Abstract
Background: Extremely low birth weight (ELBW) and extremely low gestational age (ELGA) remain major challenges in neonatology, contributing to neonatal morbidity and mortality. This study aims to examine the association between functional variants of MTHFR and PON1, genes involved in homocysteine metabolism, and the risk of ELGA, ELBW, and other complications of prematurity. A meta-analysis was also conducted to integrate literature data with the results of this study. Methods: The study included 377 premature infants, 164 mothers, and a population-based sample of 404 individuals. Genotyping was performed using TaqMan assays. Results: The fetal, but not maternal, MTHFR rs1801133 genotype was associated with ELBW (OR = 1.65; 95% CI: 1.09–2.51; p = 0.017, dominant model), bronchopulmonary dysplasia (p = 0.028), patent ductus arteriosus (p = 0.017), and neonatal mortality. The meta-analysis, which included five studies spanning 1156 cases and 1124 controls, confirmed the association between the neonatal MTHFR genotype and low birth weight (LBW), demonstrating an association of the rs1801133T allele with LBW in the TT homozygote model (vs. CT: OR = 1.41; 95% CI: 1.08–1.80; p = 0.0097). Subgroup analyses indicated that the rs1801133T allele is a protective factor against LBW in more developed countries, such as Canada and the UK (dominant model), whereas in other countries, such as China, Turkey, and Poland, it is a risk factor for LBW (recessive model). No association with PON1 variants with ELBW or ELGA was found. Conclusions: This study provides the first global evidence confirming that the neonatal MTHFR genotype contributes to LBW, underscoring the population-specific effects of this genetic variant.
1. Introduction
Advances in neonatology and obstetric care have significantly improved the survival of infants born prematurely, with low birth weight (LBW, <2500 g), or small for gestational age (SGA). It is estimated that one-quarter of liveborn infants are now affected by one or more of these three vulnerabilities. Because of their similar pathogenic mechanisms and therapeutic approaches, these newborns have recently been collectively termed small vulnerable newborns (SVNs) [1]. Among SVNs, those with extremely low birth weight (ELBW), defined as a birth weight below 1000 g, and extremely low gestational age (ELGA), characterized as a gestational age below 28 weeks, represent the phenotypes of extreme prematurity. These infants remain at the highest risk for prematurity-related morbidity and perinatal mortality and require the most intensive and specialized medical care [2,3].
ELBW and ELGA are multifactorial traits influenced by both maternal and fetal factors [4,5]. The genetic basis of these traits remains incompletely understood. Twin concordance studies suggest that maternal genetic factors have a significant influence on gestational age, a pattern consistent with mitochondrial inheritance [6,7]. By contrast, variation in birth weight is determined mainly by fetal genes, with heritability estimated at around 70% [8]. Recent genome-wide association studies have identified 60 variants associated with birth weight, showing that the fetal genotype accounts for most of the variability in birth weight in 93% of the identified loci [9]. Among the candidate genes, the methylenetetrahydrofolate reductase (MTHFR) gene is one of the most frequently analyzed in the context of low gestational age and low birth weight. Previous studies, summarized in meta-analyses, have confirmed the important role of the maternal MTHFR genotype [10,11]. However, few studies have been conducted to date to investigate the role of fetal genotype, and none have specifically addressed its association with ELBW or ELGA.
MTHFR is an enzyme involved in the metabolism of folate, which catalyzes the transfer of methyl groups necessary for DNA synthesis, methylation, and the conversion of the toxic metabolite homocysteine (Hcy) to methionine [12]. The MTHFR gene is located on chromosome 1 [1p36.22] and has a common variant rs1801133 (c.665C>T; p.Ala222Val) resulting in reduced enzyme stability and activity [13]. In rs1801133 CT heterozygotes, enzyme activity is about 64% of normal, while in TT homozygotes it decreases to ~32%. This reduction in MTHFR activity leads to elevated Hcy levels, particularly when folate intake is insufficient [14,15]. Given the crucial role of methyl groups in fetal growth and development [16,17], supplementation with folic acid or its active form, 5-methyltetrahydrofolate, is recommended both before and during pregnancy [18,19].
The toxic effects of elevated Hcy levels are exerted in part through an increase in the concentration of its reactive derivative, Hcy-thioxanthone (Hcy-TLC) [20]. This compound is detoxified by paraoxonase 1 (PON1) [21], an antioxidant enzyme encoded by the PON1 gene located on chromosome 7 [7q21.3]. Two common coding variants of PON1, rs854560 (c.163T>A; p.Leu55Met) and rs662 (c.575A>G; p.Gln192Arg), influence hydrolytic activity toward Hcy-TLC (paraoxonase/thiolactonase). The rs854560A (55Met) allele is associated with lower thiolactonase activity, while the rs662G (192Arg) allele is associated with higher thiolactonase activity, with rs662GG homozygotes showing about four-fold-higher activity compared to rs662AA individuals [22,23]. Moreover, the presence of the rs854560A allele is associated with elevated serum PON1 concentrations, which can be attributed in part to linkage disequilibrium with certain promoter variants that influence gene expression [24,25].
This study aims to comprehensively evaluate the relationship between functional variants of MTHFR and PON1, key genes involved in Hcy and Hcy–TLC metabolism, and the risk of ELGA, ELBW, and other complications of prematurity. A meta-analysis integrating literature data with the study’s results was also performed to validate the impact of the MTHFR variant on birth weight.
2. Materials and Methods
2.1. Study Population
The study included a population of 377 infants born before 32 weeks of gestation enrolled between 2009 and 2020 from the Neonatal Intensive Care Unit at the Clinical Hospital of Gynecology and Obstetrics at Poznan University of Medical Sciences. In the first stage of the study (between 2009 and 2014), 164 mothers of premature infants were also recruited. Additionally, 404 individuals (49.9% women) with mean age 49.1 ± 15.5 years from the local adult population (Great Poland region) without known signs of chronic diseases were analyzed to obtain population genotype frequencies (see Figure 1 for details). The exclusion criteria for the study included infants born from multiple pregnancies and those with chromosomal abnormalities. All participants in the study were of Caucasian origin.
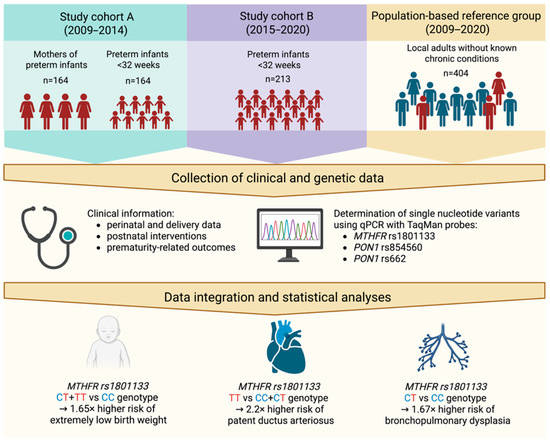
Figure 1.
Schematic overview of the study design and obtained results. Created in BioRender. Skulimowski, B. (2025) https://BioRender.com/3l5efzr (accessed on 19 August 2025).
2.2. Clinical Features and Outcomes
The following clinical factors were analyzed: gestational age (weeks), birth weight (grams), sex, preterm prelabor rupture of membranes (PPROM) duration (days), mode of delivery (vaginal delivery vs. cesarean section), Apgar scores at 1 and 5 min, blood transfusions, surfactant and oxygen therapy, and duration of mechanical ventilation (days). ELGA was diagnosed for gestational age <28 weeks and ELBW for birth weight <1000 g. The incidence of SGA and fetal growth restriction (FGR; formerly termed intrauterine growth retardation, IUGR) was assessed. The diagnosis of SGA was based on WHO criteria, defined as neonatal weight below the 10th percentile for gestational age [26]. FGR was diagnosed according to Lees et al. (2020), based on biometric and Doppler ultrasonography, with consideration of gestational age and growth dynamics [27]. The presence of BPD, intraventricular hemorrhage (IVH), patent ductus arteriosus (PDA), respiratory distress syndrome (RDS), necrotizing enterocolitis (NEC), periventricular leukomalacia (PVL), and retinopathy of prematurity (ROP), including its proliferative type (pROP), was assessed. The criteria for diagnosing BPD, IVH, RDS, NEC, PVL, and ROP have been described in detail previously [28]. The diagnosis of PDA was based on ultrasonography.
