PRC2 Diversity in Neuronal Differentiation and Developmental Disorders
Abstract
1. Introduction
2. The Discovery of Polycomb Group (PcG) Proteins as Epigenetic Guardians of Gene Repression
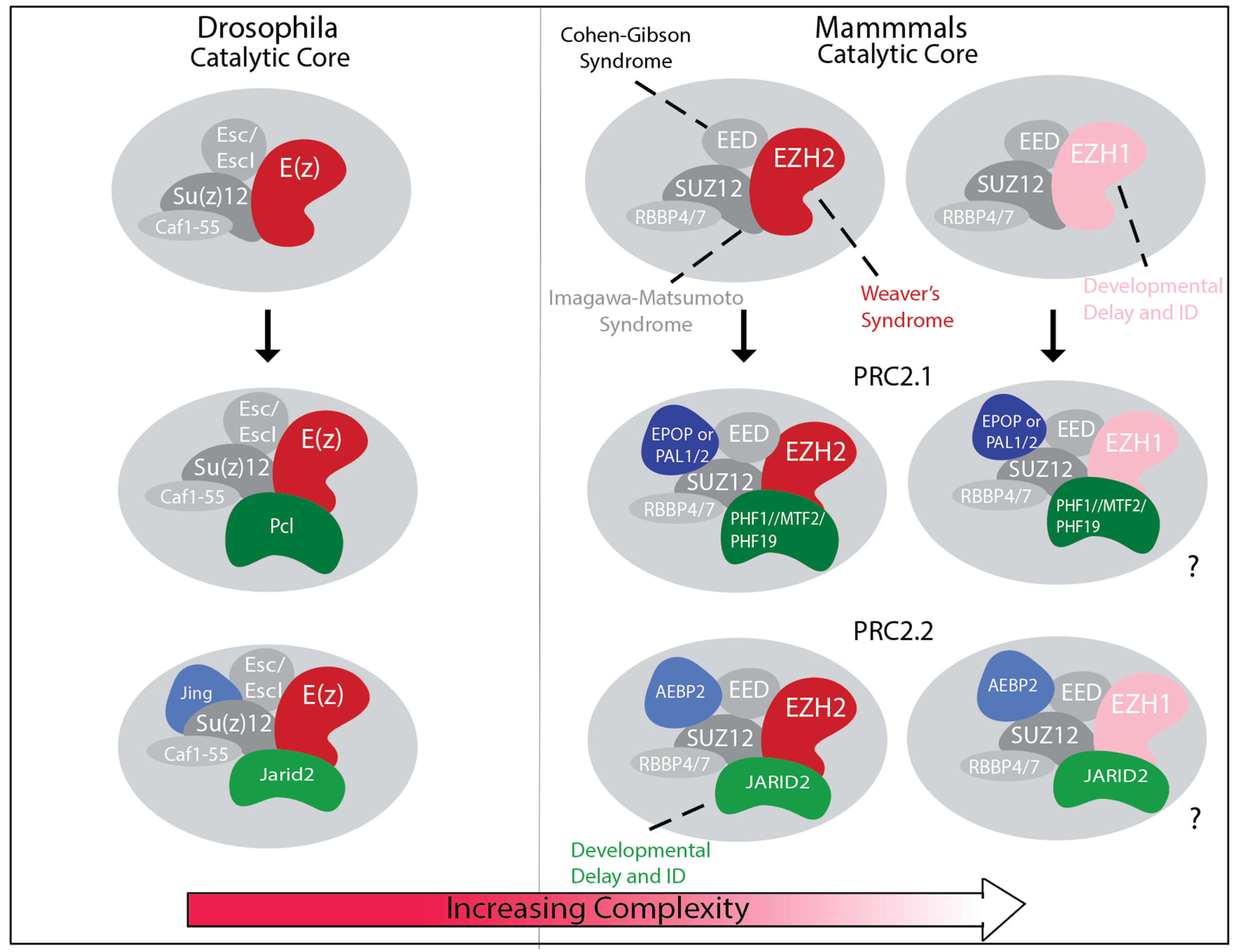
3. Diversification of PRC2 Composition
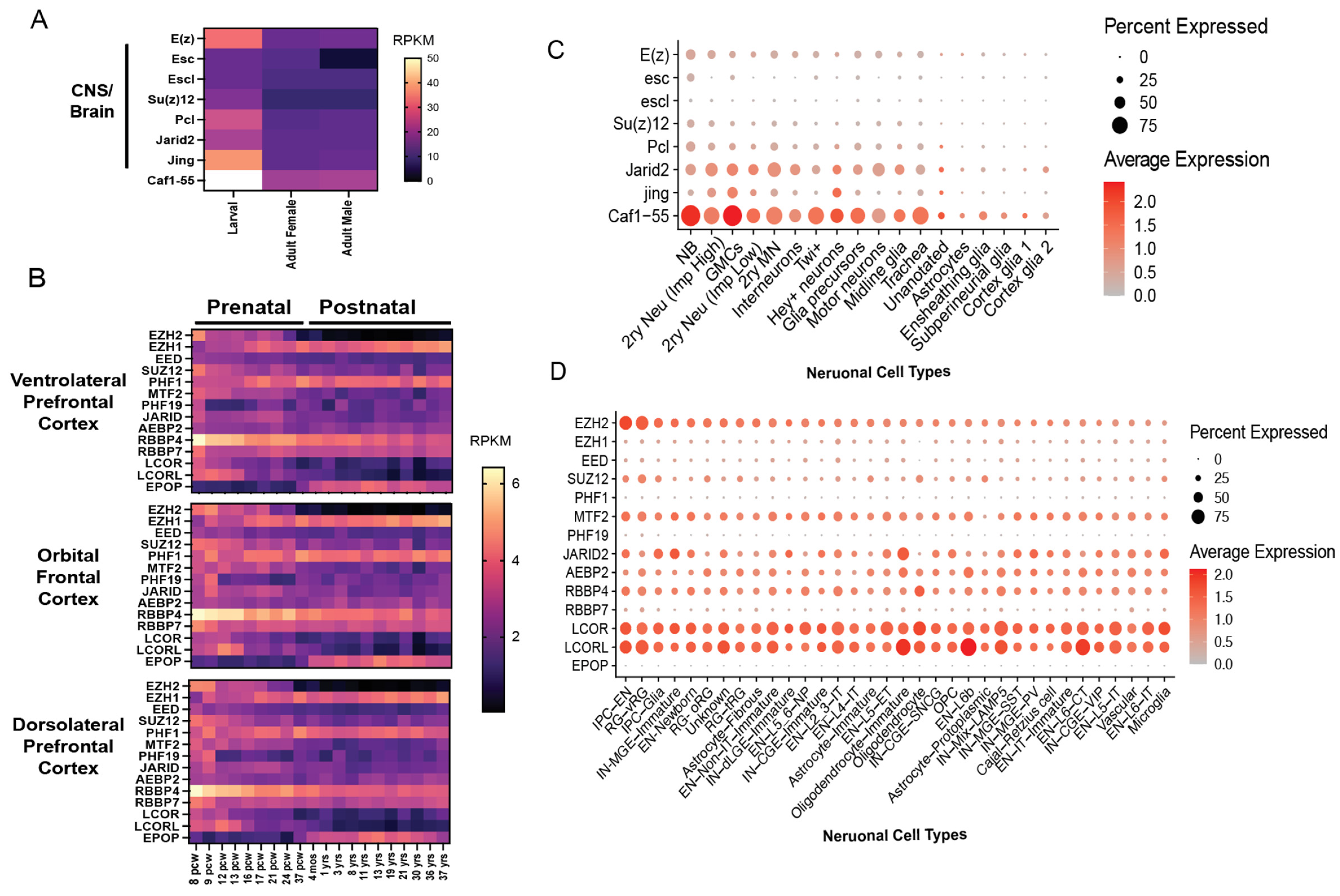
4. Context-Specific Functions of PRC2: Lessons from Development and Neurodevelopmental Disorders
4.1. EZH2, EED and SUZ12 Associated Overgrowth with Intellectual Disability (OGID) Syndromes
4.2. EZH1 Gain- and Loss-of-Function Variants in Overlapping Neurodevelopmental Syndromes
4.3. JARID2-Associated Neurodevelopmental Syndrome
4.4. PRC2 Subunits Not Yet Implicated in Human Neurodevelopmental Disease
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silbereis, J.C.; Pochareddy, S.; Zhu, Y.; Li, M.; Sestan, N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 2016, 89, 248–268. [Google Scholar] [CrossRef]
- Yoon, K.J.; Vissers, C.; Ming, G.L.; Song, H. Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. J. Cell Biol. 2018, 217, 1901–1914. [Google Scholar] [CrossRef]
- Hirabayashi, Y.; Gotoh, Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 2010, 11, 377–388. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kingston, R.E. Context-specific Polycomb mechanisms in development. Nat. Rev. Genet. 2022, 23, 680–695. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Diaz, C.; Zhou, Y.; Yang, Q.; Maroofian, R.; Espana-Bonilla, P.; Lee, C.H.; Zhang, S.; Padilla, N.; Fueyo, R.; Waxman, E.A.; et al. Gain and loss of function variants in EZH1 disrupt neurogenesis and cause dominant and recessive neurodevelopmental disorders. Nat. Commun. 2023, 14, 4109. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Shao, Z.; Li, D.; Xie, H.; Kim, W.; Huang, J.; Taylor, J.E.; Pinello, L.; Glass, K.; Jaffe, J.D.; et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 2015, 57, 304–316. [Google Scholar] [CrossRef]
- Mousavi, K.; Zare, H.; Wang, A.H.; Sartorelli, V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell 2012, 45, 255–262. [Google Scholar] [CrossRef]
- Lee, C.H.; Holder, M.; Grau, D.; Saldana-Meyer, R.; Yu, J.R.; Ganai, R.A.; Zhang, J.; Wang, M.; LeRoy, G.; Dobenecker, M.W.; et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol. Cell 2018, 70, 435–448.e5. [Google Scholar] [CrossRef]
- Lewis, P.; Mislove, R. New mutants report. Drosoph. Inf. Serv. 1947, 21, 69. [Google Scholar]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Jürgens, G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 1985, 316, 153–155. [Google Scholar] [CrossRef]
- Struhl, G.; Akam, M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985, 4, 3259–3264. [Google Scholar] [CrossRef]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 1975, 14, 9–25. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Nanney, D.L. Epigenetic Control Systems. Proc. Natl. Acad. Sci. USA 1958, 44, 712–717. [Google Scholar] [CrossRef]
- Holliday, R. Epigenetics: An overview. Dev. Genet. 1994, 15, 453–457. [Google Scholar] [CrossRef]
- Strutt, H.; Cavalli, G.; Paro, R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997, 16, 3621–3632. [Google Scholar] [CrossRef]
- Orlando, V.; Paro, R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 1993, 75, 1187–1198. [Google Scholar] [CrossRef] [PubMed]
