NKX6-3 in B-Cell Progenitor Differentiation and Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analyses
2.2. Cell Lines and Treatments
2.3. Polymerase Chain Reaction (PCR) Analyses
2.4. Genomic Profiling Analysis
2.5. Protein Analyses
3. Results
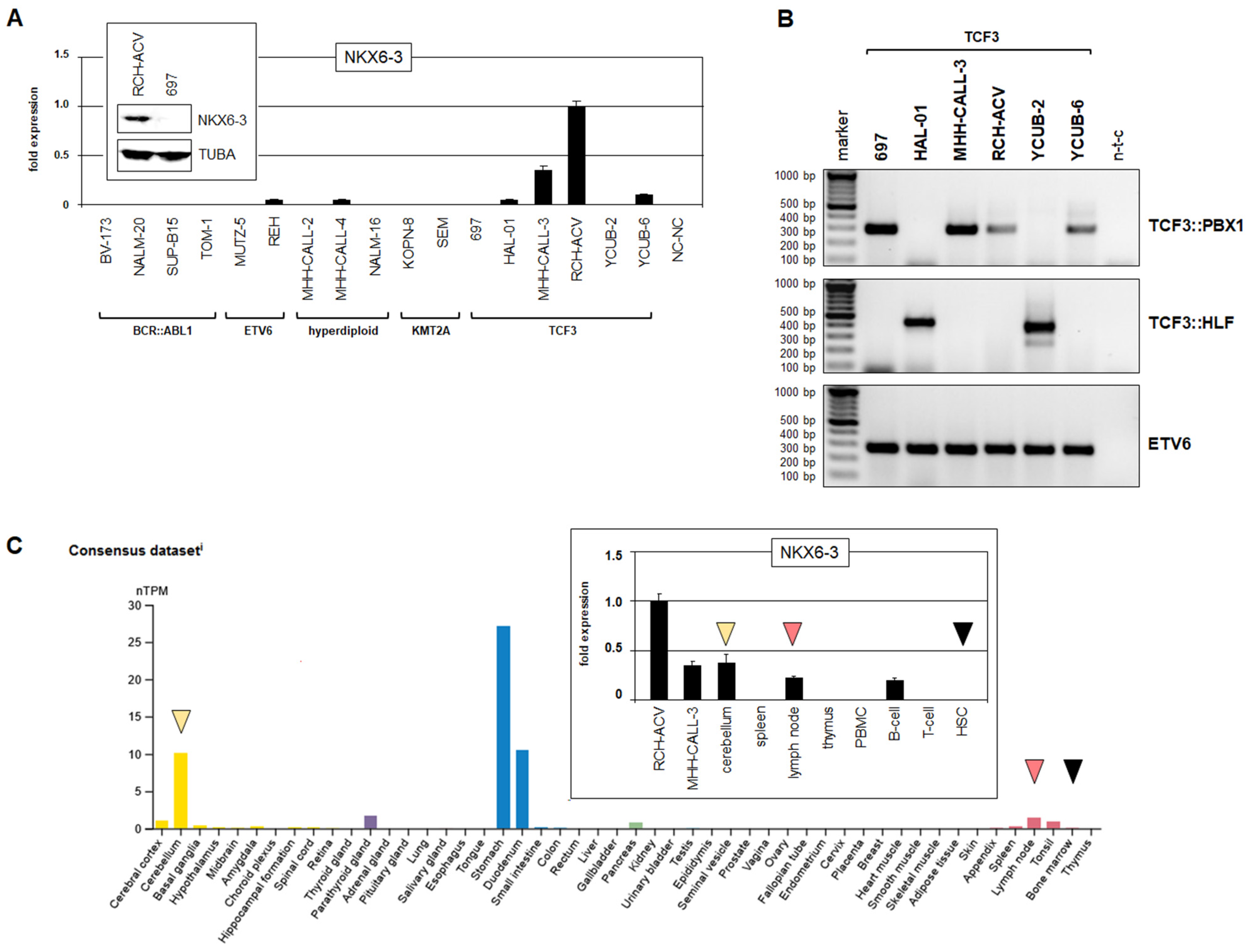
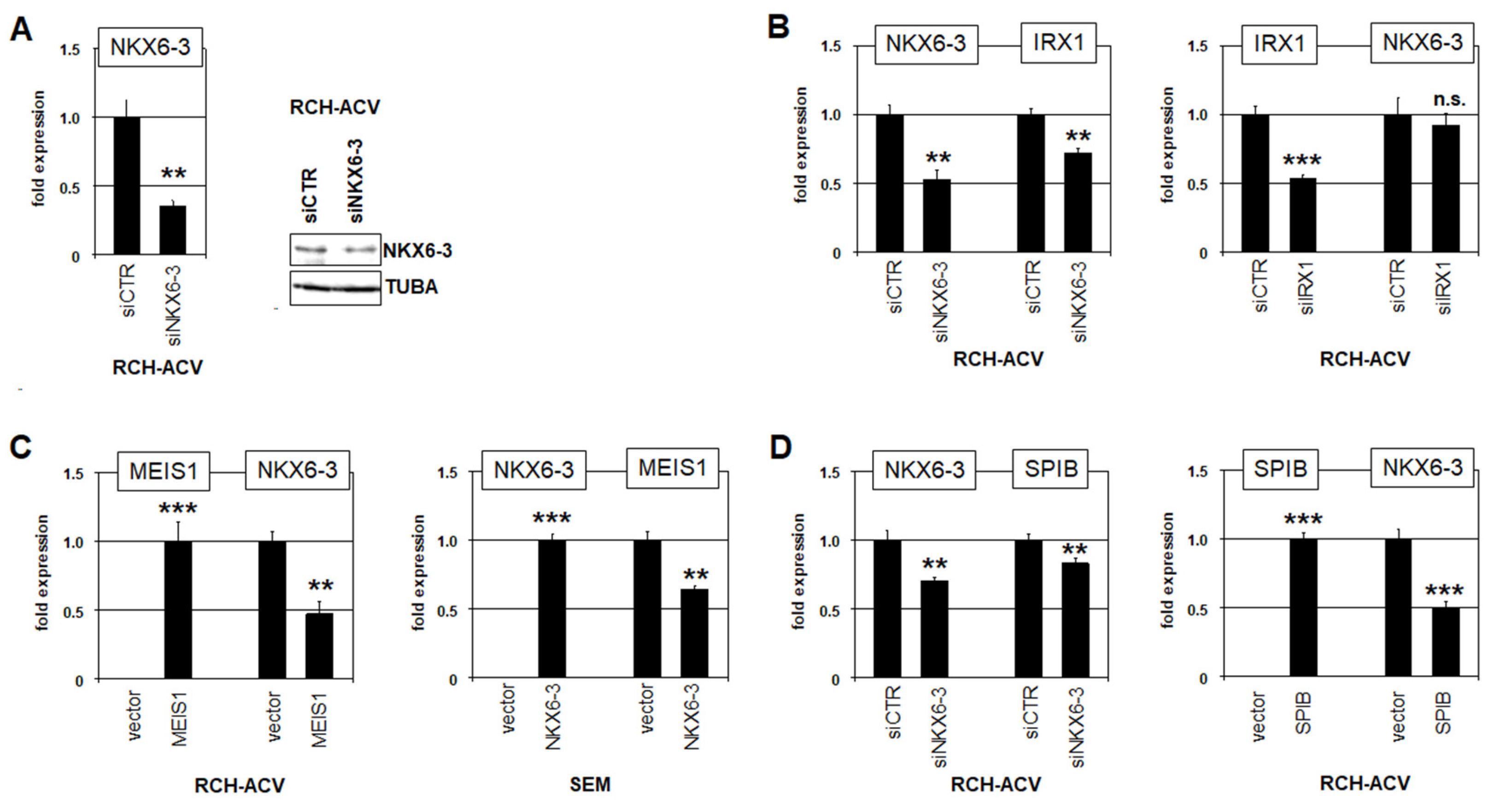
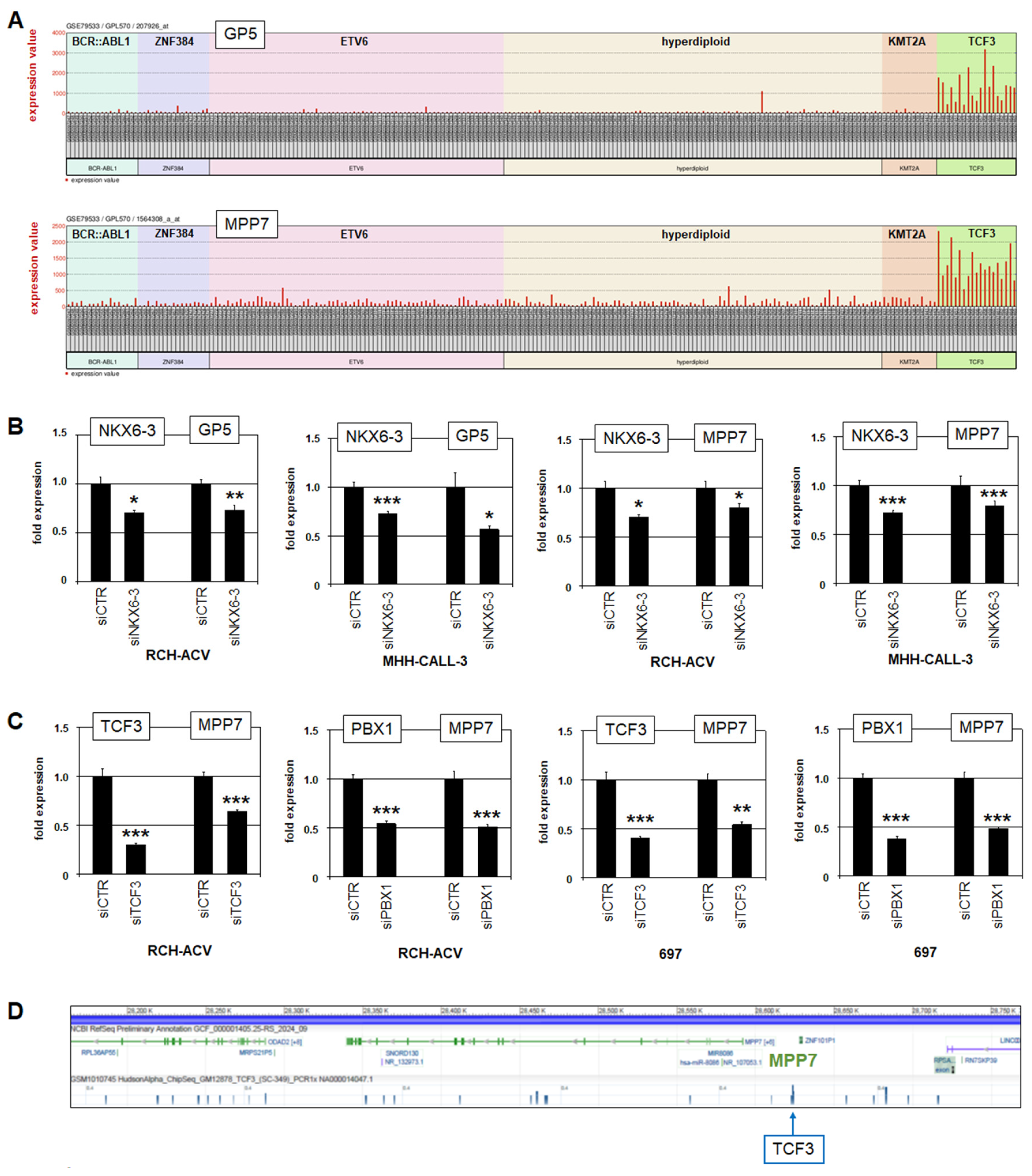
3.1. Expression and Regulation of NKX6-3 in B-Cell Progenitors
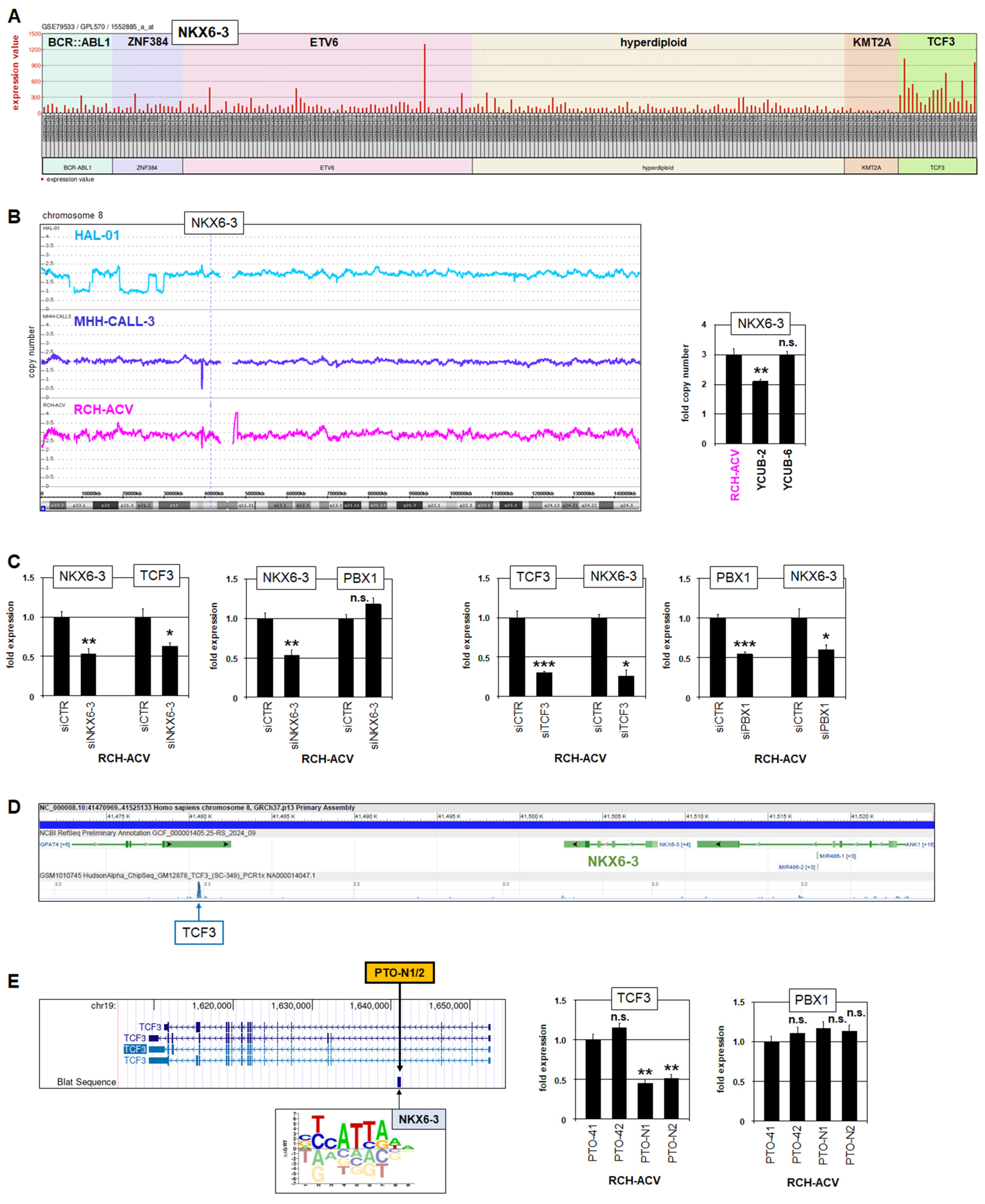
3.2. NKX6-3 Is a Target Gene of TCF3::PBX1 in BCP-ALL
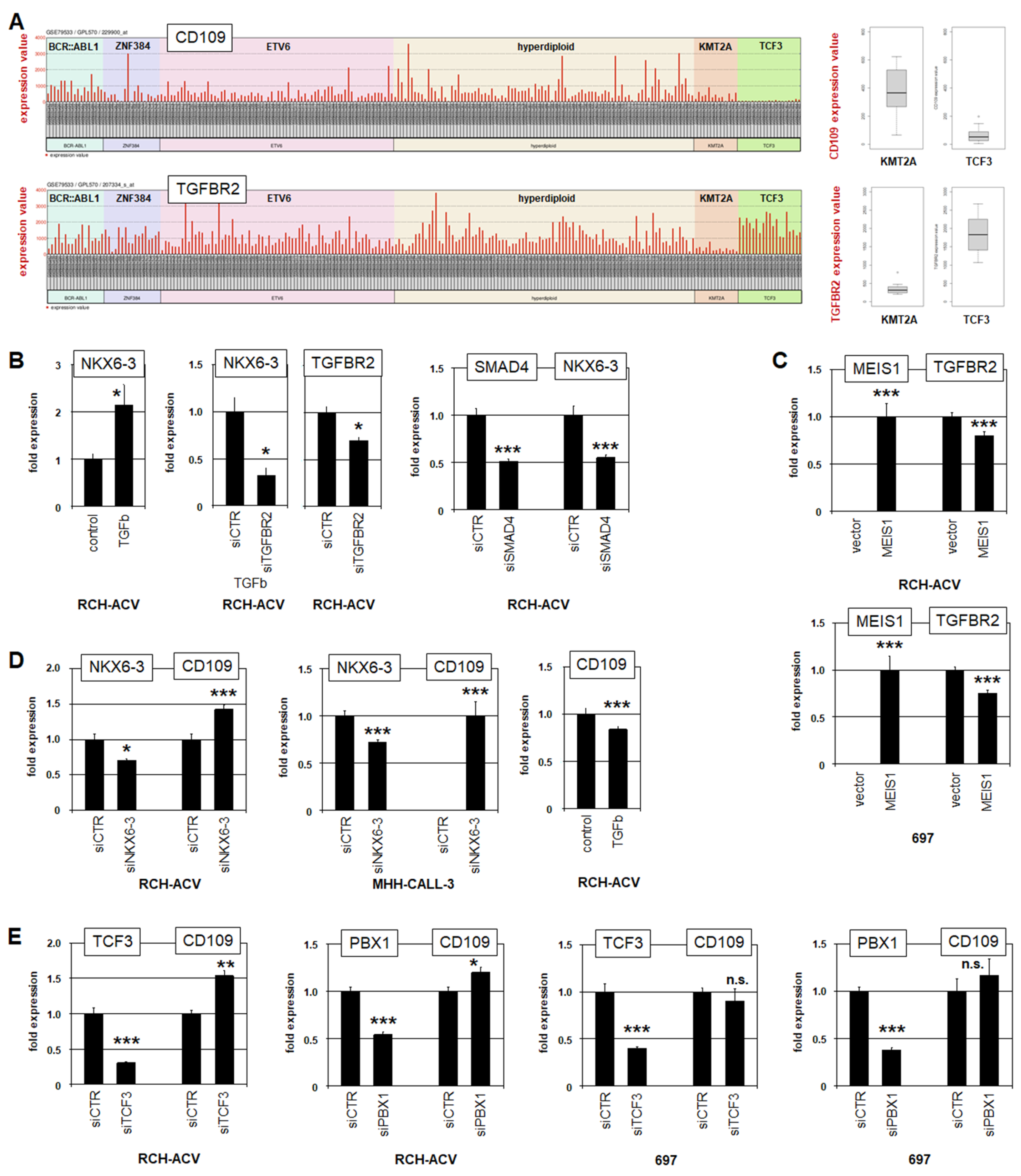
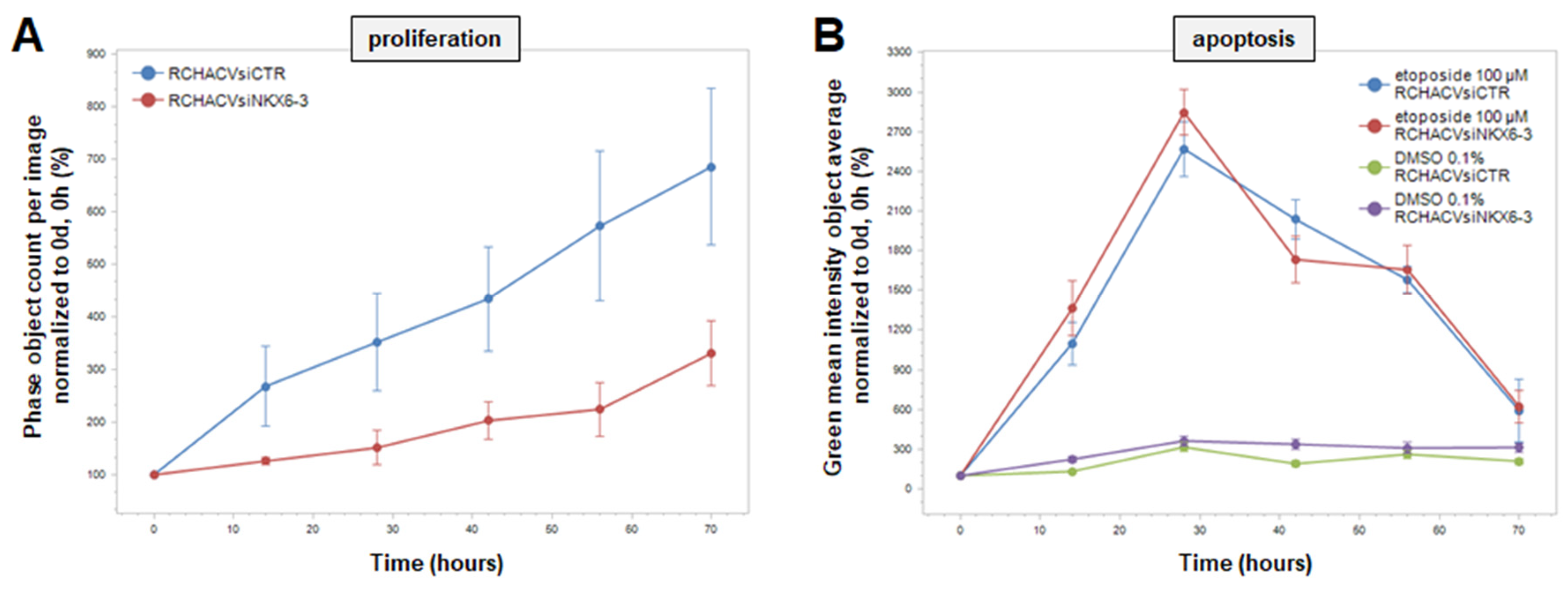
3.3. NKX6-3 Is Linked to TGFb-Signalling and the HIPPO-Pathway
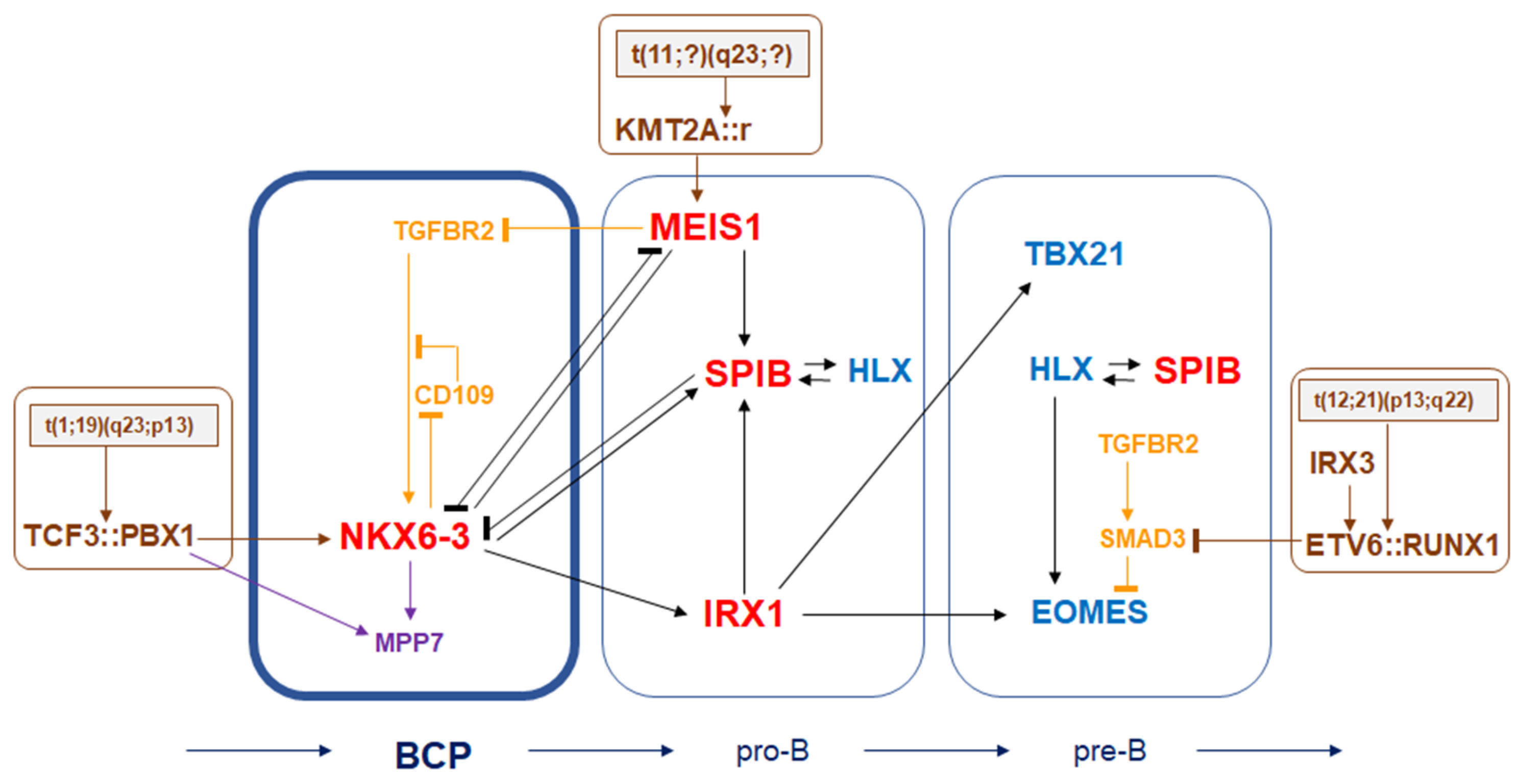
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothenberg, E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014, 32, 283–321. [Google Scholar] [CrossRef]
- Rothenberg, E.V.; Kueh, H.Y.; Yui, M.A.; Zhang, J.A. Hematopoiesis and T-cell specification as a model developmental system. Immunol. Rev. 2016, 271, 72–97. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.K.; Foster, S.D.; Wang, X.; Knezevic, K.; Schütte, J.; Kaimakis, P.; Chilarska, P.M.; Kinston, S.; Ouwehand, W.H.; Dzierzak, E.; et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010, 7, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Sigvardsson, M. Transcription factor networks link B-lymphocyte development and malignant transformation in leukemia. Genes Dev. 2023, 37, 703–723. