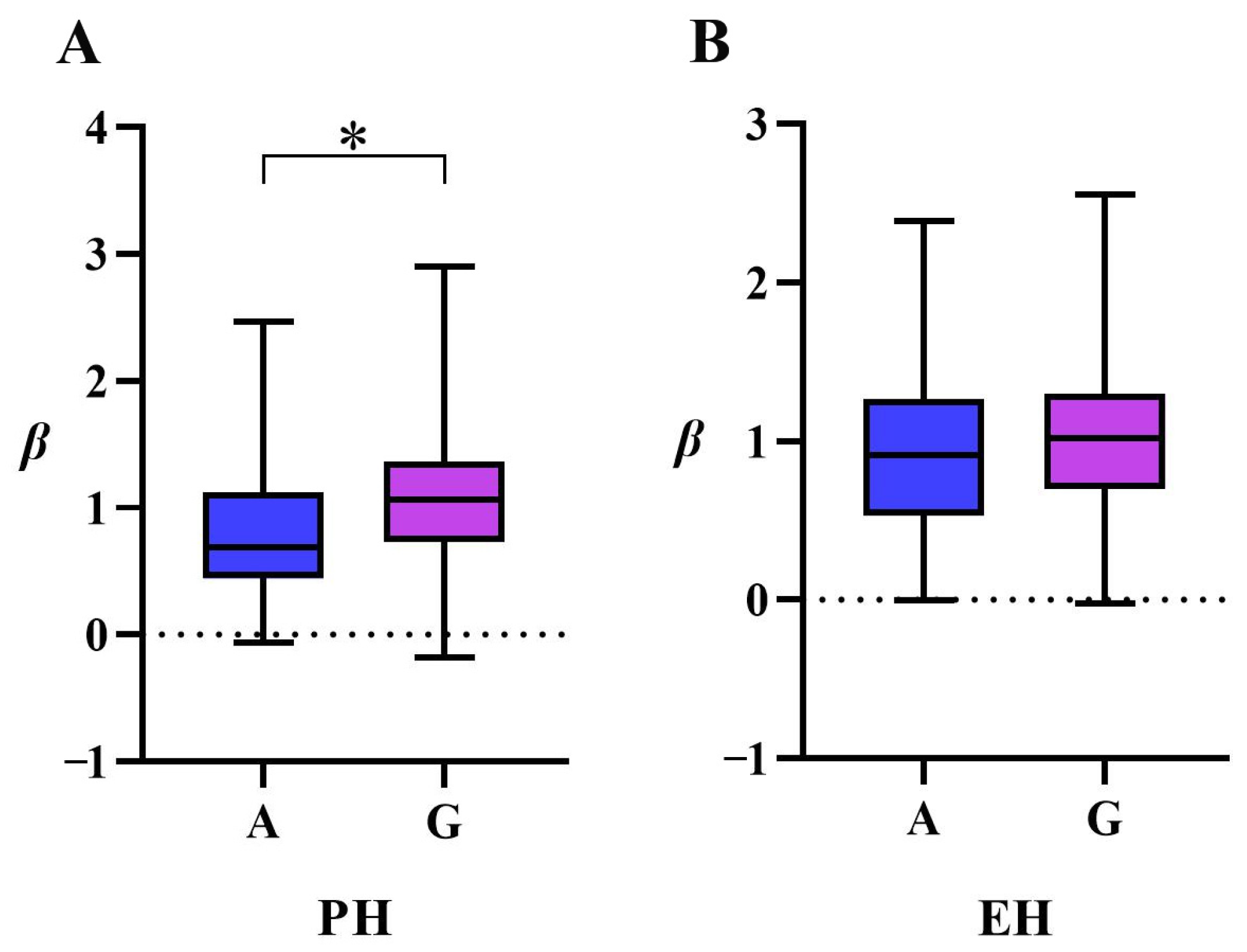
3.2. GWAS Analysis
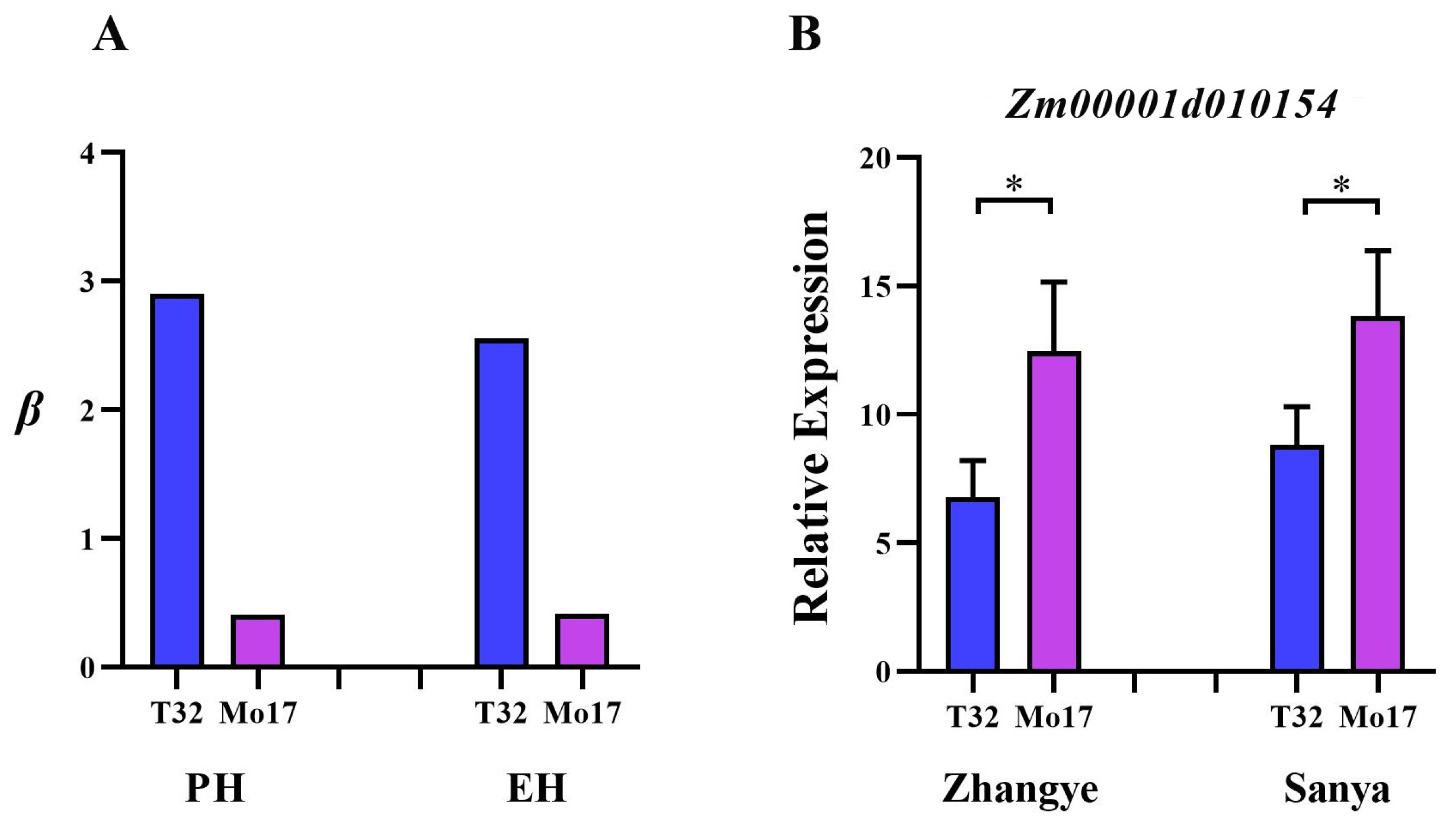
3.3. Integrating GWAS and Transcriptomics to Predict Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PH | Plant height |
| EH | Ear height |
| GWAS | Genome-wide association studies |
| β-PH | Plasticity values of plant height |
| β-EH | Plasticity values of ear height |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| DEGs | Differentially expressed genes |
| GCA | Significant general combining ability |
References
- Tanumihardjo, S.A.; McCulley, L.; Roh, R.; Lopez-Ridaura, S.; Palacios-Rojas, N.; Gunaratna, N.S. Maize agro-food systems to ensure food and nutrition security in reference to the Sustainable Development Goals. Glob. Food Secur. 2020, 25, 100327. [Google Scholar] [CrossRef]
- Peng, Q.; Shen, R.; Li, X.; Ye, T.; Dong, J.; Fu, Y.; Yuan, W. A twenty-year dataset of high-resolution maize distribution in China. Sci. Data 2023, 10, 658. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Luo, Y.; Cao, J.; Li, Z. Optimizing genotype-environment-management interactions for maize farmers to adapt to climate change in different agro-ecological zones across China. Sci. Total Environ. 2020, 728, 138614. [Google Scholar] [CrossRef]
- Harrington, L.J.; Schleussner, C.F.; Otto, F.E.L. Quantifying uncertainty in aggregated climate change risk assessments. Nat. Commun. 2021, 12, 7140. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhou, M.; Shao, J.; Chen, Z.; Wei, X.; Yang, J. Maize, wheat and rice production potential changes in china under the background of climate change. Agric. Syst. 2020, 182, 102853. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, P.; Xiao, Y.; Liu, H.J.; Ding, J.; Qu, Y.; Gao, J.; Li, W.; Guo, X.; Wang, A.; et al. GAIN: A long-term maize germplasm innovation plan. Mol. Plant 2025, 18, 915–919. [Google Scholar] [CrossRef]
