Abstract
Objectives: To provide insights for breeding high-quality rice varieties, we analyzed local rice (Oryza sativa L.) germplasm from Yunnan Province, China, focusing on the relationships among Waxy gene alleles, indica–japonica differentiation, and amylose content (AC). Methods: We examined 201 local rice accessions. Two functional molecular markers for the Waxy gene were used to detect four alleles (Wxa, Wxb, Wxin, Wxmw). Additionally, 33 InDel markers were employed to classify indica–japonica attributes, and AC was measured according to GB/T 15683-2008. Results: We detected 175 accessions with Wxa, 20 with Wxb, 4 with Wxin, and 2 with Wxmw, indicating Wxa dominance and a diverse genetic basis at the Waxy locus. Indica–japonica classification identified 180 indica-type, 19 japonica-type, and 2 intermediate-type accessions, confirming predominant indica differentiation in Yunnan rice. Integrating Waxy allele detection, indica–japonica attributes, and AC showed that Wxa occurred primarily in indica rice with higher AC (mean 22.55%), comparable to Wxin (mean 24.33%); Wxb was mainly found in japonica rice with lower AC (mean 13.46%), similar to Wxmw (mean 15.65%). Conclusions: Local Yunnan rice exhibits Wxa predominance at the Waxy locus and clear indica differentiation. The observed associations between Waxy alleles, subspecies attributes, and AC provide useful references for marker-assisted breeding of premium rice and for exploiting indica–japonica heterosis.
1. Introduction
Rice (Oryza sativa) is a cornerstone of global food security, serving as the primary staple for over 60% of the world’s population and contributing significantly to caloric intake in Asia [1]. The eating and cooking quality (ECQ) of rice grains, which encompasses traits like texture, stickiness, and flavor, is a critical factor influencing consumer preference and market value [2]. Within this, amylose content (AC) in the endosperm starch plays a pivotal role, determining gel consistency, retrogradation, and overall palatability [3]. High AC typically results in firmer, less sticky cooked rice, preferred in some cuisines, while low AC yields softer, glutinous textures [4].
The biosynthesis of amylose is primarily regulated by the Waxy (Wx) gene, located on chromosome 6, which encodes granule-bound starch synthase I (GBSSI) [5]. This enzyme catalyzes the elongation of amylose chains in the endosperm, and allelic variations at the Wx locus profoundly affect AC and ECQ [6]. The amylose content in rice endosperm is determined by the processing efficiency of Wx pre-mRNA, with the excision of intron I identified as the key regulatory step [7]. To date, at least nine Wx alleles have been identified, including Wxa, Wxb, Wxin, Wxmw, Wxlv, Wxla, Wxmq, Wxmp, and Wxop/hp, each arising from single nucleotide polymorphisms (SNPs), insertions/deletions (InDels), or splicing site mutations [8,9,10]. For instance, the G-to-T mutation at the 5′ splice site of intron 1 distinguishes Wxa (high AC, predominant in indica subspecies) from Wxb (low AC, common in japonica) by altering pre-mRNA splicing efficiency [11]. Further refinements include Wxin, derived from an A-to-C SNP in intron 6 on a Wxa background, which slightly reduces AC compared to Wxa [12], and Wxmw, a similar mutation on Wxb that maintains comparable low AC levels [13]. Recent studies have also uncovered ancestral alleles like Wxlv, which modulates grain mouthfeel by influencing amylose chain length [14], and Wxla, whose origin provides insights into domestication-driven quality improvements [15]. These variations not only explain up to 60–80% of AC phenotypic variance but also interact with other loci (e.g., ALK for alkali spreading) to fine-tune starch properties [16,17].
Beyond allelic diversity at Wx, rice germplasm exhibits profound subspecies differentiation between indica (O. sativa subsp. indica) and japonica (O. sativa subsp. japonica), shaped by independent domestication events, environmental adaptations, and human selection [18]. Indica varieties generally feature elongated grains, higher AC, and adaptation to tropical climates, while japonica types have rounder grains, lower AC, and tolerance to temperate conditions [19]. This differentiation manifests in genetic barriers, such as hybrid sterility in F1 generations, but also enables strong heterosis in inter-subspecific crosses, boosting yield by 15–30% [20]. Accurate classification of indica–japonica attributes is essential for exploiting this heterosis, and molecular markers like InDels have proven reliable, revealing fixed differences at thousands of loci [3,21].