2.3. Genotyping
For DNA isolation, 1 ml of venous blood or a swab from the inner cheek (in duplicate) was collected. The samples were frozen at −80 °C and delivered to the Institute of Human Genetics of the Polish Academy of Sciences, where genomic DNA isolation and genetic analysis were performed. Genomic DNA was extracted from circulating blood lymphocytes using the QIAamp DNA Kit (Qiagen GmbH, Hilden, Germany) or from buccal swabs using the innuPREP DNA Kit (Analytik Jena AG, Jena, Germany), according to the manufacturer’s instructions. Analysis of the three variants, MTHFR rs1801133, PON1 rs854560, and rs662, was performed using quantitative polymerase chain reaction (qPCR) with commercially available, pre-designed TaqMan genotyping assays (Thermo Fisher Scientific, Waltham, MA, USA): C___1202883_20, C___2259750_20, and C___2548962_20, respectively). Genotype analysis was performed on an ABI 7900HT Fast Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). Genotyping success rate was between 99–100%.
2.4. Ethical Statement
All procedures carried out on human participants in this study were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments (or comparable ethical standards). The study was approved by the Bioethics Committee of Poznan University of Medical Sciences (no. 1140/05, issued on 8 September 2005; no. 1117/18 from 7 November 2018). Written prior informed consent was obtained from all participants, including consent from the parents or guardians of the study infants.
2.5. Meta-Analysis of the MTHFR rs1801133 Variant and Risk of Low Birth Weight
The meta-analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [29]. Associations of the MTHFR rs1801133 variant with growth-related neonatal outcomes were examined.
2.5.1. Search Strategy
A literature search was conducted in PubMed (including MEDLINE), Cochrane Library, Embase, Scopus, Web of Science, Google Scholar, and associated tools (including LitVar2 NCBI) to identify relevant studies on the neonatal MTHFR genotype. The search included publications up to 10 August 2025. A combination of the following keywords and MeSH terms was used: (“Methylenetetrahydrofolate reductase” OR “MTHFR” OR “C677T” OR “C665T” OR “rs1801133”) AND (“variant” OR “variation” OR “polymorphism” OR “mutation”) AND (“Preterm Birth” OR “Low Birth Weight” OR “Fetal Growth Retardation” OR “Intrauterine Growth Retardation” OR “Small for Gestational Age”). References contained in the retrieved articles were also reviewed and included as additional studies when they met the eligibility criteria. For studies using overlapping data or the same study group, only the most complete or recent studies were included.
2.5.2. Inclusion and Exclusion Criteria
The following criteria were applied to include studies in the meta-analysis: (a) studies with a case–control, cohort or family-based design; (b) studies focusing on the association between MTHFR variant and LBW/FGR/IUGR/SGA; and (c) availability of sufficient genotype data in the case and control groups to calculate crude odds ratios (ORs) and 95% confidence intervals (CIs). The exclusion criteria were as follows: (a) studies not designed as case–control, cohort, or family-based; (b) lack of genotype data or inability to calculate it; (c) studies based on pedigree data, twins studies, and linkage studies; (d) case reports, review articles, posters, abstracts, and animal studies; and (e) studies with incomplete or overlapping data. In cases of overlapping or duplicate publications, only the largest or most recent dataset was included.
2.5.3. Data Extraction
Two authors independently reviewed and extracted the following information from all included studies using a structured data collection form: first author name, publication date, participant location, ethnicity, presence of premature neonates, studied outcomes, genotyping method, sample sizes of cases and controls, genotype frequency distribution, minor allele frequency (MAF), and Hardy–Weinberg equilibrium (HWE) in controls. Any discrepancies were resolved by consensus among all authors. If detailed genotype or HWE information was not reported, we calculated the necessary data and added the relevant details.
2.5.4. Quality Assessment
The quality (Q) of studies was independently assessed by the Newcastle–Ottawa quality scale (NOS) [30,31,32]. Studies with scores of 0–3, 4–6, 7–9 were, respectively, considered as low-, moderate-, and high-quality.
2.6. Statistical Analysis
Testing for HWE was performed using a χ2 test with the online tool available at https://www.had2know.org/academics/hardy-weinberg-equilibrium-calculator-2-alleles.html (accessed on 23 June 2025). For each variant, the MAF was calculated. The effect of genotypes was estimated using ORs with 95% CIs, with minor alleles considered as risk factors. Continuous variables were tested for normality using the Kolmogorov–Smirnov test. In statistical analyses, qualitative variables were analyzed using the χ2 test or Fisher’s exact test, while quantitative variables were assessed using the t-test. Variables deviating from the normal distribution were evaluated using Mann–Whitney U test.
The meta-analysis was performed using the online tool METAGENYO available at http://metagenyo.genyo.es/ (accessed on 11 August 2025) [33]. Six genetic models were examined for each variant: allele contrast, recessive, dominant, overdominant, homozygote comparison, and heterozygote comparison. Heterogeneity was evaluated using Cochran’s Q and I2 statistics [34], with FEM applied in cases of low heterogeneity and REM otherwise. For each model, ORs, p-values (95% CIs), and adjusted p-values were calculated; HWE in controls was tested with corresponding p-values and adjusted p-values. Publication bias was assessed using funnel plots and Egger’s test.
All reported tests were two-sided, with statistical significance defined as p < 0.05. Statistical analyses were performed using STATISTICA version 10.0 and GraphPad Prism version 6.04. Post hoc power analysis for associations was performed using Quanto v 1.2 software. For visualization of birth weight and gestational age distributions, a plot was generated using the “ggplot2” and “introdataviz” packages in the R environment version 4.5.1. [35,36,37].
3. Results
3.1. Demographic and Clinical Characteristics of the Study Group and Associations with ELBW and ELGA
The characteristics of the study cohort of 377 premature infants are presented in Table 1. We identified 149 infants with ELBW (39.5%) and 152 with ELGA (40.3%).

Table 1.
Demographic and clinical characteristics of premature infants and their associations with ELBW and ELGA.
Demographic and clinical characteristics of ELBW and ELGA infants were compared with those of non-ELBW and non-ELGA infants to assess associations with extreme prematurity phenotypes. Both ELBW and ELGA infants had lower birth weight and gestational age. ELBW was positively associated with SGA (OR = 2.3; p = 0.0004) and FGR/IUGR (OR = 13.0; p < 0.0001), whereas ELGA was negatively associated with SGA (OR = 0.22; p < 0.0001) and showed a nonsignificant trend toward lower incidence of FGR/IUGR. As expected, ELBW and ELGA were associated with lower Apgar scores at 1 and 5 min, respiratory failure (including more frequent surfactant therapy, resuscitation, mechanical ventilation, and prolonged oxygen supplementation), increased incidence of prematurity-related complications (except PDA, which was associated with ELBW only), and higher mortality rates. No associations with sex were observed.
In the analysis of maternal risk factors, maternal age (mean 28.3 ± 5.8 years; range 17–45) was not associated with either ELBW or ELGA. The women studied had a mean of 1.3 ± 1.8 previous pregnancies, including 24 women (14.6%) with a history of preterm birth and 28 (17.1%) with a history of spontaneous miscarriage. Delivery by cesarean section was negatively associated with ELGA (p ≤ 0.01), with no effect on ELBW, whereas PPROM and prolonged PPROM were not associated with extreme prematurity phenotypes.