- Xu, Y.; Skinner, D.J.; Wu, H.; Palacios-Rojas, N.; Araus, J.L.; Yan, J.; Gao, S.; Warburton, M.L.; Crouch, J.H. Advances in maize genomics and their value for enhancing genetic gains from breeding. Int. J. Plant Genom. 2009, 2009, 957602. [Google Scholar] [CrossRef]
- Gouesnard, B.; Rebourg, C.; Welcker, C.; Charcosset, A. Analysis of photoperiod sensitivity within a collection of tropical maize populations. Genet. Resour. Crop Evol. 2002, 49, 471–481. [Google Scholar] [CrossRef]
- Duvick, D.N. The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 2005, 86, 83–145. [Google Scholar]
- Peiffer, J.A.; Romay, M.C.; Gore, M.A.; Flint-Garcia, S.A.; Zhang, Z.; Millard, M.J.; Gardner, C.A.C.; McMullen, M.D.; Holland, J.B.; Bradbury, P.J.; et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2014, 46, 108–116. [Google Scholar]
- Liu, C.; Zhou, Q.; Dong, L.; Wang, H.; Liu, F.; Weng, J.; Li, X.; Xie, C. Genetic relationship between plant height and ear height in maize (Zea mays L.) and its genetic basis. Crop J. 2021, 9, 1104–1113. [Google Scholar]
- Zhao, Y.; Wang, H.; Bo, C.; Dai, W.; Zhang, X.; Li, J.; Dong, H. Genome-wide association study of maize plant architecture using F1 populations. Plant Mol. Biol. 2019, 99, 1–15. [Google Scholar] [CrossRef]
- Lan, J.H.; Chu, D. Study on the genetic basis of plant height and ear height in maize (Zea mays L.) by QTL dissection. Yi Chuan 2005, 27, 925–934. [Google Scholar] [PubMed]
- Wen, X.; Li, H.Y.; Song, Y.L.; Zhang, P.Y.; Zhang, Z.; Bu, H.H.; Dong, C.L.; Ren, Z.Q.; Chang, J.Z. Genome-wide association study for plant height and ear height in maize under well-watered and water-stressed conditions. BMC Genom. 2025, 26, 745. [Google Scholar] [CrossRef]