Yunnan Province in southwestern China stands out as a global hotspot for rice genetic diversity and a putative center of Asian cultivated rice origin [8,10]. As one of the world’s major centers of genetic diversity, it harbors a wide range of unique landraces with distinct genetic backgrounds [22,23]. Its complex topography, ranging from low-altitude tropics to high-elevation plateaus, coupled with diverse climates and ethnic farming practices, has fostered extensive ecotypic variation [8]. Yunnan harbors both indica and japonica landraces, along with wild relatives (e.g., O. rufipogon), and boasts over 58 cultivated variants [24]. Studies using SSR and SNP markers have documented high nucleotide diversity (π) in Yunnan rice, with indica–japonica differentiation varying along altitudinal gradients—indica dominating lowlands and japonica higher elevations [25]. A systematic characterization of Waxy (Wx) allele distribution and its relationship with amylose content and subspecies differentiation in this region is essential for germplasm conservation, breeding strategy optimization, and for complementing global rice genetic studies. This richness provides untapped potential for quality breeding, yet comprehensive analyses linking Wx alleles, subspecies attributes, and AC in local germplasm remain limited [26]. Current research on rice quality often examines the Wx gene, subspecies identity (indica/japonica), and amylose content (AC) in isolation, rather than as an integrated system [2,16]. The interaction between functional Wx alleles and their genetic backgrounds remains poorly understood, particularly in diverse germplasm. This disconnect is evident in regions like Yunnan, where rich genetic resources coexist with significant indica–japonica differentiation. The lack of combined analysis limits the understanding of how subspecies background modulates the effect of Wx alleles on AC. Such improvement is essential for effective marker-assisted breeding aimed at improving rice quality through informed utilization of genetic diversity.
Therefore, in this study, we designed experiments with the following specific objectives: genotyped 201 Yunnan local rice accessions using two functional markers to detect Wxa, Wxb, Wxin, and Wxmw alleles, classified indica–japonica attributes with 33 InDel markers, and quantified AC per national standards. This study aimed to identify key allelic distributions linked to subspecies differentiation and AC, to support practical marker-assisted selection (MAS) and the development of high-quality, high-yielding rice varieties.
2. Materials and Methods
2.1. Plant Materials
A total of 201 local rice accessions from various Yunnan regions were sourced from the Grain Crops Research Institute, Yunnan Academy of Agricultural Sciences (Table S1).
2.2. DNA Extraction
Genomic DNA was extracted from 3–4 leaf-stage seedlings using a modified rapid method: 200 μL extraction buffer (100 mmol/L Tris-HCl pH 8.7, 1 mol/L KCl, 10 mmol/L EDTA; 1:1:1 ratio) per sample, autoclaved at 101 °C for 10 min, diluted to 25 ng/μL with deionized water, and stored at −20 °C. DNA quantity and quality were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
2.3. Waxy Gene Genotyping
Two functional markers, Waxygt-ARMS and Waxyac-ARMS [10], were used (Table S2). To ensure robustness and reproducibility, preliminary optimization was performed including gradient PCR (annealing temperatures from 55 °C to 65 °C), testing of primer ratios (0.2–1.0 μM), and titration of DNA polymerase concentration. PCR reactions (10 μL) contained 5 μL 2× Taq Master Mix (Dye) (Xuzhou Probe Gene Technology Co., Ltd. No. 99 Daxue Road, Xuzhou City, Jiangsu, China), 1 μL primers (inner:outer ratio 1.5:1), 1 μL template DNA, and 3 μL ddH2O. Cycling: 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 40 s; 72 °C for 10 min. Products were separated on 8% denaturing polyacrylamide gels and silver-stained. Band patterns identified alleles: Wxa (287/143 bp, 185/113 bp); Wxb (297/198 bp, 185/113 bp); Wxin (287/143 bp, 185/126 bp); Wxmw (297/198 bp, 185/126 bp).
2.4. Indica–Japonica Classification
Thirty-three InDel markers [11] were used (Table S3). PCR conditions mirrored Waxy genotyping but without inner/outer primers; annealing temperatures varied per marker. Bands were compared to standards (9311 for indica, Nipponbare for japonica). Indica index (FA) and Japonica index (FB) were calculated as:
where XA, XH, XB are homozygous indica, heterozygous, and homozygous japonica loci; N is total loci. Classification followed Table S4.