When comparing the clinical profiles of cohorts A (n = 164) and B (n = 213) to assess the consistency of risk factors over time, we observed lower birth weight (p = 0.02) and a higher incidence of FGR/IUGR (p = 0.049) in the more recent cohort B, accompanied by more intensive respiratory support, including prolonged mechanical ventilation (p = 0.0001) and oxygen supplementation (p = 0.020; Supplemental Table S1). A higher incidence of RDS (p < 0.0001) and proliferative ROP (p = 0.041), a complication related to oxygen supplementation, was also noted in this cohort. Conversely, mechanical ventilation was less frequently initiated in this group, and the incidence of PPROM and prolonged PPROM decreased over the study period.
3.2. Frequencies of MTHFR and PON1 Variants in the Study Groups
The distribution of the studied MTHFR and PON1 genotypes was consistent with the Hardy–Weinberg equilibrium (p > 0.05) in all studied groups: infants, mothers, and the population group (Table 2). There were no differences in the distribution of studied variants between groups: the frequency of the MTHFR rs1801133T allele was between 0.293 and 0.330, the PON1 rs854560T allele between 0.314 and 0.378, and the PON1 rs662G allele between 0.265 and 0.287. For genotypes, two effects were found: the decreased frequency of MTHFR rs1801133T allele carriers in infants compared to the population group (OR = 0.75; p = 0.046 (Table 2, Figure 2)), and higher frequency of the PON1 rs854560TT homozygotes in infants compared to mothers: OR = 2.00; p = 0.029.

Table 2.
Comparison of MTHFR and PON1 genotype frequencies between study groups.
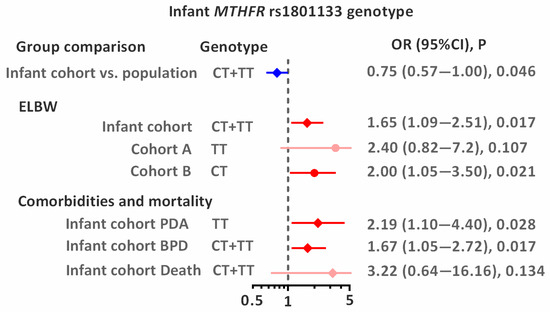
Figure 2.
Associations between infant MTHFR rs1801133 genotype and studied outcomes. Odds ratios (ORs) with 95% confidence intervals (CIs) are shown. The dashed vertical line indicates the null value (OR = 1), and horizontal lines represent 95% CIs. Blue indicates lower risk, whereas red indicates higher risk (significant association or statistical trend).
3.3. Associations of MTHFR and PON1 Genotypes with ELBW and ELGA
The significant association between infant MTHFR rs1801133 genotype and ELBW was observed in the studied population (Table 3 and Figure 2). In the combined analysis of both cohorts (A + B), carriers of the rs1801133T allele had a 1.65-fold increased risk of ELBW (p = 0.017). Although studied cohorts did not differ significantly in genotype distribution, the association of the rs1801133T allele with ELBW tended to be stronger for homozygotes (OR = 2.4; p = 0.107) in cohort A and for heterozygotes (OR = 2.0; p = 0.021) in cohort B. Analysis by sex revealed that the MTHFR genotype in newborns (heterozygotes) is a risk factor for both ELBW (OR = 1.89; p = 0.038) and ELGA (OR = 1.90; p = 0.034; Supplemental Table S2). No statistically significant association was observed between maternal genotypes and ELBW or ELGA. The distribution of birth weight and gestational age according to sex and MTHFR rs1801133 genotype is shown in Figure 3.

Table 3.
Distribution of studied genetic variants in preterm infants and their mothers from the Polish population in relation to time of enrollment, extremely low birth weight (ELBW) and extremely low gestational age (ELGA). The table presents Hardy–Weinberg equilibrium (HWE) p-values, minor allele frequencies (MAFs), and the impact of SNVs on ELBW and ELGA.
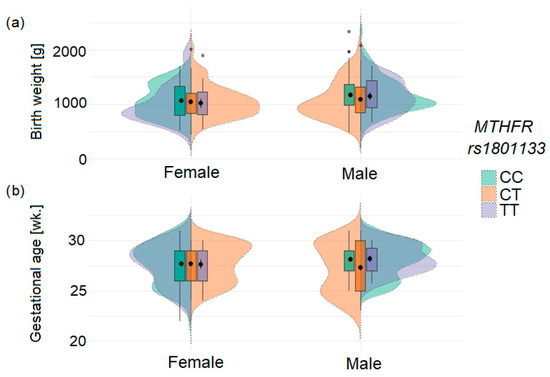
Figure 3.
Distribution of birth weight (a) and gestational age (b) in infants according to sex and MTHFR rs1801133 genotype. The plots illustrate differential effects of TT homozygotes and CT heterozygotes by sex. Black dots indicate mean values, and error bars represent standard deviations; grey dots represent outlier values.
3.4. Associations of MTHFR and PON1 Genotypes with Neonatal Comorbidities and Mortality
The study was conducted on the entire group of premature infants (n = 377; Table 4). Two statistically significant associations were observed for the MTHFR rs1801133T variant: the CT heterozygous genotype was associated with a 1.67-fold-higher risk of developing BPD (vs. CC; p = 0.017), and the TT homozygous genotype was associated with a 2.19-fold-higher risk of developing PDA (recessive model; p = 0.028). A borderline statistically significant association was also observed between the PON1 rs662AG heterozygous genotype and the risk of PDA (vs. AA homozygotes: OR = 1.80; p = 0.053). No significant associations were found between the genotypes studied and the occurrence of RDS, IVH, NEC, ROP, or pROP. No significant associations were also observed for mortality, most likely due to the limited sample size. However, carriers of the MTHFR rs1801133T allele were highly prevalent among the deceased infants (75% of the group), showing a trend toward a 3.22-fold-increased risk of death (p = 0.156). A similar trend was observed for carriers of the PON1 rs662G allele (OR = 3.19; p = 0.151).

Table 4.
Distribution of studied genetic variants in a cohort of 377 preterm infants in relation to comorbidities and mortality.
3.5. Meta-Analysis of the Effect of MTHFR Genotype on Low Birth Weight
A literature search retrieved 464 articles potentially addressing the association between the neonatal MTHFR rs1801133 genotype and low birth weight. After removing 189 duplicate records, 201 unique articles remained. Screening of titles and abstracts reduced this number to twelve studies deemed suitable for full-text evaluation. Two Chinese reports could not be retrieved and therefore ten full texts were assessed for eligibility. Two of them were excluded due to a wrong publication type (review and meta-analysis) and another four due to the absence of neonatal genotype data. Applying the predefined inclusion and exclusion criteria ultimately yielded four eligible studies, all with a case–control design, comprising a total of 1007 cases and 896 controls [38,39,40,41]. Combined with our data, the meta-analyses included a total of 1156 cases and 1124 controls (Figure 4).
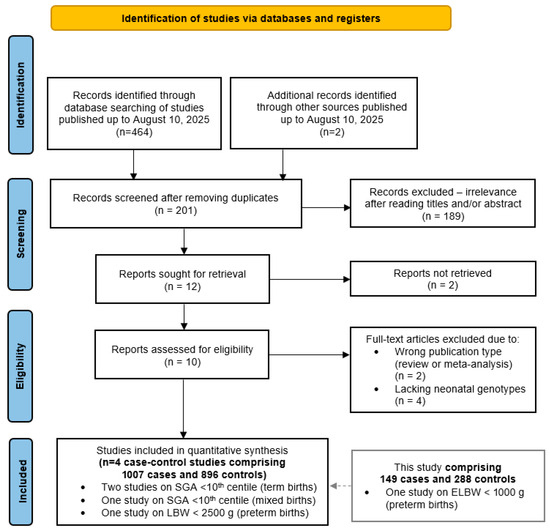
Figure 4.