- Breiling, A.; Edgar, B.; Simona, F.; Becker, B.P.; Paro, R. The Drosophila Polycomb Protein Interacts with Nucleosomal Core Particles In Vitro via Its Repression Domain. Mol. Cell. Biol. 1999, 19, 8451–8460. [Google Scholar] [CrossRef]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.-t.; Bender, W.; Kingston, R.E. Stabilization of Chromatin Structure by PRC1, a Polycomb Complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.A.; Ng, J.; Peterson, A.J.; Morgan, K.; Simon, J.; Jones, R.S. The Drosophila esc and E(z) Proteins Are Direct Partners in Polycomb Group-Mediated Repression. Mol. Cell. Biol. 1998, 18, 2825–2834. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Furuyama, T.; Harte, P.J. The Drosophila Polycomb Group proteins ESC and E (Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 1998, 125, 3483–3496. [Google Scholar] [CrossRef] [PubMed]
- Platero, J.S.; Sharp, E.J.; Adler, P.N.; Eissenberg, J.C. In vivo assay for protein-protein interactions using Drosophila chromosomes. Chromosoma 1996, 104, 393–404. [Google Scholar] [CrossRef]
- Rastelli, L.; Chan, C.S.; Pirrotta, V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993, 12, 1513–1522. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes. Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef]
- Uckelmann, M.; Davidovich, C. Chromatin compaction by Polycomb group proteins revisited. Curr. Opin. Struct. Biol. 2024, 86, 102806. [Google Scholar] [CrossRef]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Kasinath, V.; Beck, C.; Sauer, P.; Poepsel, S.; Kosmatka, J.; Faini, M.; Toso, D.; Aebersold, R.; Nogales, E. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 2021, 371, 362. [Google Scholar] [CrossRef]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Muller, C.W.; Vermeulen, M.; Muller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef]
- Tamburri, S.; Lavarone, E.; Fernandez-Perez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874.e9. [Google Scholar] [CrossRef] [PubMed]
- Youmans, D.T.; Gooding, A.R.; Dowell, R.D.; Cech, T.R. Competition between PRC2.1 and 2.2 subcomplexes regulates PRC2 chromatin occupancy in human stem cells. Mol. Cell 2021, 81, 488–501.e9. [Google Scholar] [CrossRef]
- Glancy, E.; Wang, C.; Tuck, E.; Healy, E.; Amato, S.; Neikes, H.K.; Mariani, A.; Mucha, M.; Vermeulen, M.; Pasini, D.; et al. PRC2.1- and PRC2.2-specific accessory proteins drive recruitment of different forms of canonical PRC1. Mol. Cell 2023, 83, 1393–1411.e7. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Gorchakov, A.A.; Kharchenko, P.V.; Kuroda, M.I. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. USA 2014, 111, 2488–2493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jones, A.; Sun, C.W.; Li, C.; Chang, C.W.; Joo, H.Y.; Dai, Q.; Mysliwiec, M.R.; Wu, L.C.; Guo, Y.; et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 2011, 29, 229–240. [Google Scholar] [CrossRef]
- Ryan, C.W.; Peirent, E.R.; Regan, S.L.; Guxholli, A.; Bielas, S.L. H2A monoubiquitination: Insights from human genetics and animal models. Hum. Genet. 2024, 143, 511–527. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Wang, G.G. Polycomb Gene Silencing Mechanisms: PRC2 Chromatin Targeting, H3K27me3 ‘Readout’, and Phase Separation-Based Compaction. Trends Genet. 2021, 37, 547–565. [Google Scholar] [CrossRef]