- Sultan, S.E. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 2000, 5, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Pigliucci, M. Evolution of phenotypic plasticity: Where are we going now? Trends Ecol. Evol. 2005, 20, 481–486. [Google Scholar] [CrossRef]
- Li, P.; An, Z.; Xu, N.; Li, J.; Li, Q.; He, C. Phenotypic plasticity and stability in plants: Genetic mechanisms, environmental adaptation, evolutionary implications, and future directions. Plant Cell Environ. 2025, 48, 5847–5860. [Google Scholar] [CrossRef] [PubMed]
- Laitinen, R.A.E.; Nikoloski, Z. Genetic basis of plasticity in plants. J. Exp. 2019, 70, 739–745. [Google Scholar] [CrossRef]
- Guo, T.; Mu, Q.; Wang, J.; Vanous, A.E.; Onogi, A.; Iwata, H.; Li, X.; Yu, J. Dynamic effects of interacting genes underlying rice flowering-time phenotypic plasticity and global adaptation. Genome Res. 2020, 30, 673–683. [Google Scholar] [CrossRef]
- Ma, Y. Genetic Dissection and Candidate Mining of Theplasticity of Maize Plant Height and Flowering Time. Ph.D. Thesis, Shenyang Agricultural University, Shenyang, China, 2024; pp. 1–121. [Google Scholar]
- Wu, X.; Wang, A.; Guo, X.; Liu, P.; Zhu, Y.; Li, X.; Chen, Z. Genetic characterization of maize germplasm derived from Suwan population and temperate resources. Hereditas 2019, 156, 2. [Google Scholar] [CrossRef]
- Finlay, K.W.; Wilkinson, G.N. The analysis of adaptation in a plant-breeding programme. Crop Pasture Sci. 1963, 14, 742–754. [Google Scholar] [CrossRef]