PRISMA diagram showing the flow of study selection for inclusion in the meta-analysis of the MTHFR rs1801133 variant and adverse neonatal outcomes.
Table 5 summarizes the main characteristics of the studies included in the meta-analysis. Research on the MTHFR rs1801133 variant covered diverse geographic regions and ethnicities, including Asian (China), Caucasian (the UK, Turkey, and Poland), and mixed-ethnicity (Canada) populations. In all studies, genotyping was performed in newborns, with case definitions based on anthropometric or birth weight thresholds: SGA (<10th percentile), LBW (<2500 g), or ELBW (<1000 g). Two investigations, Chen et al. (2004) [39] and the present study (2025), included only preterm infants, whereas the others enrolled either mixed preterm and term groups or exclusively term neonates.

Table 5.
Characteristics of studies included in the meta-analysis of the association between neonatal MTHFR rs1801133 genotype and LBW.
Study sizes varied widely, from the smallest dataset of 55 cases and 55 controls [41] to the largest with 467 cases and 461 controls [38]. The frequency of the T allele ranged from 0.255 in the Turkish population to 0.604 in the Chinese population. All control groups adhered to Hardy–Weinberg equilibrium. Quality assessment using the NOS assigned scores of 6 to 8 across all included studies, consistent with moderate to high methodological rigor.
The pooled analysis of five studies demonstrated a significant association between the homozygous TT genotype of the MTHFR variant and an increased risk of low birth weight. The best-fitting model was comparison of TT vs. CT genotypes (OR = 1.40; 95% CI: 1.80–1.71; p = 0.0097; FEM; Figure 5a). In a recessive model (TT vs. CT + TT) this association was also significant in FEM; however moderate heterogeneity was observed in this model (I2 = 60.4%, Figure 5b).
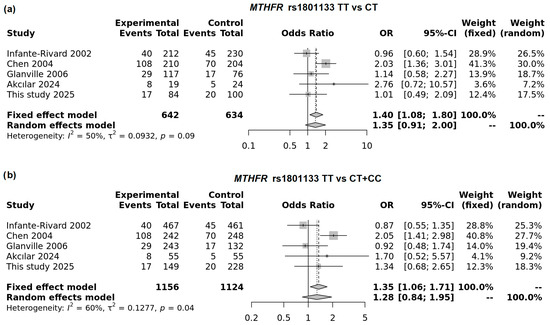
Figure 5.
Forest plot on the association of the MTHFR rs1801133T allele with LBW in TT vs. CT model (a) and recessive model (b).
We performed subgroup analyses and found that in the UK and Canada—the most developed countries included in this meta-analysis—the T allele in the dominant model was associated with a reduced risk of LBW (OR = 0.79; 95% CI: 0.63–0.99; p = 0.038; FEM). In contrast, in countries such as China and Turkey, the T allele was associated with increased risk (Figure 6). In this meta-analysis, Poland also clustered with countries showing increased risk for the MTHFR variant. The strongest association was observed in the recessive model (OR = 1.85; 95% CI: 1.34–2.53; p < 0.0001; FEM). A summary of the overall and subgroup meta-analyses is presented in Supplemental Table S3.
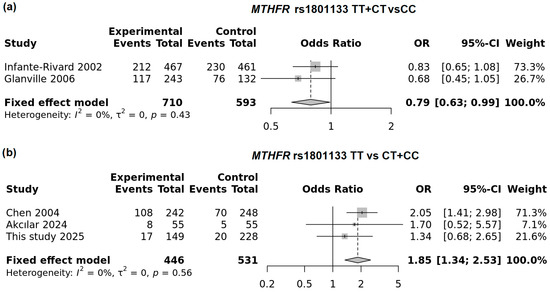
Figure 6.
Subgroup analysis of the neonatal MTHFR rs1801133T allele: (a) dominant model in developed countries and (b) recessive model in other countries (including this study).
The funnel plots for the studied associations (Figure 7a–d) showed no asymmetry, suggesting the absence of publication bias in performed analyses. This was further supported by Egger’s test, which revealed no statistically significant regression intercepts for the studied models (with the exception of the subgroup of developed countries, where calculation was not possible).
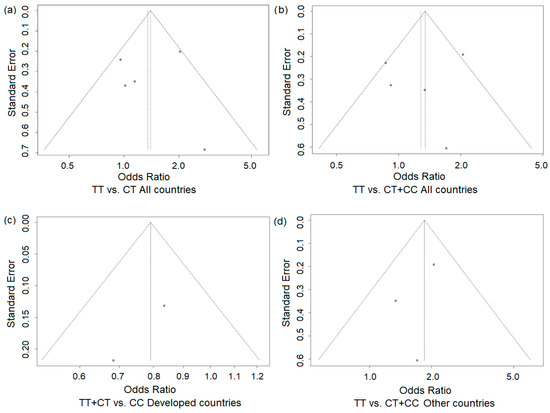
Figure 7.
Funnel plots for the association of the MTHFR rs1801133 T allele across genetic models and study populations: all countries (a,b) and developed (Canada and the UK (c)) and other countries (d). Each dot represents an individual study included in the meta-analysis.
4. Discussion
Despite advances in perinatal care, ELGA and ELBW remain serious challenges in neonatology, contributing to severe short- and long-term health complications, reduced quality of life, and increased mortality [42,43]. In this study, we investigated the role of functional variants in two candidate genes involved in Hcy and TLC-Hcy metabolism, MTHFR and PON1, in relation to ELBW, ELGA, and neonatal complications of prematurity. The genetic analysis was conducted in infants, mothers, and a population sample, as well as complemented by a meta-analysis of available studies, thus allowing us to position our findings within a broader clinical and genetic context.
In the clinical evaluation, we confirmed previously described maternal risk factors for preterm delivery, including PPROM, short interpregnancy interval, and history of prior preterm delivery [44,45,46]. Approximately 15% of mothers reported a history of preterm birth, which suggests persistence of chronic or recurrent risk factors across pregnancies. High frequency of delivery by cesarean section (50.9%) was also noted, consistent with earlier reports [47,48]. Over the study period, we observed a decline in the frequency and duration of PPROM, reflecting improvements in obstetric care. However, neonatal outcomes such as Apgar scores remained stable, while the survival of infants with lower birth weight, FGR/IUGR, and RDS increased, leading to higher rates of complications such as pROP.
The distribution of MTHFR and PON1 genotypes differed between infants, their mothers, and the reference population. Preterm infants had a lower frequency of carriers of the MTHFR rs1801133T allele compared to the general population, which may indicate a selective loss of fetuses carrying this allele under adverse intrauterine conditions. Conversely, homozygotes for the PON1 rs854560T allele were observed more frequently among preterm infants than in mothers, suggesting a role of the decreased concentration of PON1 in the fetus in susceptibility to preterm birth or adverse neonatal outcomes. No significant differences were found for PON1 rs662, indicating that this variant has a limited or no contribution to the risk of preterm birth in our population.
We identified several significant associations between infant genotypes and neonatal outcomes. The MTHFR rs1801133T allele increased the risk of ELBW (OR = 1.65) with sex-dependent trends: the TT genotype was particularly detrimental in female infants, while the CT genotype conferred risk for both ELBW and ELGA in males. This allele was also associated with increased risks of BPD and PDA. Also, the PON1 rs662AG genotype showed a borderline association with PDA, suggesting possible involvement of Hcy-TLC-related pathways. Interestingly, carriers of the MTHFR rs1801133T allele and PON1 rs662G allele were more common among infants who died in the early postnatal period. However, statistical significance was not reached, likely due to limited case numbers. No associations were observed with RDS, IVH, NEC, or ROP. Importantly, maternal genotypes were not associated with extreme prematurity, pointing to a predominant role of fetal genetic factors.