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Mandel, E.M.; Pott, S.; Györy, I.; Firner, S.; Liu, E.T.; Grosschedl, R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 2010, 32, 714–725. [Google Scholar] [CrossRef]
- Holland, P.W.; Booth, H.A.; Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 47. [Google Scholar] [CrossRef]
- Nagel, S.; Pommerenke, C.; Meyer, C.; MacLeod, R.A.F.; Drexler, H.G. Establishment of the TALE-code reveals aberrantly activated homeobox gene PBX1 in Hodgkin lymphoma. PLoS ONE 2021, 16, e0246603. [Google Scholar] [CrossRef]
- Nagel, S.; Meyer, C. Normal and Aberrant TALE-Class Homeobox Gene Activities in Pro-B-Cells and B-Cell Precursor Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022, 23, 11874. [Google Scholar] [CrossRef]
- Nagel, S. NKL-Code in Normal and Aberrant Hematopoiesis. Cancers 2021, 13, 1961. [Google Scholar] [CrossRef]
- Nagel, S.; MacLeod, R.A.F.; Meyer, C.; Kaufmann, M.; Drexler, H.G. NKL homeobox gene activities in B-cell development and lymphomas. PLoS ONE 2018, 13, e0205537. [Google Scholar] [CrossRef]
- Nagel, S.; Meyer, C.; Pommerenke, C. Establishment of the lymphoid ETS-code reveals deregulated ETS genes in Hodgkin lymphoma. PLoS ONE 2023, 18, e0288031. [Google Scholar] [CrossRef]
- Nagel, S.; Meyer, C. Identification of Gene Regulatory Networks in B-Cell Progenitor Differentiation and Leukemia. Genes 2024, 15, 978. [Google Scholar] [CrossRef]