2.5. Amylose Content Measurement
The amylose content of rice grain samples was determined using the iodine colorimetric method as specified in GB/T 15683-2025 [27]. Rice grains were ground into a fine powder, passed through a 0.5 mm sieve, and dried at 40 °C to constant weight. A 100 mg sample (weighed to 0.1 mg accuracy) was dispersed in 1 mL of 95% ethanol and 9 mL of 1 mol/L NaOH in a 100 mL volumetric flask. The mixture was heated in a boiling water bath for 10 min to gelatinize the starch, cooled to room temperature, and diluted to 100 mL with distilled water. A 5 mL aliquot was transferred to a 50 mL volumetric flask, neutralized with 1 mL of 1 mol/L acetic acid, and mixed with 2 mL of iodine solution (0.2% I2 in 2% KI). The solution was diluted to 50 mL with distilled water and allowed to stand for 20 min to develop a stable blue color. Absorbance was measured at 620 nm using a spectrophotometer, with distilled water as the blank. Each sample was analyzed in three technical replicates. A standard curve, prepared with known amylose standards, was used to calculate the amylose content (% dry weight). For detailed procedures, including reagent preparation and calculation formulas, refer to GB/T 15683-2025 (https://openstd.samr.gov.cn/, accessed on 1 August 2025).
3. Results
3.1. Geographic and Ecological Diversity of Sampled Germplasm
Of the 201 rice accessions, 179 were sourced from 35 counties across 12 of Yunnan’s 16 prefecture-level administrative divisions, encompassing approximately 27% of the province’s 129 counties and achieving a coverage rate of 75% at the prefectural level. The remaining 22 accessions lacked confirmed sources of origin. This sampling strategy captures the province’s complex topography and climatic variability, spanning from high-altitude plateaus in the northwest, such as Ninglang County (elevations exceeding 3000 m), to low-elevation tropical regions in the southeast, like Jinghong County in Xishuangbanna Prefecture (average elevation around 550 m). The sampled counties include highland areas (e.g., Ninglang and Jianchuan Counties), mountainous terrains (e.g., Yun and Yongde Counties), and river valleys (e.g., Yuanyang and Jinghong Counties), representing diverse ecosystems from subtropical forests to alpine meadows and supporting a wide array of agricultural environments that enhance the representativeness of the germplasm for studying rice genetic diversity (Figure 1, Table 1).
Figure 1.
Sampling map of Yunnan Province depicting the distribution of sampled counties, derived from administrative boundary data provided by the National Geomatics Center of China under map review number YunS(2021)46 (http://bzdt.ch.mnr.gov.cn/, accessed on 20 July 2025). Counties with sampling sites are marked by red inverted triangles.

Table 1.
Geographic origin and ecological distribution of the 201 Yunnan local rice accessions.
3.2. Allelic Variation Statistics of the Waxy Gene in 201 Yunnan Local Rice Germplasm Accessions
Waxygt-ARMS amplified 287/143 bp in 180 accessions (G allele) and 297/198 bp in 21 (T allele) (Figure 2). Waxyac-ARMS yielded 185/113 bp in 195 (A allele) and 185/126 bp in 6 (C allele). Combined, alleles were: 175 Wxa, 20 Wxb, 4 Wxin, 2 Wxmw (Figure 2, Table S5).
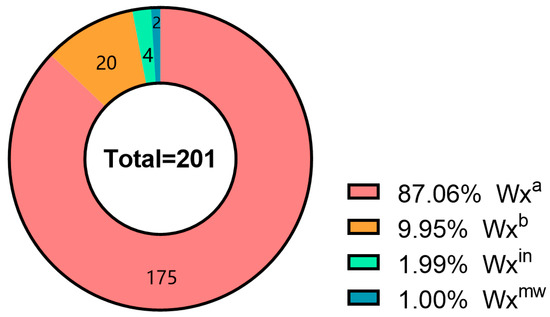
Figure 2.
Allelic Variation Statistics of the Waxy Gene in 201 Yunnan Local Rice Germplasm Accessions. Four colors represent distinct genotypes: Wxa (n = 175), Wxb (n = 20), Wxin (n = 4), and Wxmw (n = 2).
3.3. Distribution of Indica and Japonica Characteristics Among Yunnan Local Rice Germplasm Accessions with Different Waxy Genotypes
InDel markers classified 180 indica (121 typical indica, 53 indica, 6 indica-leaning), 2 intermediate, 19 japonica (2 japonica-leaning, 9 japonica, and 8 typical japonica). Wxa occurred in 175 indica types; Wxb in 15 japonica and 5 indica; Wxin in 2 indica and 2 japonica; Wxmw in 2 japonica. A chi-square test showed a highly significant association between Waxy genotype and rice type (χ2 = 197.3, df = 18, p < 0.001). Wxa was mainly found in Indica (95.4%, including both typical indca and indca), while Wxb was more common in Japonica (75.0%, including both typical japonica and japonica), indicating a strong correlation between Waxy genotype and rice type (Figure 3, Table 2 and Table S6).