- Oksuz, O.; Narendra, V.; Lee, C.H.; Descostes, N.; LeRoy, G.; Raviram, R.; Blumenberg, L.; Karch, K.; Rocha, P.P.; Garcia, B.A.; et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol. Cell 2018, 70, 1149–1162.e5. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Zylicz, J.J.; Bousard, A.; Zumer, K.; Dossin, F.; Mohammad, E.; da Rocha, S.T.; Schwalb, B.; Syx, L.; Dingli, F.; Loew, D.; et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 2019, 176, 182–197.e3. [Google Scholar] [CrossRef]
- Nekrasov, M.; Klymenko, T.; Fraterman, S.; Papp, B.; Oktaba, K.; Kocher, T.; Cohen, A.; Stunnenberg, H.G.; Wilm, M.; Muller, J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007, 26, 4078–4088. [Google Scholar] [CrossRef]
- Herz, H.M.; Mohan, M.; Garrett, A.S.; Miller, C.; Casto, D.; Zhang, Y.; Seidel, C.; Haug, J.S.; Florens, L.; Washburn, M.P.; et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol. Cell Biol. 2012, 32, 1683–1693. [Google Scholar] [CrossRef]
- Wen, C.; Margolis, M.; Dai, R.; Zhang, P.; Przytycki, P.F.; Vo, D.D.; Bhattacharya, A.; Matoba, N.; Tang, M.; Jiao, C.; et al. Cross-ancestry atlas of gene, isoform, and splicing regulation in the developing human brain. Science 2024, 384, eadh0829. [Google Scholar] [CrossRef] [PubMed]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- Paulsen, B.; Velasco, S.; Kedaigle, A.J.; Pigoni, M.; Quadrato, G.; Deo, A.J.; Adiconis, X.; Uzquiano, A.; Sartore, R.; Yang, S.M.; et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 2022, 602, 268–273. [Google Scholar] [CrossRef]
- Villa, C.E.; Cheroni, C.; Dotter, C.P.; Lopez-Tobon, A.; Oliveira, B.; Sacco, R.; Yahya, A.C.; Morandell, J.; Gabriele, M.; Tavakoli, M.R.; et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 2022, 39, 110615. [Google Scholar] [CrossRef]
- Ciceri, G.; Baggiolini, A.; Cho, H.S.; Kshirsagar, M.; Benito-Kwiecinski, S.; Walsh, R.M.; Aromolaran, K.A.; Gonzalez-Hernandez, A.J.; Munguba, H.; Koo, S.Y.; et al. An epigenetic barrier sets the timing of human neuronal maturation. Nature 2024, 626, 881–890. [Google Scholar] [CrossRef]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.E.; Gillis, J.; Kramer, M.; Lihm, J.; Yoon, S.; Berstein, Y.; Mistry, M.; Pavlidis, P.; Solomon, R.; Ghiban, E.; et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 2014, 19, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Kochinke, K.; Zweier, C.; Nijhof, B.; Fenckova, M.; Cizek, P.; Honti, F.; Keerthikumar, S.; Oortveld, M.A.; Kleefstra, T.; Kramer, J.M.; et al. Systematic Phenomics Analysis Deconvolutes Genes Mutated in Intellectual Disability into Biologically Coherent Modules. Am. J. Hum. Genet. 2016, 98, 149–164. [Google Scholar] [CrossRef]
- Cyrus, S.; Burkardt, D.; Weaver, D.D.; Gibson, W.T. PRC2-complex related dysfunction in overgrowth syndromes: A review of EZH2, EED, and SUZ12 and their syndromic phenotypes. Am. J. Med. Genet. Part C Semin. Med. Genet. 2019, 181, 519–531. [Google Scholar] [CrossRef]
- Verberne, E.A.; Goh, S.; England, J.; van Ginkel, M.; Rafael-Croes, L.; Maas, S.; Polstra, A.; Zarate, Y.A.; Bosanko, K.A.; Pechter, K.B.; et al. JARID2 haploinsufficiency is associated with a clinically distinct neurodevelopmental syndrome. Genet. Med. 2021, 23, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, P.; Yin, Y.; Bates, A.S.; Dorkenwald, S.; Eichler, K.; Brooks, P.; Han, D.S.; Gkantia, M.; Dos Santos, M.; Munnelly, E.J.; et al. Whole-brain annotation and multi-connectome cell typing of Drosophila. Nature 2024, 634, 139–152. [Google Scholar] [CrossRef]