- Kaler, A.S.; Gillman, J.D.; Beissinger, T.; Purcell, L.C. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 2020, 10, 1794. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Lu, X.; Tu, L.; Gao, Y.; Wang, D.; Guo, S.; Xiao, Y.; Xiao, P.; Guo, X.; et al. Integration of GWAS, linkage analysis and transcriptome analysis to reveal the genetic basis of flowering time-related traits in maize. Front. Plant Sci. 2023, 14, 1145327. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Jiang, Y.; Guo, S.; Wang, D.; Tu, L.; Liu, P.; Guo, X.; Wang, A.; Zhu, Y.; Lu, X.; Chen, Z.; et al. Integrated GWAS, linkage, and transcriptome analysis to identify genetic loci and candidate genes for photoperiod sensitivity in maize. Front. Plant Sci. 2024, 15, 1441288. [Google Scholar] [CrossRef]
- Lu, X.; Liu, P.; Tu, L.; Guo, X.; Wang, A.; Zhu, Y.; Jiang, Y.; Zhang, C.; Xu, Y.; Chen, Z.; et al. Joint-GWAS, linkage mapping, and transcriptome analysis to reveal the genetic basis of plant architecture-related traits in maize. Int. J. Mol. Sci. 2024, 25, 2694. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, Z.; Guo, X.; Wang, A.; Wu, X.; Liu, P. Breeding and application of inbred line QB446 based on Suwan-Lancaster population improvement of maize. Guizhou Agric. Sci. 2020, 48, 1–5. [Google Scholar]
- Liu, N.; Du, Y.; Warburton, M.L.; Xiao, Y.; Yan, J. Phenotypic Plasticity Contributes to Maize Adaptation and Heterosis. Mol. Biol. Evol. 2021, 38, 1262–1275. [Google Scholar] [CrossRef]
- Jin, M.; Liu, H.; Liu, X.; Guo, T.; Guo, J.; Yin, Y.; Ji, Y.; Li, Z.; Zhang, J.; Wang, X.; et al. Complex genetic architecture underlying the plasticity of maize agronomic traits. Plant Commun. 2023, 4, 100473. [Google Scholar] [CrossRef]