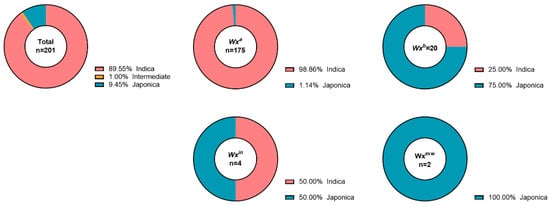
Figure 3.
Distribution of Indica and Japonica Characteristics among Yunnan Local Rice Germplasm Accessions with Different Waxy Genotypes. Indica categories include Typical Indica, Indica, and Indica-Leaning; Japonica categories include Japonica-Leaning, Japonica, and Typical Japonica.

Table 2.
Analysis of the Association between Waxy Genotype and Indica–Japonica type.
3.4. Statistical Analysis Between Different Waxy Genotypes and Their Amylose Content
Amylose content was determined for 201 Yunnan local rice germplasm accessions, revealing significant variation across different Waxy genotypes. AC ranged from 3.74% (Ma Xian Gu(2), ID 139, Wxb) to 29.84% (Chi Bai Gu, ID 27, Wxa), with an overall mean of 21.70% (standard deviation: 5.74%). Among the genotypes, Wxa (175 accessions) exhibited the highest average AC at 22.55% (range: 8.26–29.84%), followed by Wxin (4 accessions) at 24.33% (range: 21.36–28.39%). In contrast, Wxb (20 accessions) and Wxmw (2 accessions) showed lower mean AC values of 13.46% (range: 3.74–24.82%) and 15.65% (range: 12.30–19.00%), respectively. Statistical analysis confirmed that accessions with Wxa and Wxin genotypes had significantly higher AC compared to those with Wxb (Figure 4, Table S7).
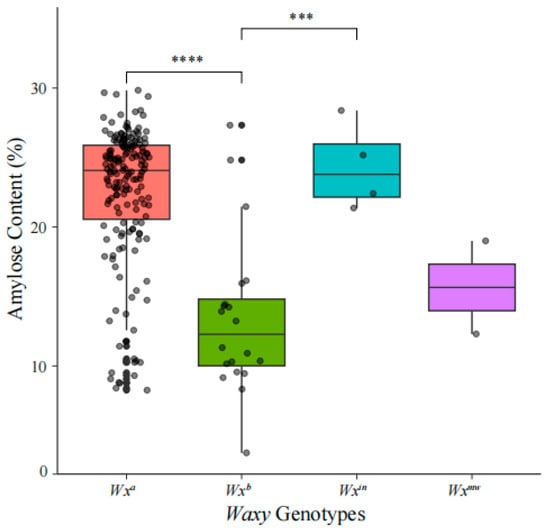
Figure 4.
Significant differences among genotypes were assessed by one-way ANOVA followed by Tukey’s HSD test (*** p < 0.001; **** p < 0.0001).
4. Discussion
Our analysis of 201 Yunnan local rice accessions revealed a diverse yet Wxa-dominant allelic landscape at the Waxy locus, with 175 accessions carrying Wxa (87.06%), 20 Wxb (9.95%), 4 Wxin (1.99%), and 2 Wxmw (1.00%). This predominance of Wxa aligns with global patterns in indica-heavy germplasm [6,14] and underscores Yunnan’s role as an indica-rich diversity center [28]. The detection of rarer alleles like Wxin and Wxmw, though limited in number, highlights untapped variation potentially useful for fine-tuning AC [12,13]. Methodologically, the ARMS-PCR approach proved efficient for multiplex allele discrimination, confirming band patterns consistent with prior validations [11]. However, occasional non-specific bands suggest optimizations in primer ratios or enzyme activity could enhance robustness [17].