Our meta-analysis of five studies, including our own, confirmed the contribution of the neonatal MTHFR rs1801133 genotype to the determination of lower birth weight. Homozygous TT carriers had a significantly higher risk of low birth weight, with the TT vs. CT model yielding the most significant association (OR = 1.41; p = 0.0097). However, in the recessive model (TT vs. CT + CC), moderate heterogeneity (60%) was observed between populations. The analyses of subgroups revealed trends: in developed countries such as the UK and Canada, the T allele was protective, whereas in China, Turkey, and Poland, it was associated with increased risk. These findings suggest population-specific modifiers influencing the risk. Maternal nutritional status appears to play a crucial role in determining LBW across different countries, while in developed countries the widespread use of folic acid may mitigate the risk associated with MTHFR variants.
Our results contrast with earlier meta-analyses, which emphasized the role of maternal rather than infant genotypes. Wu et al. (2017) identified maternal T allele carriers as a risk factor for preterm birth and LBW, particularly in Asian and Caucasian cohorts [10]. Similarly, the most recent meta-analysis by Vafapour et al. (2025) confirmed maternal associations, though limited to Asian and Indian populations [11]. In contrast, our data support the importance of fetal genotypes, particularly in extreme prematurity. These discrepancies may reflect ethnic differences, methodological variation, or the limited availability of neonatal genotype data in prior studies.
Several studies on the neonatal rs1801133 variant and LBW were also excluded from our meta-analysis due to insufficient data. Yang et al. (2010) [49] and Mei et al. (2015) [50] could not be accessed in full text, while Sukla et al. (2013) [51] and Horikoshi et al. (2016) [9] lacked detailed genotype data. Similarly, Kordas et al. (2009) [52] and Obeid et al. (2023) [53] did not report infant genotypes, with the latter explicitly finding no associations. This raises a risk of publication bias. Moreover, none of the included studies distinguished pathological FGR/IUGR from constitutionally small SGA newborns [54]. We observed that ELBW infants were more than twice as likely to be SGA compared to preterm infants with higher birth weight, suggesting that ELBW may be partially attributable to SGA, whereas no such association was found for ELGA, indicating distinct risk profiles for ELGA and ELBW/SGA [26]. Although our meta-analysis is limited to the available data set, it is noteworthy that the results of our subgroup analysis mirror previous reports on maternal MTHFR genotype, with differences between developed and developing countries [10].
The observed associations may be explained by several mechanisms. The MTHFR rs1801133 variant reduces enzyme activity, leading to hyperhomocysteinemia, which results in endothelial dysfunction and impaired placental perfusion [55]. This may selectively influence fetal survival and growth, consistent with our observation of a lower T allele frequency in surviving preterm infants. For instance, in a Spanish population, the rs1801133T allele was shown to affect fetal growth parameters even under adequate maternal folate intake, with opposing effects depending on fetal sex [56]. Furthermore, animal studies demonstrated that mouse offspring exposed in utero to diet-induced hyperhomocysteinemia exhibited reduced birth weight, increased perinatal mortality, and impaired brain and muscle development [57]. Notably, several of these effects were sex-specific, highlighting differential fetal susceptibility to disturbances in one-carbon metabolism—a pattern consistent with our findings of sex-specific effects of the studied variants. These mechanisms may also contribute to PDA, as previously reported [58], as well as to the risk of BPD, a severe respiratory complication of prematurity. Notably, our finding represents a novel association in the context of BPD. However, previous studies have linked the MTHFR rs1801133 variant to the occurrence of childhood asthma, suggesting its broader role in susceptibility to lung diseases [59].
Second, observed sex-specific associations with ELBW and ELGA suggest differential epigenetic regulation, which is supported by recent evidence of sexually dimorphic placental methylation patterns [60] and different susceptibility of male and female fetuses to Hcy-related disturbances in one-carbon metabolism [56,57]. An epigenome-wide study of pregnancy-related methylation patterns has shown that female fetal plasma DNA is generally less methylated compared to male fetal plasma DNA. This pattern was observed in cells derived from the placenta, with much weaker associations observed in cells from cord blood [60]. These findings underscore the potential role of placental methylation in the observed sex differences. Furthermore, the researchers show that some genomic regions of cord blood DNA were more methylated in the group of women who took vitamin supplements, but this effect was not observed in mothers who were carriers of the rs1801133T allele [61]. In another study, the MTHFR rs1801133TT genotype was not associated with a higher global plasma DNA methylation ratio. However, pyrosequencing of a selected locus in the MTHFR gene showed that DNA in this region was more methylated in TT homozygotes compared with the CC homozygotes, 16.5% vs. 13.8%, respectively [62,63]. These epigenetic effects may also be influenced by environmental factors related to the mother’s health. For example, placental cells from pregnancies affected by gestational obesity were found to contain more methylated DNA than cells from healthy control women [64]. Different methylation patterns and different maternal allele expression ratios were also observed in placental imprinted genes in preeclampsia and placenta accreta spectrum disorders compared with control samples, as well as a correlation between increased oxidative stress and changes in gene methylation was reported [65].
Third, PON1 variants may influence antioxidant capacity and vulnerability to oxidative stress, a known contributor to PPROM and neonatal morbidity [66,67,68,69]. The borderline associations with PDA, and the literature linking PON1 to reduced antioxidant activity, give some support to this hypothesis.
In addition to MTHFR, our previous studies linked infant genotypes in selenoprotein P [70] and angiotensin receptor 1 [71] to ELBW and ELGA, suggesting that extreme prematurity may result from multiple genetic factors acting through oxidative stress and impaired placental vascular regulation.
The strengths of this study include analysis of both infant and maternal genotypes, the use of a reference population, and integration of findings into a meta-analysis. Limitations include modest subgroup sizes, particularly for rare outcomes such as mortality, limited maternal genetic data, and restriction to a single ethnic group. Post hoc power analysis for the study of the Polish population showed 70% power for detecting the association between the MTHFR rs1801133T allele and ELBW (dominant model) and approximately 55% power for comparing the frequency of carriers of this allele between preterm infants and the population sample, indicating moderate statistical power for detecting these major effects. The meta-analysis may, on the other hand, be affected by publication bias, as studies without significant results are less likely to be published. Finally, methodological variability across studies, including inconsistent definitions of SGA and LBW, complicates interpretation.
Our findings suggest that the infant, rather than the maternal, MTHFR genotype plays an important role in susceptibility to extreme prematurity and its complications. They also highlight possible sex-specific vulnerability, which warrants further research. Future studies should include larger, ethnically diverse cohorts, rigorous phenotyping distinguishing pathological FGR/IUGR from constitutional SGA, and integration of genetic, epigenetic, and biochemical markers.
5. Conclusions
In summary, we provide novel evidence that the fetal MTHFR rs1801133 genotype contributes to the risk of ELBW and BPD and extend existing data regarding PDA and perinatal mortality. This is the first study to demonstrate such associations in ELBW infants and to integrate them within a broader meta-analysis. Our findings emphasize the need for comprehensive, multifactorial models of extreme prematurity that combine genetic, environmental, and epigenetic factors and point toward the potential of genetic profiling in risk stratification for the most vulnerable neonatal populations.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes16101192/s1, Table S1: Comparison of clinical parameters between Study Cohort A and B; Table S2: Comparison of the impact of neonatal MTHFR rs1801133 genotypes on ELBW and ELGA in males and females; Table S3: Meta-analysis results of association of MTHFR variant with LBW.