- Rieu, P.; Turchi, L.; Thévenon, E.; Zarkadas, E.; Nanao, M.; Chahtane, H.; Tichtinsky, G.; Lucas, J.; Blanc-Mathieu, R.; Zubieta, C.; et al. The F-box protein UFO controls flower development by redirecting the master transcription factor LEAFY to new cis-elements. Nat. Plants 2023, 9, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lin, B.; Wang, H.; Li, X.; Yang, F.; Ding, X.; Yan, J.; Chu, Z. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 2019, 51, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Xie, Y.; Yu, S.; Yang, J.; Chen, S.; Yuan, X.; Guo, T.; Wang, H.; Liu, Y.; Chen, C.; et al. The DnaJ domain-containing heat-shock protein NAL11 determines plant architecture by mediating gibberellin homeostasis in rice (Oryza sativa). New Phytol. 2023, 237, 2163–2179. [Google Scholar] [CrossRef] [PubMed]
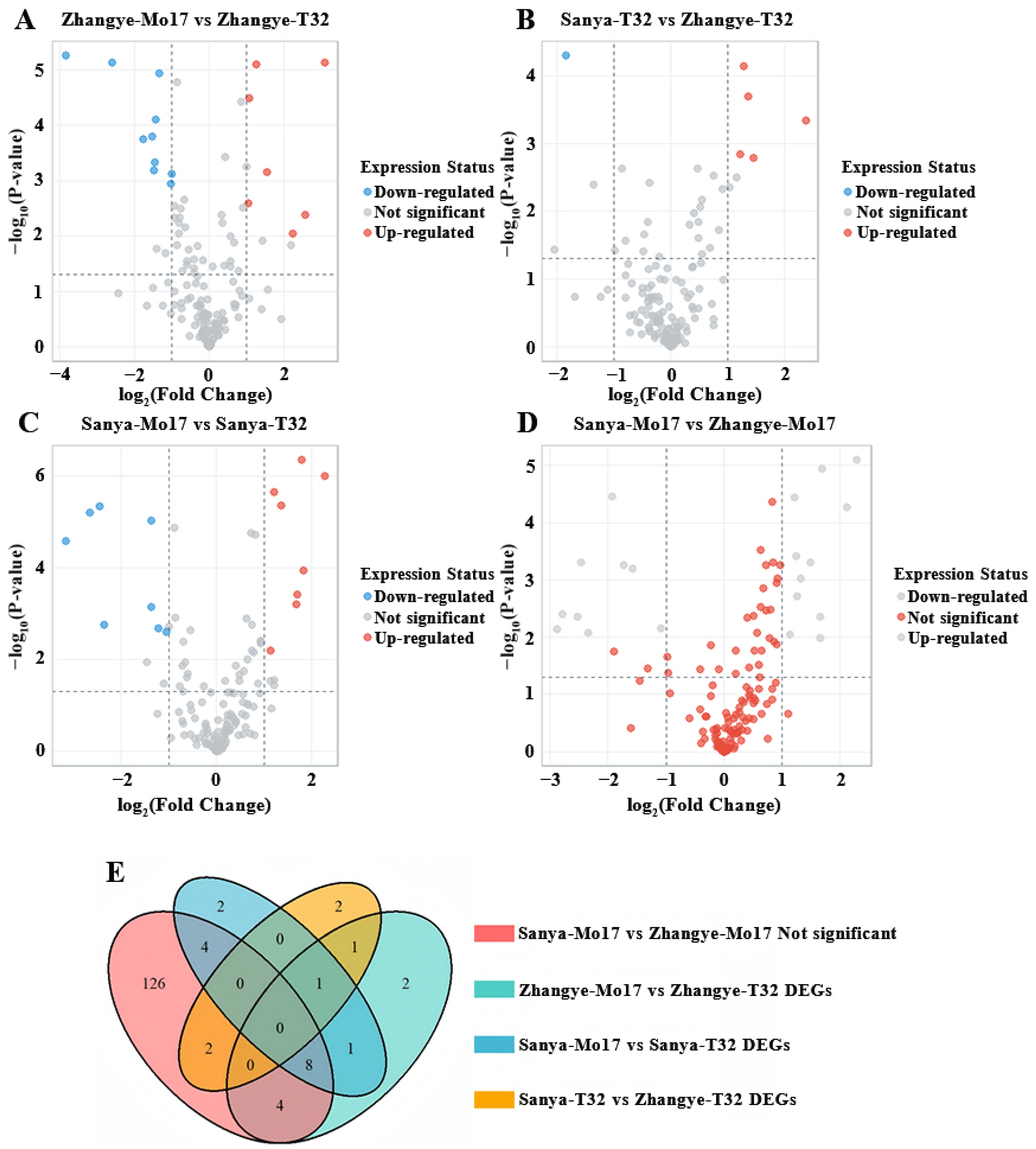
| Gene | Comparison | Difference Expressed | Function |
|---|---|---|---|
| Zm00001d003607 | Sanya-Mo17 vs. Zhangye-Mo17 | No | F-box domain-containing protein |
| Zhangye-Mo17 vs. Zhangye-T32 | Down | ||
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Zm00001d023682 | Sanya-Mo17 vs. Zhangye-Mo17 | No | ABC transporter F family member 1 |
| Zhangye-Mo17 vs. Zhangye-T32 | Up | ||
| Sanya-Mo17 vs. Sanya-T32 | Up | ||
| Zm00001d026647 | Sanya-Mo17 vs. Zhangye-Mo17 | No | DNA mismatch repair protein MSH3 |
| Zhangye-Mo17 vs. Zhangye-T32 | Down | ||
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Zm00001d026651 | Sanya-Mo17 vs. Zhangye-Mo17 | No | Expressed protein%3B protein |
| Zhangye-Mo17 vs. Zhangye-T32 | Up | ||
| Sanya-Mo17 vs. Sanya-T32 | Up | ||
| Zm00001d043110 | Zhangye-Mo17 vs. Zhangye-T32 | Down | Beta-glucosidase 11 |
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Sanya-T32 vs. Zhangye-T32 | Up | ||
| Zm00001d044542 | Sanya-Mo17 vs. Zhangye-Mo17 | No | F-box domain-containing protein |
| Zhangye-Mo17 vs. Zhangye-T32 | Down | ||
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Zm00001d045384 | Sanya-Mo17 vs. Zhangye-Mo17 | No | Superoxide dismutase16 |