Integrating Wx genotyping with AC measurements demonstrated clear allele-specific effects: Wxa and Wxin accessions exhibited high mean AC (22.55% and 24.33%, respectively), while Wxb and Wxmw showed lower values (13.46% and 15.65%). These findings corroborate literature indicating Wxa drives high amylose synthesis via efficient splicing, resulting in firmer grains [3,11], whereas Wxb reduces it through splicing defects [4]. The comparable AC between Wxin and Wxa supports subtle modulatory roles of the intron 6 SNP [12], and similarly for Wxmw on Wxb [13]. Intriguingly, our data did not fully replicate reports of Wxin elevating AC on Wxa backgrounds or lowering it on Wxb [6], possibly due to small sample sizes for rare alleles or interactions with background loci like ALK or SSIIa [15,16]. Future studies with sufficiently large sample sizes should be conducted using balanced designs to validate these findings. In addition, potential interactions between ALK and SSIIa alleles should be carefully examined to clarify their confounding effects. Broader comparisons reveal parallels with Korean rice collections, where Wx haplotypes explained ECQ variance [20], and Vietnamese cultivars, where SNPs at Wx modulated glycemic index via AC [21]. In Yunnan context, these alleles may interact with environmental factors (e.g., altitude), as high-elevation landraces often exhibit altered starch profiles [10].
It should be specifically noted that the apparent amylose content of 3.74% measured by iodine colorimetry in this study may have limitations. This value could partially originate from non-specific binding of iodine to long-chain amylopectin rather than true amylose, a phenomenon particularly significant in low-amylose samples [1]. Although the iodine colorimetric method offers advantages of operational simplicity and low cost, its accuracy is highly dependent on the quality of standard curve construction [2]. When using potato amylose for calibration, the iodine-binding capacity of amylopectin may lead to false positive values in waxy rice samples [3]. In contrast, physical analysis methods such as Size Exclusion Chromatography (SEC) and Differential Scanning Calorimetry (DSC) can directly resolve starch molecular structure, providing more accurate results for waxy samples [4]. While the standard curve in this study was strictly prepared according to GB/T 15683-2008, future research will consider adopting mixed standard (amylose-amylopectin) calibration curves to further improve detection accuracy in the low concentration range [29].
Subspecies classification via InDel markers identified 180 indica-types (89.55%), 19 japonica-types (9.45%), and 2 intermediates (1.00%), confirming pronounced indica–japonica differentiation with indica dominance [8,30]. This mirrors altitudinal patterns in Yunnan, where indica prevails in warmer lowlands and japonica in cooler highlands [31]. Wxa was overwhelmingly associated with indica (173/175), reinforcing subspecies-specific distributions [6,18], while Wxb appeared in 15 japonica but also 5 indica, suggesting occasional introgression [28]. Rare alleles like Wxin spanned both subspecies, hinting at ancestral polymorphisms predating differentiation [14,15]. The low frequency of intermediates (1%) contrasts with some Yunnan studies reporting higher admixture [32], potentially due to our germplasm’s bias toward cultivated landraces. Nonetheless, these intermediates could facilitate indica–japonica hybrids, enhancing fertility and yield via heterosis [19,20].
From a breeding perspective, our results advocate MAS using Wx functional markers to tailor AC for premium varieties—e.g., incorporating Wxin into Wxa backgrounds for moderate AC and improved palatability [14]. The identified Wxin and Wxmw donors could adjust starch viscosity and fissure resistance [16], addressing quality issues in high-yielding hybrids. Exploiting Yunnan’s diversity for heterosis, as in indica–japonica crosses, may boost grain quality traits [33], especially with intermediates bridging compatibility [34]. Limitations include the focus on four alleles; future work should screen for emerging variants like Wxlv or Wxla using whole-genome sequencing to capture epistatic interactions [35].
In this study, although our findings are consistent with previously reported associations between Waxy (Wx) alleles, amylose content, and indica–japonica differentiation, the work provides an important regional contribution. The predominance of the Wxa allele (87.06%) in Yunnan aligns with its high frequency in indica-rich regions like Vietnam (83.5%) but contrasts sharply with temperate japonica collections from Korea where Wxb predominates (91.3%) [14]. Yunnan Province is recognized as one of the global centers of rice genetic diversity, harboring a wide range of landraces with unique genetic backgrounds. Systematically characterizing the distribution of Wx alleles in this germplasm not only confirms global patterns but also offers critical baseline data for future research and breeding [22,23].
Unlike many previous studies that focused primarily on identifying novel Wx alleles, our analysis emphasizes the allele frequency distribution and its association with subspecies identity in a large, representative set of Yunnan rice germplasm. This provides valuable information for germplasm conservation, selection of parental lines for breeding, and for monitoring allele shifts under modern breeding and climate change. Even though no completely new alleles were detected, these findings strengthen the understanding of the genetic structure of local rice populations and complement global research on rice quality improvement.
In conclusion, this study elucidates Wx allelic dynamics in Yunnan rice, linking them to subspecies attributes and AC. These insights pave the way for targeted breeding, leveraging local germplasm to develop high-quality, resilient varieties amid climate challenges.