Author Contributions
Conceptualization, E.S.; methodology, E.S. and A.G.-W.; software, E.S.; validation, A.D.; investigation, E.S., A.G.-W., A.S. and B.S.; data curation, E.S., A.G.-W. and A.S.; writing—original draft preparation E.S., B.S. and A.D.; writing—review and editing, A.G.-W. and A.S.; visualization, B.S., E.S. and A.D.; supervision, E.S.; funding acquisition, E.S. and A.G.-W. All authors have read and agreed to the published version of the manuscript.
Funding
The work has been supported by the Scientific Research Committee in Poland under grant no. 2 P05E 098 30 (A.G.-W. and E.S.).
Institutional Review Board Statement
All procedures carried out on human participants in this study were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments (or comparable ethical standards). The study was approved by the Bioethics Committee of the Poznan University of Medical Sciences (no. 1140/05, issued on 8 September 2005; no. 1117/18 from 7 November 2018).
Informed Consent Statement
Written prior informed consent was obtained from all participants, including consent from the parents or guardians of the study infants.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors acknowledge the use of Grammarly (Grammarly Inc., San Francisco, CA, USA) for assistance in language editing. All scientific content and conclusions are the sole responsibility of the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BPD | Bronchopulmonary dysplasia |
| BW | Birth weight |
| CS | Cesarean section |
| ELBW | Extremely low birth weight |
| ELGA | Extremely low gestational age |
| FGA | Fetal growth restriction |
| GA | Gestational age |
| Gln192Arg | Glutamine 192 Arginine (PON1 p.Gln192Arg) |
| Hcy | Homocysteine |
| Hcy-TLC | Homocysteine-thiolactone |
| HWE | Hardy–Weinberg equilibrium |
| IUGR | Intrauterine growth restriction |
| IVH | Intraventricular hemorrhage |
| Leu55Met | Leucine 55 Methionine (PON1 p.Leu55Met) |
| MAF | Minor allele frequency |
| MTHFR | Methylenetetrahydrofolate reductase |
| NEC | Necrotizing enterocolitis |
| PDA | Patent ductus arteriosus |
| PON1 | Paraoxonase 1 |
| PPROM | Preterm prelabor rupture of membranes |
| pROP | Proliferative retinopathy of prematurity |
| PVL | Periventricular leukomalacia |
| qPCR | Quantitative polymerase chain reaction |
| RDS | Respiratory distress syndrome |
| ROP | Retinopathy of prematurity |
| ROS | Reactive oxygen species |
| SGA | Small for gestational age |
| SVN | Small vulnerable newborn |
References
- Ashorn, P.; Ashorn, U.; Muthiani, Y.; Aboubaker, S.; Askari, S.; Bahl, R.; Black, R.E.; Dalmiya, N.; Duggan, C.P.; Hofmeyr, G.J.; et al. Small vulnerable newborns-big potential for impact. Lancet 2023, 401, 1692–1706. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, J.; Sun, Y.; Li, N.; Li, X.; Song, X.; Zhang, P.; Li, J.; Huang, K. A clinical analysis of very and extremely low birth weight preterm infants. Am. J. Transl. Res. 2021, 13, 9395–9403. [Google Scholar]
- Nosherwan, A.; Cheung, P.-Y.; Schmölzer, G.M. Management of Extremely Low Birth Weight Infants in Delivery Room. Clin. Perinatol. 2017, 44, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Buchole, M.; Oleszczuk, J.; Leszczyńska-Gorzelak, B. Selected determinants of body mass in premature infants. Ginekol. Pol. 2010, 81, 37–40. [Google Scholar]
- Ji, X.; Wu, C.; Chen, M.; Wu, L.; Li, T.; Miao, Z.; Lv, Y.; Ding, H. Analysis of risk factors related to extremely and very preterm birth: A retrospective study. BMC Pregnancy Childbirth 2022, 22, 818. [Google Scholar] [CrossRef]
- Clausson, B.; Lichtenstein, P.; Cnattingius, S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG 2000, 107, 375–381. [Google Scholar] [CrossRef]
- Kistka, Z.A.F.; DeFranco, E.A.; Ligthart, L.; Willemsen, G.; Plunkett, J.; Muglia, L.J.; Boomsma, D.I. Heritability of parturition timing: An extended twin design analysis. Am. J. Obstet. Gynecol. 2008, 199, 43.e41–43.e45. [Google Scholar] [CrossRef]
- Magnus, P. Further evidence for a significant effect of fetal genes on variation in birth weight. Clin. Genet. 1984, 26, 289–296. [Google Scholar] [CrossRef]
- Horikoshi, M.; Beaumont, R.N.; Day, F.R.; Warrington, N.M.; Kooijman, M.N.; Fernandez-Tajes, J.; Feenstra, B.; van Zuydam, N.R.; Gaulton, K.J.; Grarup, N.; et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 2016, 538, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhu, P.; Geng, X.; Liu, Z.; Cui, L.; Gao, Z.; Jiang, B.; Yang, L. Genetic polymorphism of MTHFR C677T with preterm birth and low birth weight susceptibility: A meta-analysis. Arch. Gynecol. Obs. 2017, 295, 1105–1118. [Google Scholar] [CrossRef]
- Vafapour, M.; Talebi, H.; Danaei, M.; Yeganegi, M.; Azizi, S.; Dastgheib, S.A.; Bahrami, R.; Pourkazemi, M.; Jayervand, F.; Shahbazi, A.; et al. Global and population-specific association of MTHFR polymorphisms with preterm birth risk: A consolidated analysis of 44 studies. BMC Pregnancy Childbirth 2025, 25, 230. [Google Scholar] [CrossRef]
- Araszkiewicz, A.F.; Jańczak, K.; Wójcik, P.; Białecki, B.; Kubiak, S.; Szczechowski, M.; Januszkiewicz-Lewandowska, D. MTHFR Gene Polymorphisms: A Single Gene with Wide-Ranging Clinical Implications—A Review. Genes 2025, 16, 441. [Google Scholar] [CrossRef]
- Frosst, P.; Blom, H.J.; Milos, R.; Goyette, P.; Sheppard, C.A.; Matthews, R.G.; Boers, G.J.; den Heijer, M.; Kluijtmans, L.A.; van den Heuvel, L.P.; et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995, 10, 111–113. [Google Scholar] [CrossRef]
- Chango, A.; Boisson, F.; Barbé, F.; Quilliot, D.; Droesch, S.; Pfister, M.; Fillon-Emery, N.; Lambert, D.; Frémont, S.; Rosenblatt, D.S.; et al. The effect of 677C → T and 1298A → C mutations on plasma homocysteine and 5,10-methylenetetrahydrofolate reductase activity in healthy subjects. Br. J. Nutr. 2000, 83, 593–596. [Google Scholar] [CrossRef]
- Crider, K.S.; Zhu, J.-H.; Hao, L.; Yang, Q.-H.; Yang, T.P.; Gindler, J.; Maneval, D.R.; Quinlivan, E.P.; Li, Z.; Bailey, L.B.; et al. MTHFR 677C→T genotype is associated with folate and homocysteine concentrations in a large, population-based, double-blind trial of folic acid supplementation. Am. J. Clin. Nutr. 2011, 93, 1365–1372. [Google Scholar] [CrossRef]
- Plumptre, L.; Tammen, S.A.; Sohn, K.-J.; Masih, S.P.; Visentin, C.E.; Aufreiter, S.; Malysheva, O.; Schroder, T.H.; Ly, A.; Berger, B.; et al. Maternal and Cord Blood Folate Concentrations Are Inversely Associated with Fetal DNA Hydroxymethylation, but Not DNA Methylation, in a Cohort of Pregnant Canadian Women. J. Nutr. 2020, 150, 202–211. [Google Scholar] [CrossRef]