- Tijchon, E.; Havinga, J.; van Leeuwen, F.N.; Scheijen, B. B-lineage transcription factors and cooperating gene lesions required for leukemia development. Leukemia 2013, 27, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Yoshida, K.; Shiozawa, Y.; Nannya, Y.; Iijima-Yamashita, Y.; Kiyokawa, N.; Shiraishi, Y.; Chiba, K.; Tanaka, H.; Isobe, T.; et al. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020, 4, 5165–5173. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wang, J.; Tan, Y.; Jiang, M.; Zhang, H.; Meng, G. Transcription factor abnormalities in B-ALL leukemogenesis and treatment. Trends Cancer 2023, 9, 855–870. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.W.; Roberts, K.G.; Gu, Z.; Shi, L.; Pounds, S.; Pei, D.; Cheng, C.; Dai, Y.; Devidas, M.; Qu, C.; et al. The genomic landscape of pediatric acute lymphoblastic leukemia. Nat. Genet. 2022, 54, 1376–1389. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef]
- Den Boer, M.L.; van Slegtenhorst, M.; De Menezes, R.X.; Cheok, M.H.; Buijs-Gladdines, J.G.; Peters, S.T.; Van Zutven, L.J.; Beverloo, H.B.; Van der Spek, P.J.; Escherich, G.; et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009, 10, 125–134. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumour: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Ferrando, A.A.; Armstrong, S.A.; Neuberg, D.S.; Sallan, S.E.; Silverman, L.B.; Korsmeyer, S.J.; Look, A.T. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood 2003, 102, 262–268. [Google Scholar] [CrossRef]