5. Conclusions
Yunnan rice exhibits Waxy allelic diversity with Wxa predominance, linked to indica types and higher AC. These insights facilitate breeding for quality and heterosis utilization.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16101198/s1, Table S1. Sources of 201 Yunnan Local Rice Germplasm Accessions. Table S2. Information of molecular markers of Waxy gene alleles. Table S3. Information of molecular markers of Indica-Japonica Classification. Table S4. Criteria for Classification of Indica and Japonica Rice. Table S5. Details of Allelic Variations in the Waxy Gene across 201 Yunnan Local Rice Germplasm Accessions. Table S6. Amylose Content Associated with Different Waxy Genotypes in 201 Yunnan Local Rice Germplasm Accessions. Table S7. Indica-Japonica Attributes Associated with Different Waxy Genotypes in 201 Yunnan Local Rice Germplasm Accessions.
Author Contributions
Conceptualization, Y.L. and X.L.; methodology, Y.L.; software, X.Z. (Xueqian Zuo); validation, D.L., Y.D. and Y.X.; formal analysis, Y.L.; investigation, J.Z. (Jianhua Zhang), X.Z. (Xiao Zhang), J.Z. (Jinwen Zhang), J.T. (Jian Tu), L.K. and A.G.; resources, X.S., W.D. and X.L.; data curation, X.Z. (Xueqian Zuo) and Y.X.; writing—original draft preparation, Y.L.; writing—review and editing, J.T. (Jing Tan), W.D. and X.L.; visualization, Y.L.; supervision, X.L.; project administration, W.D.; funding acquisition, X.L., W.D. and J.Z. (Jianhua Zhang). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Yunnan Seed Industry Joint Laboratory Program (202205AR070001-6) and Yunnan Province Major Science and Technology (202402AE090036, 202402AE090010, and 202302AE090003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Acknowledgments
We thank the Yunnan Academy of Agricultural Sciences for materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| AC | Amylose content |
| ALK | Gene associated with alkali spreading value/gelatinization temperature (linked with SSIIa) |
| ARMS-PCR | Amplification Refractory Mutation System PCR |
| bp | Base pair |
| ECQ | Eating and cooking quality |
| FA | Indica index |
| FB | Japonica index |
| GBSSI | Granule-bound starch synthase I |
| InDel | Insertion–deletion polymorphism |
| MAS | Marker-assisted selection |
| PCR | Polymerase chain reaction |
| SD | Standard deviation |
| SNP | Single-nucleotide polymorphism |
| SSR | Simple sequence repeat |
| SSIIa | Soluble starch synthase IIa |
| Wx | Waxy gene (granule-bound starch synthase locus) |
References
- Balindong, J.L.; Ward, R.M.; Liu, L.; Rose, T.J.; Pallas, L.A.; Ovenden, B.W.; Snell, P.J.; Waters, D.L.E. Rice grain protein composition influences instrumental measures of rice cooking and eating quality. J. Cereal Sci. 2018, 79, 35–42. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, L.; Feng, L.; Jiang, J.; Huang, L.; Liu, Q.; Zhang, Y.; Zhang, C.; Liu, Q. Deciphering the Role of Waxy Gene Mutations in Enhancing Rice Grain Quality. Foods 2024, 13, 1624. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Cai, X.L.; Li, Q.F.; Tang, S.Z.; Gong, Z.-Y.; Yu, H.-X.; Yan, C.-J.; Wang, Z.-Y.; Gu, M.H. Molecular marker-assisted selection for improving cooking and eating quality in Teqing and its hybid rice. J. Acta Agron. Sin. 2006, 32, 64–69. [Google Scholar]
- Biselli, C.; Cavalluzzo, D.; Perrini, R.; Gianinetti, A.; Bagnaresi, P.; Urso, S.; Orasen, G.; Desiderio, F.; Lupotto, E.; Cattivelli, L.; et al. Improvement of marker-based predictability of Apparent Amylose Content in japonica rice through GBSSI allele mining. Rice 2014, 7, 1. [Google Scholar] [CrossRef]