- Azevedo, F.A.; Carvalho, L.R.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.; Leite, R.E.; Jacob Filho, W.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009, 513, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, C.; Moriano, J.A.; Chen, S.; Zuo, G.; Cebrian-Silla, A.; Zhang, S.; Mukhtar, T.; Wang, S.; Song, M.; et al. Molecular and cellular dynamics of the developing human neocortex. Nature 2025, 1–10. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Vicidomini, R.; Choudhury, S.D.; Han, T.H.; Maric, D.; Brody, T.; Serpe, M. scRNA-seq data from the larval Drosophila ventral cord provides a resource for studying motor systems function and development. Dev. Cell 2024, 59, 1210–1230.e9. [Google Scholar] [CrossRef]
- Gibson, W.T.; Hood, R.L.; Zhan, S.H.; Bulman, D.E.; Fejes, A.P.; Moore, R.; Mungall, A.J.; Eydoux, P.; Babul-Hirji, R.; An, J.; et al. Mutations in EZH2 cause Weaver syndrome. Am. J. Hum. Genet. 2012, 90, 110–118. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Hanks, S.; Ruark, E.; Zachariou, A.; Duarte Sdel, V.; Ramsay, E.; Snape, K.; Murray, A.; Perdeaux, E.R.; Seal, S.; et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2011, 2, 1127–1133. [Google Scholar] [CrossRef]
- Weaver, D.D.; Graham, C.B.; Thomas, I.T.; Smith, D.W. A new overgrowth syndrome with accelerated skeletal maturation, unusual facies, and camptodactyly. J. Pediatr. 1974, 84, 547–552. [Google Scholar] [CrossRef]
- Deevy, O.; Li, J.; Monger, C.; Matra, F.; Tuck, E.; Davies, M.; Badonyi, M.; Boyce, M.; Doyle, E.J.; Hokamp, K.; et al. Dominant-negative effects of Weaver syndrome-associated EZH2 variants. Genes. Dev. 2025. [Google Scholar] [CrossRef]
- Lui, J.C.; Barnes, K.M.; Dong, L.; Yue, S.; Graber, E.; Rapaport, R.; Dauber, A.; Nilsson, O.; Baron, J. Ezh2 Mutations Found in the Weaver Overgrowth Syndrome Cause a Partial Loss of H3K27 Histone Methyltransferase Activity. J. Clin. Endocrinol. Metab. 2018, 103, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.S.; Yap, D.B.; Lewis, M.E.; Chijiwa, C.; Ramos-Arroyo, M.A.; Tkachenko, N.; Milano, V.; Fradin, M.; McKinnon, M.L.; Townsend, K.N.; et al. Weaver Syndrome-Associated EZH2 Protein Variants Show Impaired Histone Methyltransferase Function In Vitro. Hum. Mutat. 2016, 37, 301–307. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Murray, A.; Hanks, S.; Douglas, J.; Armstrong, R.; Banka, S.; Bird, L.M.; Clericuzio, C.L.; Cormier-Daire, V.; Cushing, T.; et al. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am. J. Med. Genet. A 2013, 161, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Al-Salem, A.; Alshammari, M.J.; Hassan, H.; Alazami, A.M.; Alkuraya, F.S. Weaver syndrome and defective cortical development: A rare association. Am. J. Med. Genet. A 2013, 161, 225–227. [Google Scholar] [CrossRef]
- Ren, L.; Jiang, L.; Jiang, X.; Wang, H.; Li, Y.; Ren, X. Clinical and genetic characterization of Weaver syndrome: A case report of an EZH2 mutation and review of the literature. Medicine 2025, 104, e44080. [Google Scholar] [CrossRef]
- Choufani, S.; Gibson, W.T.; Turinsky, A.L.; Chung, B.H.Y.; Wang, T.; Garg, K.; Vitriolo, A.; Cohen, A.S.A.; Cyrus, S.; Goodman, S.; et al. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020, 106, 596–610. [Google Scholar] [CrossRef]