- Orlovsky, K.; Kalinkovich, A.; Rozovskaia, T.; Shezen, E.; Itkin, T.; Alder, H.; Ozer, H.G.; Carramusa, L.; Avigdor, A.; Volinia, S.; et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc. Natl. Acad. Sci. USA 2011, 108, 7956–7961. [Google Scholar]
- Hirabayashi, S.; Ohki, K.; Nakabayashi, K.; Ichikawa, H.; Momozawa, Y.; Okamura, K.; Yaguchi, A.; Terada, K.; Saito, Y.; Yoshimi, A.; et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 2017, 102, 118–129. [Google Scholar] [PubMed]
- Meyer, L.H.; Eckhoff, S.M.; Queudeville, M.; Kraus, J.M.; Giordan, M.; Stursberg, J.; Zangrando, A.; Vendramini, E.; Möricke, A.; Zimmermann, M.; et al. Early relapse in ALL is identified by time to leukemia in NOD/SCID mice and is characterized by a gene signature involving survival pathways. Cancer Cell 2011, 19, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef]
- Pommerenke, C.; Geffers, R.; Bunk, B.; Bhuju, S.; Eberth, S.; Drexler, H.G.; Quentmeier, H. Enhanced whole exome sequencing by higher DNA insert lengths. BMC Genom. 2016, 17, 399. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Mudge, J.M.; Carbonell-Sala, S.; Diekhans, M.; Martinez, J.G.; Hunt, T.; Jungreis, I.; Loveland, J.E.; Arnan, C.; Barnes, I.; Bennett, R.; et al. GENCODE 2025, Reference gene annotation for human and mouse. Nucleic Acids Res. 2025, 53, D966–D975. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID Gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene list. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Nagel, S.; Scherr, M.; Kel, A.; Hornischer, K.; Crawford, G.E.; Kaufmann, M.; Meyer, C.; Drexler, H.G.; MacLeod, R.A. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007, 67, 1461–1471. [Google Scholar] [CrossRef]
- van Dongen, J.J.; Macintyre, E.A.; Gabert, J.A.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and Validation of Housekeeping Genes for Gene Expression Analysis of Cancer Stem Cells. PLoS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.G.; Varma, N.; Abdulkadir, S.A.; Singh, P.; Sachdeva, M.U.S.; Naseem, S.; Siddiqui, M.R.; Bose, P.; Binota, J.; Malhotra, P.; et al. Identification and validation of the optimal reference genes for standardizing the gene expression profiling diagnostic panel of Ph-like B-lineage acute lymphoblastic leukemia. Clin. Exp. Med. 2023, 23, 4539–4551. [Google Scholar] [CrossRef] [PubMed]
- Gertz, J.; Savic, D.; Varley, K.E.; Partridge, E.C.; Safi, A.; Jain, P.; Cooper, G.M.; Reddy, T.E.; Crawford, G.E.; Myers, R.M. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 2013, 52, 25–36. [Google Scholar] [CrossRef] [PubMed]