- Bekdash, R.A. Methyl Donors, Epigenetic Alterations, and Brain Health: Understanding the Connection. Int. J. Mol. Sci. 2023, 24, 2346. [Google Scholar] [CrossRef] [PubMed]
- Paduch-Jakubczyk, W.; Dubińska, M. The Role of Folic Acid and Its Derivatives in Reproductive Health. Actual Gynecol. Obstet./Aktuaální Gynekol. A Porod. 2024, 16, 65–69. [Google Scholar]
- Tate, C.; Shuman, A.; Nice, S.; Salehi, P. The Critical Role of Folate in Prenatal Health and a Proposed Shift from Folic Acid to 5-Methyltetrahydrofolate Supplementation. Georget. Med. Rev. 2024, 8. [Google Scholar] [CrossRef]
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol. Rev. 2019, 99, 555–604. [Google Scholar] [CrossRef] [PubMed]
- Perła-Kaján, J.; Jakubowski, H. Paraoxonase 1 and homocysteine metabolism. Amino Acids 2012, 43, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Mackness, B.; Mackness, M.I.; Arrol, S.; Turkie, W.; Durrington, P.N. Effect of the molecular polymorphisms of human paraoxonase (PON1) on the rate of hydrolysis of paraoxon. Br. J. Pharmacol. 1997, 122, 265–268. [Google Scholar] [CrossRef]
- Jakubowski, H.; Ambrosius, W.T.; Pratt, J.H. Genetic determinants of homocysteine thiolactonase activity in humans: Implications for atherosclerosis. FEBS Lett. 2001, 491, 35–39. [Google Scholar] [CrossRef]
- Leviev, I.; James, R.W. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arter. Thromb. Vasc. Biol. 2000, 20, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Brophy, V.H.; Jampsa, R.L.; Clendenning, J.B.; McKinstry, L.A.; Jarvik, G.P.; Furlong, C.E. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am. J. Hum. Genet. 2001, 68, 1428–1436. [Google Scholar] [CrossRef]
- Fenton, T.R.; Elmrayed, S.; Alshaikh, B.N. Fenton Third-Generation Growth Charts of Preterm Infants Without Abnormal Fetal Growth: A Systematic Review and Meta-Analysis. Paediatr. Perinat. Epidemiol. 2025, 29, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Lees, C.C.; Stampalija, T.; Baschat, A.; da Silva Costa, F.; Ferrazzi, E.; Figueras, F.; Hecher, K.; Kingdom, J.; Poon, L.C.; Salomon, L.J.; et al. ISUOG Practice Guidelines: Diagnosis and management of small-for-gestational-age fetus and fetal growth restriction. Ultrasound Obs. Gynecol. 2020, 56, 298–312. [Google Scholar] [CrossRef]
- Kosik, K.; Szpecht, D.; Al-Saad, S.R.; Karbowski, L.M.; Kurzawińska, G.; Szymankiewicz, M.; Drews, K.; Wolski, H.; Seremak-Mrozikiewicz, A. Single nucleotide vitamin D receptor polymorphisms (FokI, BsmI, ApaI, and TaqI) in the pathogenesis of prematurity complications. Sci. Rep. 2020, 10, 21098. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement. Open Med. 2009, 3, e123–e130. [Google Scholar]
- Wells, G.A.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. 2025. Available online: https://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 10 August 2025).
- Luchini, C.; Stubbs, B.; Solmi, M.; Veronese, N. Assessing the quality of studies in meta-analyses: Advantages and limitations of the Newcastle Ottawa Scale. World J. Meta-Anal. 2017, 5, 80–84. [Google Scholar] [CrossRef]
- Luchini, C.; Veronese, N.; Nottegar, A.; Shin, J.I.; Gentile, G.; Granziol, U.; Soysal, P.; Alexinschi, O.; Smith, L.; Solmi, M. Assessing the quality of studies in meta-research: Review/guidelines on the most important quality assessment tools. Pharm. Stat. 2021, 20, 185–195. [Google Scholar] [CrossRef]
- Martorell-Marugan, J.; Toro-Dominguez, D.; Alarcon-Riquelme, M.E.; Carmona-Saez, P. MetaGenyo: A web tool for meta-analysis of genetic association studies. BMC Bioinform. 2017, 18, 563. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Healy, K. Data Visualization: A Practical Introduction; Princeton University Press: Princeton, NJ, USA, 2018. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025. [Google Scholar]
- Infante-Rivard, C.; Rivard, G.E.; Yotov, W.V.; Génin, E.; Guiguet, M.; Weinberg, C.; Gauthier, R.; Feoli-Fonseca, J.C. Absence of association of thrombophilia polymorphisms with intrauterine growth restriction. N. Engl. J. Med. 2002, 347, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.F.; Hu, Y.H.; Yang, F.; Wu, B.Y.; Chen, L.; Fang, Z.A.; Wang, L.H. Mother’s and child’s methylenetetrahydrofolate reductase C677T polymorphism is associated with preterm delivery and low birth weight. Beijing Da Xue Xue Bao Yi Xue Ban 2004, 36, 248–253. [Google Scholar]
- Glanville, T.; Yates, Z.; Ovadia, L.; Walker, J.J.; Lucock, M.; Simpson, N.A. Fetal folate C677T methylenetetrahydrofolate reductase gene polymorphism and low birth weight. J. Obs. Gynaecol. 2006, 26, 11–14. [Google Scholar] [CrossRef]
- Akcılar, R.; Yalınbaş, E.E.; Mutlu, F. MTHFR 677 C > T Gene Polymorphism is Associated with Large for Gestational Age Infants. Fetal Pediatr. Pathol. 2024, 43, 234–245. [Google Scholar] [CrossRef]
- Moreira, A.; Benvenuto, D.; Fox-Good, C.; Alayli, Y.; Evans, M.; Jonsson, B.; Hakansson, S.; Harper, N.; Kim, J.; Norman, M.; et al. Development and Validation of a Mortality Prediction Model in Extremely Low Gestational Age Neonates. Neonatology 2022, 119, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Varshney, P.G.; Sinharay, M.; Gupta, A.; Pandey, M. Extremely low birth weight (ELBW) neonates for emergency surgery—A challenge for the Anesthesiologist: A case series. J. Neonatal Surg. 2024, 13, 14. [Google Scholar] [CrossRef]
- Deer, E.; Herrock, O.; Campbell, N.; Cornelius, D.; Fitzgerald, S.; Amaral, L.M.; LaMarca, B. The role of immune cells and mediators in preeclampsia. Nat. Rev. Nephrol. 2023, 19, 257–270. [Google Scholar] [CrossRef]
- DeFranco, E.; Ehrlich, S.; Muglia, L. Influence of interpregnancy interval on birth timing. BJOG 2014, 121, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Lelic, M.; Bogdanovic, G.; Ramic, S.; Brkicevic, E. Influence of Maternal Anemia During Pregnancy on Placenta and Newborns. Med. Arh. 2014, 68, 184. [Google Scholar] [CrossRef]
- Higgins, B.V.; Baer, R.J.; Steurer, M.A.; Karvonen, K.L.; Oltman, S.P.; Jelliffe-Pawlowski, L.L.; Rogers, E.E. Resuscitation, survival and morbidity of extremely preterm infants in California 2011–2019. J. Perinatol. 2024, 44, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Adane, A.A.; Shepherd, C.C.J.; Farrant, B.M.; White, S.W.; Bailey, H.D. Patterns of recurrent preterm birth in Western Australia: A 36-year state-wide population-based study. Aust. N. Z. J. Obs. Gynaecol. 2022, 62, 494–499. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, J.; Wu, D.; Shang, Q.; Wu, L.; Han, X.; Liang, B.; Xu, L.; Tang, J. Association of Genetic Polymorphisms in Methylenetetrahydrofolate Reductase Gene, Plasminogen Activator Inhibitor-1 Gene with Preterm Birth and Spastic Cerebral Palsy. Chin. J. Appl. Clin. Pediatr. 2010, 25, 1580–1582. [Google Scholar]