- Shao, Y.; Peng, Y.; Mao, B.; Lv, Q.; Yuan, D.; Liu, X.; Zhao, B. Allelic variations of the Wx locus in cultivated rice and their use in the development of hybrid rice in China. PLoS ONE 2020, 15, e0232279. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Sheng, W.-T.; Wu, J.; Zhang, C.-Q.; Liu, Q.-Q.; Deng, Q.-Y. Differential expressions among five Waxy alleles their effects on the eating cooking qualities in specialty rice cultivars. J. Integr. Agric. 2015, 14, 1153–1162. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zheng, F.Q.; Shen, G.Z.; Gao, J.P.; Snustad, D.P.; Li, M.G.; Zhang, J.L.; Hong, M.M. The amylose content in rice endosperm is related to the post–transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar]
- Wang, F.; Sun, T.; Yu, S.; Liu, C.; Cheng, Z.; Xia, J.; Han, L. Ethnobotanical studies on rice landraces under on-farm conservation in Xishuangbanna of Yunnan Province, China. J. Ethnobiol. Ethnomed. 2024, 20, 45. [Google Scholar] [CrossRef]
- Ma, M.; Lei, E.; Wang, T.; Meng, H.; Zhang, W.; Lu, B. Genetic Diversity and Association Mapping of Grain-Size Traits in Rice Landraces from the Honghe Hani Rice Terraces System in Yunnan Province. Plants 2023, 12, 1678. [Google Scholar] [CrossRef]
- Zeng, Y.; Shen, S.; Li, Z.; Yang, Z.; Wang, X.; Zhang, H.; Wen, G. Ecogeographic and genetic diversity based on morphological characters of indigenous rice (Oryza sativa L.) in Yunnan, China. J. Genet. Resour. 2003, 50, 567–577. [Google Scholar]
- Liu, Y.; Zhang, A.; Wang, F.; Wang, J.; Bi, J.; Kong, D.; Zhang, F.; Luo, L.; Liu, G.; Yu, X. Development and validation of a PCR-based functional marker system for identifying the low amylose content-associated gene Wx (hp) in rice. Breed. Sci. 2019, 69, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ye, J.; Zhai, R.; Wu, M.; Yu, F.; Zhang, X.; Zhu, G.; Han, J.; Ye, S. Identification of Indica–Japonica Attributes and Analysis of Heterosis Using InDel Markers. Agronomy 2024, 14, 2832. [Google Scholar] [CrossRef]
- Teng, B.; Zeng, R.; Wang, Y.; Liu, Z.; Zhang, Z.; Zhu, H.; Ding, X.; Li, W.; Zhang, G. Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol. Breed. 2011, 30, 583–595. [Google Scholar] [CrossRef]
- Zhou, H.; Xia, D.; Zhao, D.; Li, Y.; Li, P.; Wu, B.; Gao, G.; Zhang, Q.; Wang, G.; Xiao, J.; et al. The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 2021, 63, 878–888. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, J.; Chen, S.; Fan, X.; Li, Q.; Lu, Y.; Wang, M.; Yu, H.; Yi, C.; Tang, S.; et al. Wxlv, the Ancestral Allele of Rice Waxy Gene. Mol. Plant 2019, 12, 1157–1166. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Y.; Chen, S.; Liu, X.; Zhu, J.; Zhou, L.; Lu, Y.; Li, Q.; Fan, X.; Tang, S.; et al. A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 2021, 63, 889–901. [Google Scholar] [CrossRef]
- Medrano, R.F.; de Oliveira, C.A. Guidelines for the tetra-primer ARMS-PCR technique development. Mol. Biotechnol. 2014, 56, 599–608. [Google Scholar] [CrossRef]
- Sano, Y. Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 1984, 68, 467–473. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, F.; Zafar, S.; Wang, J.; Lu, H.; Naveed, S.; Lou, J.; Xu, J. Genetic dissection of heterosis of indica-japonica by introgression line, recombinant inbred line and their testcross populations. Sci. Rep. 2021, 11, 10265. [Google Scholar] [CrossRef]
- Maung, T.Z.; Yoo, J.M.; Chu, S.H.; Kim, K.W.; Chung, I.M.; Park, Y.J. Haplotype Variations and Evolutionary Analysis of the Granule-Bound Starch Synthase I Gene in the Korean World Rice Collection. Front. Plant Sci. 2021, 12, 707237. [Google Scholar] [CrossRef]