| Zhangye-Mo17 vs. Zhangye-T32 | Down | ||
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Zm00001d045386 | Sanya-Mo17 vs. Zhangye-Mo17 | No | Ethylene-responsive transcription factor RAP2-2 |
| Zhangye-Mo17 vs. Zhangye-T32 | Down | ||
| Sanya-Mo17 vs. Sanya-T32 | Down | ||
| Zm00001d053972 | Sanya-Mo17 vs. Zhangye-Mo17 | No | DNAJ heat shock family protein |
| Zhangye-Mo17 vs. Zhangye-T32 | Up | ||
| Sanya-Mo17 vs. Sanya-T32 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, L.; Liu, P.; Wang, A.; Wang, D.; Zhu, Y.; Li, G.; Jiang, Y.; Wu, X.; Zhang, Z.; Chen, Z.; et al. Integrated GWAS and Transcriptome Analysis to Identify Genes Underlying Plant Height and Ear Height Plasticity in Maize Germplasm. Genes 2025, 16, 1225. https://doi.org/10.3390/genes16101225
Tu L, Liu P, Wang A, Wang D, Zhu Y, Li G, Jiang Y, Wu X, Zhang Z, Chen Z, et al. Integrated GWAS and Transcriptome Analysis to Identify Genes Underlying Plant Height and Ear Height Plasticity in Maize Germplasm. Genes. 2025; 16(10):1225. https://doi.org/10.3390/genes16101225
Chicago/Turabian StyleTu, Liang, Pengfei Liu, Angui Wang, Dong Wang, Yunfang Zhu, Gang Li, Yulin Jiang, Xun Wu, Zhiming Zhang, Zehui Chen, and et al. 2025. "Integrated GWAS and Transcriptome Analysis to Identify Genes Underlying Plant Height and Ear Height Plasticity in Maize Germplasm" Genes 16, no. 10: 1225. https://doi.org/10.3390/genes16101225
APA StyleTu, L., Liu, P., Wang, A., Wang, D., Zhu, Y., Li, G., Jiang, Y., Wu, X., Zhang, Z., Chen, Z., & Guo, X. (2025). Integrated GWAS and Transcriptome Analysis to Identify Genes Underlying Plant Height and Ear Height Plasticity in Maize Germplasm. Genes, 16(10), 1225. https://doi.org/10.3390/genes16101225