- Mei, M.; Xi, H.; Ren, H.; Guo, H.; Wang, Y.; Li, J. The relationship between MTHFR gene, PAI gene polymorphism and preterm birth. Prog. Mod. Biomed. 2015, 15, 6435–6438. [Google Scholar] [CrossRef]
- Sukla, K.K.; Tiwari, P.K.; Kumar, A.; Raman, R. Low birthweight (LBW) and neonatal hyperbilirubinemia (NNH) in an Indian cohort: Association of homocysteine, its metabolic pathway genes and micronutrients as risk factors. PLoS ONE 2013, 8, e71587. [Google Scholar] [CrossRef]
- Kordas, K.; Ettinger, A.S.; Lamadrid-Figueroa, H.; Tellez-Rojo, M.M.; Hérnandez-Avila, M.; Hu, H.; Wright, R.O. Methylenetetrahydrofolate reductase (MTHFR) C677T, A1298C and G1793A genotypes, and the relationship between maternal folate intake, tibia lead and infant size at birth. Br. J. Nutr. 2009, 102, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Warnke, I.; Bendik, I.; Troesch, B.; Schoop, R.; Chenal, E.; Koletzko, B. Infants’ Folate Markers and Postnatal Growth in the First 4 Months of Life in Relation to Breastmilk and Maternal Plasma Folate. Nutrients 2023, 15, 1495. [Google Scholar] [CrossRef]
- Figueras, F.; Gratacós, E. Update on the diagnosis and classification of fetal growth restriction and proposal of a stage-based management protocol. Fetal Diagn. Ther. 2014, 36, 86–98. [Google Scholar] [CrossRef]
- Chakraborty, P.; Goswami, S.K.; Rajani, S.; Sharma, S.; Kabir, S.N.; Chakravarty, B.; Jana, K. Recurrent Pregnancy Loss in Polycystic Ovary Syndrome: Role of Hyperhomocysteinemia and Insulin Resistance. PLoS ONE 2013, 8, e64446. [Google Scholar] [CrossRef]
- Aguilar-Lacasaña, S.; López-Flores, I.; González-Alzaga, B.; Giménez-Asensio, M.J.; Carmona, F.D.; Hernández, A.F.; López Gallego, M.F.; Romero-Molina, D.; Lacasaña, M. Methylenetetrahydrofolate Reductase (MTHFR) Gene Polymorphism and Infant’s Anthropometry at Birth. Nutrients 2021, 13, 831. [Google Scholar] [CrossRef]
- Suszyńska-Zajczyk, J.; Witucki, Ł.; Perła-Kaján, J.; Jakubowski, H. Diet-induced hyperhomocysteinemia causes sex-dependent deficiencies in offspring musculature and brain function. Front. Cell Dev. Biol. 2024, 12, 1322844. [Google Scholar] [CrossRef]
- Dagle, J.M.; Ryckman, K.K.; Spracklen, C.N.; Momany, A.M.; Cotten, C.M.; Levy, J.; Page, G.P.; Bell, E.F.; Carlo, W.A.; Shankaran, S.; et al. Genetic variants associated with patent ductus arteriosus in extremely preterm infants. J. Perinatol. 2019, 39, 401–408. [Google Scholar] [CrossRef]
- Li, M.; Tang, Y.; Zhao, E.-Y.; Chen, C.-H.; Dong, L.-L. Relationship between MTHFR gene polymorphism and susceptibility to bronchial asthma and glucocorticoid efficacy in children. Zhongguo Dang Dai Er Ke Za Zhi 2021, 23, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Czamara, D.; Dieckmann, L.; Lahti-Pulkkinen, M.; Cruceanu, C.; Henrich, W.; Plagemann, A.; Räikkönen, K.; Braun, T.; Binder, E.B.; Lahti, J.; et al. Sex differences in DNA methylation across gestation: A large scale, cross-cohort, multi-tissue analysis. Cell. Mol. Life Sci. 2024, 81, 177. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Dou, J.F.; Feinberg, J.I.; Brieger, K.K.; Croen, L.A.; Hertz-Picciotto, I.; Newschaffer, C.J.; Schmidt, R.J.; Fallin, M.D. Prenatal Multivitamin Use and MTHFR Genotype Are Associated with Newborn Cord Blood DNA Methylation. Int. J. Environ. Res. Public Health 2020, 17, 9190. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Garcia, H.; Vilchis-Gil, J.; Castro-Cerritos, K.V.; Rivera-Susunaga, L.E.; Klünder-Klünder, M.; Granados-Riveron, J.T.; Gómez-López, J.; López-Torres, A.; Sánchez-Urbina, R. Association between maternal diet, smoking, and the placenta MTHFR 677C/T genotype and global placental DNA methylation. Placenta 2024, 146, 17–24. [Google Scholar] [CrossRef]
- Van Otterdijk, S.D.; Klett, H.; Boerries, M.; Michels, K.B. The impact of pre-pregnancy folic acid intake on placental DNA methylation in a fortified cohort. FASEB J. 2023, 37, e22698. [Google Scholar] [CrossRef]
- Nomura, Y.; Lambertini, L.; Rialdi, A.; Lee, M.; Mystal, E.Y.; Grabie, M.; Manaster, I.; Huynh, N.; Finik, J.; Davey, M.; et al. Global Methylation in the Placenta and Umbilical Cord Blood From Pregnancies with Maternal Gestational Diabetes, Preeclampsia, and Obesity. Reprod. Sci. 2014, 21, 131–137. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Wu, Y.; Lin, C.; Yang, S.; Yang, Y.; Chen, D.; Yu, B. Methylation alterations of imprinted genes in different placental diseases. Clin. Epigenetics 2024, 16, 132. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Gaunt, T.R.; Hinks, L.J.; Davey Smith, G.; Timpson, N.; Day, I.N.M.; Ebrahim, S. The association of the PON1 Q192R polymorphism with complications and outcomes of pregnancy: Findings from the British Women’s Heart and Health cohort study. Paediatr. Perinat. Epid 2006, 20, 244–250. [Google Scholar] [CrossRef]
- Chen, D.; Hu, Y.; Chen, C.; Yang, F.; Fang, Z.; Wang, L.; Li, J. Polymorphisms of the paraoxonase gene and risk of preterm delivery. Epidemiology 2004, 15, 466–470. [Google Scholar] [CrossRef]
- Soydinc, H.E.; Sak, M.E.; Evliyaoglu, O.; Evsen, M.S.; Turgut, A.; Özler, A.; Yıldız, İ.; Gul, T. Prolidase, Matrix Metalloproteinases 1 and 13 Activity, Oxidative-Antioxidative Status as a Marker of Preterm Premature Rupture of Membranes and Chorioamnionitis in Maternal Vaginal Washing Fluids. Int. J. Med. Sci. 2013, 10, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, V.O.; Pereira, R.A.; Amantéa, S.L.; Rhoden, C.R.; Colvero, M.O. Neonatal diseases and oxidative stress in premature infants: An integrative review. J. Pediatr. 2022, 98, 455–462. [Google Scholar] [CrossRef]
- Strauss, E.; Januszkiewicz-Lewandowska, D.; Sobaniec, A.; Gotz-Więckowska, A. SELENOP rs3877899 Variant Affects the Risk of Developing Advanced Stages of Retinopathy of Prematurity (ROP). Int. J. Mol. Sci. 2023, 24, 7570. [Google Scholar] [CrossRef] [PubMed]
- Durska, A.; Szpecht, D.; Gotz-Więckowska, A.; Strauss, E. Association of ACE and AGTR1 variants with retinopathy of prematurity: A case-control study and meta-analysis. J. Appl. Genet. 2024. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).