- McWhirter, J.R.; Neuteboom, S.T.; Wancewicz, E.V.; Monia, B.P.; Downing, J.R.; Murre, C. Oncogenic homeodomain transcription factor E2A-Pbx1 activates a novel WNT gene in pre-B acute lymphoblastoid leukemia. Proc. Natl. Acad. Sci. USA 1999, 96, 11464–11469. [Google Scholar] [CrossRef]
- Ali, A.; Vaikari, V.P.; Alachkar, H. CD99 in malignant hematopoiesis. Exp. Hematol. 2022, 106, 40–46. [Google Scholar] [CrossRef]
- Bizet, A.A.; Liu, K.; Tran-Khanh, N.; Saksena, A.; Vorstenbosch, J.; Finnson, K.W.; Buschmann, M.D.; Philip, A. The TGF-β co-receptor, CD109, promotes internalization and degradation of TGF-β receptors. Biochim. Biophys. Acta 2011, 1813, 742–753. [Google Scholar] [CrossRef]
- Ford, A.M.; Palmi, C.; Bueno, C.; Hong, D.; Cardus, P.; Knight, D.; Cazzaniga, G.; Enver, T.; Greaves, M. The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells. J. Clin. Investig. 2009, 119, 826–836. [Google Scholar] [PubMed]
- Kimmerlin, Q.; Moog, S.; Ravanat, C.; Strassel, C.; Lanza, F. Glycoprotein V: The unsolved GPV puzzle. Platelets 2022, 33, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Shao, A.; Kissil, J.L.; Fan, C.M. The L27 domain of MPP7 enhances TAZ-YY1 cooperation to renew muscle stem cells. EMBO Rep. 2024, 25, 5667–5686. [Google Scholar] [CrossRef]
- Zhang, F.; Issah, M.A.; Fu, H.Y.; Zhou, H.R.; Liu, T.B.; Shen, J.Z. LATS1 Promotes B-ALL Tumorigenesis by Regulating YAP1 Phosphorylation and Subcellular Localization. Curr. Med. Sci. 2024, 44, 81–92. [Google Scholar] [CrossRef]