- Hoai, T.T.; Matsusaka, H.; Toyosawa, Y.; Suu, T.D.; Satoh, H.; Kumamaru, T. Influence of single-nucleotide polymorphisms in the gene encoding granule-bound starch synthase I on amylose content in Vietnamese rice cultivars. Breed. Sci. 2014, 64, 142–148. [Google Scholar] [CrossRef]
- Chen, H.; Shan, J.; Yang, K.; Wang, Y.Y.; Lu, C.M. Abundant variation of Waxy gene in Yunnan rice landraces and molecular characterization of a novel Wxzm allele. Crop Sci. 2014, 54, 2152–2159. [Google Scholar] [CrossRef]
- Liu, L.; Ma, X.; Liu, S.; Zhu, C.; Jiang, L.; Wang, Y.; Shen, Y.; Ren, Y.; Dong, H.; Chen, L.; et al. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 2009, 71, 609–626. [Google Scholar] [CrossRef]
- Zeng, Y.W.; Li, Z.C.; Shen, S.Q.; Wang, X.K.; Yang, Z.Y.; Zhang, H.; Chen, Y.M. Diversity and Good Germplasm of Indigenous Rice Varieties in Yunnan Province. J. Chin. J. Rice Sci. 2001, 15, 169–174. [Google Scholar]
- Zeng, Y.; Zhang, H.; Li, Z.; Shen, S.; Sun, J.; Wang, M.; Liao, D.; Liu, X.; Wang, X.; Xiao, F.; et al. Evaluation of Genetic Diversity of Rice Landraces (Oryza sativa L.) in Yunnan, China. Breed. Sci. 2007, 57, 91–99. [Google Scholar] [CrossRef]
- Ma, M.; Liu, Y.; Meng, H.; Lu, B. Genetic Diversity Analysis of Red Rice from Hani’s Terraced Fields in Yunnan Province. In Proceedings of the 2016 International Conference on Artificial Intelligence and Engineering Applications, Hong Kong, China, 12–13 November 2016. [Google Scholar]
- GB/T 15683-2025; Inspection of Grain and Oils—Determination of amylose content in rice. Standards Press of China: Beijing, China, 2025.
- Cheng, Z.-Q.; Huang, X.-Q.; Ying, F.-Y.; Li, D.-Q.; Yu, T.-Q.; Fu, J.; Yan, H.-J.; Zhong, Q.-F.; Zhang, D.-Y.; Li, W.-J. Genetic Diversity of Wild Rice Species in Yunnan Province of China. Rice Sci. 2012, 19, 21–28. [Google Scholar] [CrossRef]
- Fitzgerald, M.A.; Bergman, C.J.; Resurreccion, A.P.; Möller, J.; Jimenez, R.; Reinke, R.F.; Martin, M.; Blanco, P.; Molina, F.; Chen, M.-H.; et al. Addressing the dilemmas of measuring amylose in rice. Cereal Chemistry. Cereal Chem. 2009, 86, 492–498. [Google Scholar] [CrossRef]
- Cui, D.; Tang, C.; Lu, H.; Li, J.; Ma, X.; A, X.; Han, B.; Yang, Y.; Dong, C.; Zhang, F.; et al. Genetic differentiation and restricted gene flow in rice landraces from Yunnan, China: Effects of isolation-by-distance and isolation-by-environment. Rice 2021, 14, 54. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhang, S.; Wang, Y.; Ford-Lloyd, B.V.; Tu, M.; Jin, X.; Wu, Y.; Yan, H.; Yang, X.; Liu, P.; et al. Differentiation and distribution of indica and japonica rice varieties along the altitude gradients in Yunnan Province of China as revealed by InDel molecular markers. Genet. Resour. Crop. Evol. 2010, 57, 891–902. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, J.; Wang, M.; Liao, D.; Zeng, Y.; Shen, S.; Yu, P.; Mu, P.; Wang, X.; Li, Z. Genetic structure and phylogeography of rice landraces in Yunnan, China, revealed by SSR. Genome 2007, 50, 72–83. [Google Scholar] [CrossRef]
- Feng, L.; Lu, C.; Yang, Y.; Lu, Y.; Li, Q.; Huang, L.; Fan, X.; Liu, Q.; Zhang, C. The Physicochemical Properties of Starch Are Affected by Wxlv in Indica Rice. Foods 2021, 10, 3089. [Google Scholar] [CrossRef]
- Hori, K.; Suzuki, K.; Ishikawa, H.; Nonoue, Y.; Nagata, K.; Fukuoka, S.; Tanaka, J. Genomic Regions Involved in Differences in Eating and Cooking Quality Other than Wx and Alk Genes between indica and japonica Rice Cultivars. Rice 2021, 14, 8. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, T.; Hua, Y.; Yan, X.; Liu, X.; Liu, Y.; Liu, Y.; Zhang, B.; Liu, R.; Zhu, Z.; et al. Assessment of the Characteristics of Waxy Rice Mutants Generated by CRISPR/Cas9. Front. Plant Sci. 2022, 13, 881964. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).