- Gao, C.W.; Lin, W.; Riddle, R.C.; Kushwaha, P.; Boukas, L.; Bjornsson, H.T.; Hansen, K.D.; Fahrner, J.A. A mouse model of Weaver syndrome displays overgrowth and excess osteogenesis reversible with KDM6A/6B inhibition. JCI Insight 2024, 9, e173392. [Google Scholar] [CrossRef]
- Gibson, W.T.; Lengyell, T.C.; Korecki, A.J.; Janssen, S.M.; Adair, B.A.; Gamu, D.; Lorincz, M.C.; Simpson, E.M. Minimally Humanized Ezh2 Exon-18 Mouse Cell Lines Validate Preclinical CRISPR/Cas9 Approach to Treat Weaver Syndrome. Hum. Gene Ther. 2025, 36, 618–627. [Google Scholar] [CrossRef]
- Imagawa, E.; Seyama, R.; Aoi, H.; Uchiyama, Y.; Marcarini, B.G.; Furquim, I.; Honjo, R.S.; Bertola, D.R.; Kim, C.A.; Matsumoto, N. Imagawa-Matsumoto syndrome: SUZ12-related overgrowth disorder. Clin. Genet. 2023, 103, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Loveday, C.; Zachariou, A.; Behan, L.A.; Chandler, K.; Cole, T.; D’Arrigo, S.; Dieckmann, A.; Foster, A.; Gibney, J.; et al. EED and EZH2 constitutive variants: A study to expand the Cohen-Gibson syndrome phenotype and contrast it with Weaver syndrome. Am. J. Med. Genet. A 2019, 179, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Imagawa, E.; Higashimoto, K.; Sakai, Y.; Numakura, C.; Okamoto, N.; Matsunaga, S.; Ryo, A.; Sato, Y.; Sanefuji, M.; Ihara, K.; et al. Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum. Mutat. 2017, 38, 637–648. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015, 350, aac4383. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Lazzerini Denchi, E.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.D.; Sansom, S.N.; Smith, J.; Dobenecker, M.W.; Tarakhovsky, A.; Livesey, F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 15957–15962. [Google Scholar] [CrossRef]
- Miro, X.; Zhou, X.; Boretius, S.; Michaelis, T.; Kubisch, C.; Alvarez-Bolado, G.; Gruss, P. Haploinsufficiency of the murine polycomb gene Suz12 results in diverse malformations of the brain and neural tube. Dis. Model. Mech. 2009, 2, 412–418. [Google Scholar] [CrossRef]
- Prokopuk, L.; Stringer, J.M.; White, C.R.; Vossen, R.; White, S.J.; Cohen, A.S.A.; Gibson, W.T.; Western, P.S. Loss of maternal EED results in postnatal overgrowth. Clin. Epigenet. 2018, 10, 95. [Google Scholar] [CrossRef]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.; et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Kahn, T.G.; Nix, D.A.; Li, X.Y.; Bourgon, R.; Biggin, M.; Pirrotta, V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006, 38, 700–705. [Google Scholar] [CrossRef]
- Tolhuis, B.; de Wit, E.; Muijrers, I.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 694–699. [Google Scholar] [CrossRef]
- Chamberlain, S.J.; Yee, D.; Magnuson, T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 2008, 26, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Thornton, S.R.; Butty, V.L.; Levine, S.S.; Boyer, L.A. Polycomb Repressive Complex 2 regulates lineage fidelity during embryonic stem cell differentiation. PLoS ONE 2014, 9, e110498. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Molina, S.; Respuela, P.; Tebartz, C.; Kolovos, P.; Nikolic, M.; Fueyo, R.; van Ijcken, W.F.J.; Grosveld, F.; Frommolt, P.; Bazzi, H.; et al. PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell 2017, 20, 689–705.e9. [Google Scholar] [CrossRef]
- Miller, S.A.; Damle, M.; Kim, J.; Kingston, R.E. Full methylation of H3K27 by PRC2 is dispensable for initial embryoid body formation but required to maintain differentiated cell identity. Development 2021, 148, dev196329. [Google Scholar] [CrossRef]