- Calses, P.C.; Crawford, J.J.; Lill, J.R.; Dey, A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer 2019, 5, 297–307. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, Y.; Zhao, S.; Li, J.; Li, C.; Mao, B. Xenopus Nkx6.3 is a neural plate border specifier required for neural crest development. PLoS ONE 2014, 9, e115165. [Google Scholar] [CrossRef]
- Hafler, B.P.; Choi, M.Y.; Shivdasani, R.A.; Rowitch, D.H. Expression and function of Nkx6.3 in vertebrate hindbrain. Brain Res. 2008, 1222, 42–50. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kim, O.; Eun, J.W.; Choi, S.S.; Ashktorab, H.; Smoot, D.T.; Nam, S.W.; Park, W.S. Multiple genetic mutations caused by NKX6.3 depletion contribute to gastric tumorigenesis. Sci. Rep. 2018, 8, 17609. [Google Scholar] [CrossRef]
- Yoon, J.H.; Eun, J.W.; Ashktorab, H.; Smoot, D.T.; Kim, J.K.; Nam, S.W.; Park, W.S. Depletion of NK6 Homeobox 3 (NKX6.3) causes gastric carcinogenesis through copy number alterations by inducing impairment of DNA replication and repair regulation. Oncogenesis 2021, 10, 85. [Google Scholar] [CrossRef]
- Sokalski, K.M.; Li, S.K.; Welch, I.; Cadieux-Pitre, H.A.; Gruca, M.R.; DeKoter, R.P. Deletion of genes encoding PU.1 and Spi-B in B cells impairs differentiation and induces pre-B cell acute lymphoblastic leukemia. Blood 2011, 118, 2801–2808. [Google Scholar] [CrossRef]