- Collinson, A.; Collier, A.J.; Morgan, N.P.; Sienerth, A.R.; Chandra, T.; Andrews, S.; Rugg-Gunn, P.J. Deletion of the Polycomb-Group Protein EZH2 Leads to Compromised Self-Renewal and Differentiation Defects in Human Embryonic Stem Cells. Cell Rep. 2016, 17, 2700–2714. [Google Scholar] [CrossRef]
- Shan, Y.; Liang, Z.; Xing, Q.; Zhang, T.; Wang, B.; Tian, S.; Huang, W.; Zhang, Y.; Yao, J.; Zhu, Y.; et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 2017, 8, 672. [Google Scholar] [CrossRef]
- van Mierlo, G.; Dirks, R.A.M.; De Clerck, L.; Brinkman, A.B.; Huth, M.; Kloet, S.L.; Saksouk, N.; Kroeze, L.I.; Willems, S.; Farlik, M.; et al. Integrative Proteomic Profiling Reveals PRC2-Dependent Epigenetic Crosstalk Maintains Ground-State Pluripotency. Cell Stem Cell 2019, 24, 123–137.e8. [Google Scholar] [CrossRef] [PubMed]
- Zijlmans, D.W.; Talon, I.; Verhelst, S.; Bendall, A.; Van Nerum, K.; Javali, A.; Malcolm, A.A.; van Knippenberg, S.; Biggins, L.; To, S.K.; et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 2022, 24, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.J.; Brody, L.C.; Valdes, J.M.; Erdos, M.R.; McKinley, D.R.; Castilla, L.H.; Merajver, S.D.; Couch, F.J.; Friedman, L.S.; Ostermeyer, E.A.; et al. Characterization of EZH1, a human homolog of Drosophila Enhancer of zeste near BRCA1. Genomics 1996, 37, 161–171. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.L.; Son, J.; Drury, W.J., 3rd; Voigt, P.; Martin, S.R.; Taylor, W.R.; De Marco, V.; et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009, 461, 762–767. [Google Scholar] [CrossRef]
- Grau, D.; Zhang, Y.; Lee, C.H.; Valencia-Sanchez, M.; Zhang, J.; Wang, M.; Holder, M.; Svetlov, V.; Tan, D.; Nudler, E.; et al. Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction. Nat. Commun. 2021, 12, 714. [Google Scholar] [CrossRef]
- Hu, H.; Kahrizi, K.; Musante, L.; Fattahi, Z.; Herwig, R.; Hosseini, M.; Oppitz, C.; Abedini, S.S.; Suckow, V.; Larti, F.; et al. Genetics of intellectual disability in consanguineous families. Mol. Psychiatry 2019, 24, 1027–1039. [Google Scholar] [CrossRef]
- Okamoto, N.; Yoshida, S.; Ogitani, A.; Etani, Y.; Yanagi, K.; Kaname, T. Biallelic loss-of-function variants of EZH1 cause a novel developmental disorder with central precocious puberty. Am. J. Med. Genet. A 2024, 194, e63726. [Google Scholar] [CrossRef]
- von Schimmelmann, M.; Feinberg, P.A.; Sullivan, J.M.; Ku, S.M.; Badimon, A.; Duff, M.K.; Wang, Z.; Lachmann, A.; Dewell, S.; Ma’ayan, A.; et al. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 2016, 19, 1321–1330. [Google Scholar] [CrossRef]
- Volkel, P.; Bary, A.; Raby, L.; Chapart, A.; Dupret, B.; Le Bourhis, X.; Angrand, P.O. Ezh1 arises from Ezh2 gene duplication but its function is not required for zebrafish development. Sci. Rep. 2019, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiao, L.; Liu, X.; Yang, X.; Liu, X. A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin. Mol. Cell 2020, 77, 1265–1278.e7. [Google Scholar] [CrossRef] [PubMed]
- Viitasalo, L.; Kettunen, K.; Kankainen, M.; Niemela, E.H.; Kiiski, K. A novel partial de novo duplication of JARID2 gene causing a neurodevelopmental phenotype. Mol. Genet. Genom. Med. 2022, 10, e2037. [Google Scholar] [CrossRef]
- Cadieux-Dion, M.; Farrow, E.; Thiffault, I.; Cohen, A.S.A.; Welsh, H.; Bartik, L.; Schwager, C.; Engleman, K.; Zhou, D.; Zhang, L.; et al. Phenotypic expansion and variable expressivity in individuals with JARID2-related intellectual disability: A case series. Clin. Genet. 2022, 102, 136–141. [Google Scholar] [CrossRef]
- Baroy, T.; Misceo, D.; Stromme, P.; Stray-Pedersen, A.; Holmgren, A.; Rodningen, O.K.; Blomhoff, A.; Helle, J.R.; Stormyr, A.; Tvedt, B.; et al. Haploinsufficiency of two histone modifier genes on 6p22.3, ATXN1 and JARID2, is associated with intellectual disability. Orphanet J. Rare Dis. 2013, 8, 3. [Google Scholar] [CrossRef]
- Miller, J.A.; Ding, S.L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional landscape of the prenatal human brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Gachechiladze, M.A.; Ludwig, C.H.; Laurie, M.T.; Hong, J.Y.; Nathaniel, D.; Prabhu, A.V.; Fernandopulle, M.S.; Patel, R.; Abshari, M.; et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 2019, 104, 239–255.e2. [Google Scholar] [CrossRef]
- Yu, J.R.; Lee, C.H.; Oksuz, O.; Stafford, J.M.; Reinberg, D. PRC2 is high maintenance. Genes Dev. 2019, 33, 903–935. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Watanabe, Y.; Takano-Shimizu, T.; Kondo, S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 2006, 235, 2449–2459. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef]
- Shen, X.; Kim, W.; Fujiwara, Y.; Simon, M.D.; Liu, Y.; Mysliwiec, M.R.; Yuan, G.C.; Lee, Y.; Orkin, S.H. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009, 139, 1303–1314. [Google Scholar] [CrossRef]
- Landeira, D.; Sauer, S.; Poot, R.; Dvorkina, M.; Mazzarella, L.; Jorgensen, H.F.; Pereira, C.F.; Leleu, M.; Piccolo, F.M.; Spivakov, M.; et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 2010, 12, 618–624. [Google Scholar] [CrossRef]
- Pasini, D.; Cloos, P.A.; Walfridsson, J.; Olsson, L.; Bukowski, J.P.; Johansen, J.V.; Bak, M.; Tommerup, N.; Rappsilber, J.; Helin, K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010, 464, 306–310. [Google Scholar] [CrossRef]
- Petracovici, A.; Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 2021, 81, 2625–2639.e5. [Google Scholar] [CrossRef]
- Gracia-Diaz, C.; Perdomo, J.E.; Khan, M.E.; Roule, T.; Disanza, B.L.; Cajka, G.G.; Lei, S.; Gagne, A.L.; Maguire, J.A.; Shalem, O.; et al. KOLF2.1J iPSCs carry CNVs associated with neurodevelopmental disorders. Cell Stem Cell 2024, 31, 288–289. [Google Scholar] [CrossRef]
- Pantazis, C.B.; Yang, A.; Lara, E.; McDonough, J.A.; Blauwendraat, C.; Peng, L.; Oguro, H.; Kanaujiya, J.; Zou, J.; Sebesta, D.; et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell Stem Cell 2022, 29, 1685–1702.e2. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Isono, K.; Yamada, D.; Endo, T.A.; Endoh, M.; Shinga, J.; Mizutani-Koseki, Y.; Otte, A.P.; Casanova, M.; Kitamura, H.; et al. Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the HOX gene cluster and at Cdkn2a genes. Mol. Cell Biol. 2011, 31, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Grijzenhout, A.; Godwin, J.; Koseki, H.; Gdula, M.R.; Szumska, D.; McGouran, J.F.; Bhattacharya, S.; Kessler, B.M.; Brockdorff, N.; Cooper, S. Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 2016, 143, 2716–2723. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, K.; Ekram, M.B.; Roh, T.Y.; Kim, J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS ONE 2011, 6, e25174. [Google Scholar] [CrossRef]