- Halder, S.K.; Cho, Y.J.; Datta, A.; Anumanthan, G.; Ham, A.J.; Carbone, D.P.; Datta, P.K. Elucidating the mechanism of regulation of transforming growth factor β Type II receptor expression in human lung cancer cell lines. Neoplasia 2011, 13, 912–922. [Google Scholar] [CrossRef]
- Bagherzadeh Yazdchi, S.; Witalis, M.; Meli, A.P.; Leung, J.; Li, X.; Panneton, V.; Chang, J.; Li, J.; Nutt, S.L.; Johnson, R.L.; et al. Hippo Pathway Kinase Mst1 Is Required for Long-Lived Humoral Immunity. J. Immunol. 2019, 202, 69–78. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Z.; Kashiwagi, M.; Yoshida, T.; Joshi, I.; Jena, N.; Somasundaram, R.; Emmanuel, A.O.; Sigvardsson, M.; Fitamant, J.; et al. Superenhancer reprogramming drives a B-cell-epithelial transition and high-risk leukemia. Genes Dev. 2016, 30, 1971–1990. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagel, S.; Meyer, C.; Pommerenke, C. NKX6-3 in B-Cell Progenitor Differentiation and Leukemia. Genes 2025, 16, 1199. https://doi.org/10.3390/genes16101199
Nagel S, Meyer C, Pommerenke C. NKX6-3 in B-Cell Progenitor Differentiation and Leukemia. Genes. 2025; 16(10):1199. https://doi.org/10.3390/genes16101199
Chicago/Turabian StyleNagel, Stefan, Corinna Meyer, and Claudia Pommerenke. 2025. "NKX6-3 in B-Cell Progenitor Differentiation and Leukemia" Genes 16, no. 10: 1199. https://doi.org/10.3390/genes16101199
APA StyleNagel, S., Meyer, C., & Pommerenke, C. (2025). NKX6-3 in B-Cell Progenitor Differentiation and Leukemia. Genes, 16(10), 1199. https://doi.org/10.3390/genes16101199