| Drosophila | |||
| Complex | Subunit | Molecular Function | Mutant Phenotype |
| PRC2 | E(z) | Catalyzes H3K27 methylation | Improper body segmentation; additional sex combs; lethality |
| PRC2 | Esc/Escl | Facilitates histone methyltransferase activity | Improper body segmentation; additional sex combs; lethality |
| PRC2 | Su(z)12 | Facilitates histone methyltransferase activity | Improper body segmentation; additional sex combs; lethality |
| PRC2 | Caf1-55 | Enhances PRC2 activity | Lethality |
| PRC2.1 | Pcl | Enhances the generation of H3K27me3 at target loci | Improper body segmentation; additional sex combs; lethality |
| PRC2.2 | Jing | Modulates PRC2 methyltransferase activity | Homeotic transformations; Tracheal and nervous system defects; lethality |
| PRC2.2 | Jarid2 | Modulates PRC2 methyltransferase activity | Improper body segmentation; lethality |
| Humans | |||
| Complex | Subunit | Molecular Function | Neurodevelopmental Disease |
| PRC2.1/PRC2.2 | EZH2 | Catalyzes H3K27 methylation | Weaver syndrome: overgrowth, moderate intellectual disability (in about 80% of patients), distinctive facial appearance with macrocephaly and occasionally polymicrogyria |
| PRC2.1/PRC2.2 | EZH1 | Catalyzes H3K27 methylation | NDD with developmental delay, mild-to severe intellectual disability and atypical facial features |
| PRC2.1/PRC2.2 | EED | Facilitates and enhances H3K27 methylation and propagation | Cohen–Gibson Syndrome: Overgrowth, mild-to-severe intellectual disability (in 100% of patients), distinctive facial appearance with macrocephaly |
| PRC2.1/PRC2.2 | SUZ12 | Facilitates H3K27 methylation and stabilizes PRC2 complexes and interactions | Imagawa–Matsumoto syndrome: overgrowth, mild intellectual disability (in ~50% of patients). |
| PRC2.1/PRC2.2 | RBBP4/7 | Facilitates PRC2 recruitment to target loci | N/A |
| PRC2.1 | PCL1/PHF1 | Enhances H3K27 methylation and promotes recruitment to target loci | N/A |
| PRC2.1 | PCL2/MTF2 | Promotes recruitment to target loci and maintains H3K27 methylation | N/A |
| PRC2.1 | PCL2/PHF19 | Enhances H3K27 methylation and promotes recruitment to target loci | N/A |
| PRC2.1 | EPOP | Modulates and promotes PRC2 methyltransferase activity | N/A |
| PRC2.1 | PAL1/2 | Modulates and promotes PRC2 methyltransferase activity | N/A |
| PRC2.2 | JARID2 | Facilitates recruitment of PRC2 to target loci and enhances deposition of H3K27 methylation | Developmental delay and intellectual disability with dysmorphic facies. |
| PRC2.2 | AEBP2 | Stabilizes PRC2 and helps stimulate H3K27 methylation activity | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akoto, J.; Roule, T.; Akizu, N. PRC2 Diversity in Neuronal Differentiation and Developmental Disorders. Genes 2025, 16, 1191. https://doi.org/10.3390/genes16101191
Akoto J, Roule T, Akizu N. PRC2 Diversity in Neuronal Differentiation and Developmental Disorders. Genes. 2025; 16(10):1191. https://doi.org/10.3390/genes16101191
Chicago/Turabian StyleAkoto, Jasmine, Thomas Roule, and Naiara Akizu. 2025. "PRC2 Diversity in Neuronal Differentiation and Developmental Disorders" Genes 16, no. 10: 1191. https://doi.org/10.3390/genes16101191
APA StyleAkoto, J., Roule, T., & Akizu, N. (2025). PRC2 Diversity in Neuronal Differentiation and Developmental Disorders. Genes, 16(10), 1191. https://doi.org/10.3390/genes16101191





