Association Mapping for Biomass and Kernel Traits in Doubled-Haploid Population Derived from Texas Wheat Cultivars
Abstract
1. Introduction
2. Materials and Methods
2.1. Association-Mapping Panel
Development of Doubled Haploids (DH)
2.2. Kernel Phenotyping, Biomass and Statistical Analysis
2.2.1. Experimental Layouts
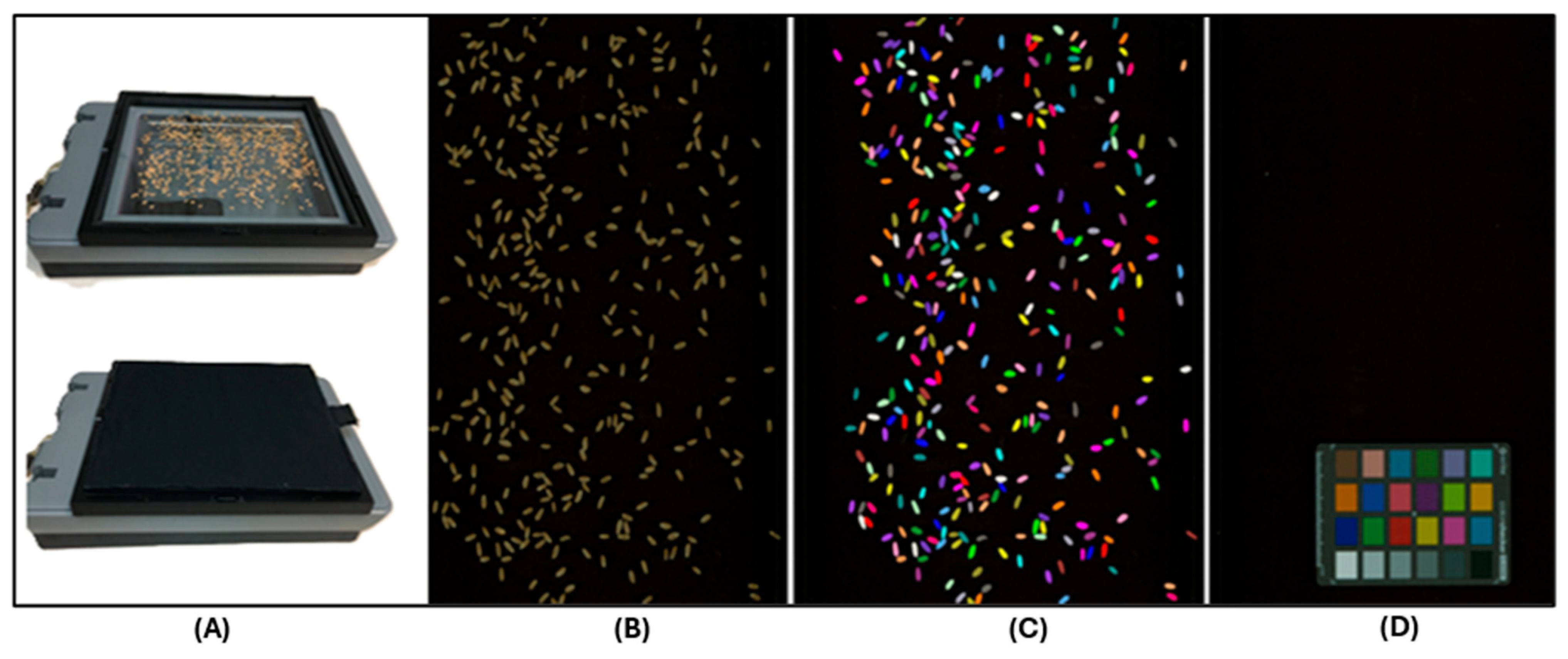
2.2.2. Kernel Image Capturing
2.2.3. Image Analysis and Data Generation
- G (x, y): Gaussian value at (x, y) coordinates.
- σ (sigma): Standard deviation, controlling the width or blurriness of the Gaussian.
- x and y: Horizontal and vertical distances from the center of the Gaussian kernel.
- e: Base of the natural logarithm.
- π: Mathematical constant (Pi).
2.2.4. Biomass Traits
- Plant weight (P. Wt, g): A half-meter-long single row representing the plot was harvested from the 2nd or 3rd row of the plot at the time of physiological maturity, and the weight was measured including the stem, leaves, and heads.
- Head count (H. Count): The number of heads were counted and recorded from the same plant.
- Head weight (H. Wt, g): Heads were separated from the stems at the base of the spike and all heads were weighed together from the same plant.
2.2.5. Data Analysis
2.3. Genotyping and SNP Calling
2.4. Association-Mapping Analysis
3. Results
3.1. Phenotyping and Statistical Analysis
3.2. Genomic Libraries and SNP Calling
3.3. Population Structure, Maker Heterozygosity and Kinship Matrix
3.4. Genome Wide Association Mapping
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| MAF | Minor Allele Frequency |
| PVE | Phenotypic Variance Explained |
| QTL | Quantitative Trail Loci |
| GWAS | Genome-Wide Association Study |
| DH | Doubled Haploid |
| GAPIT | Genome Association and Prediction Integrated Tool |
| MLM | Mixed Linear Model |
| MLMM | Multiple-Locus Mixed Linear Model |
| BLINK | Bayesian information and Linkage-disequilibrium Iteratively Nested Keyway |
| FarmCPU | Fixed and random model Circulating Probability Unification |
| DPI | Dots Per Inch |
| BD | Bushland Dryland |
| BI | Bushland Irrigated |
| MTA | Marker–Trait Association |
| PCA | Principal Component Analysis |
| UPGMA | Unweighted Pair Group Method with Arithmetic Mean |
References
- Rabieyan, E.; Alipour, H. NGS-based multiplex assay of trait-linked molecular markers revealed the genetic diversity of Iranian bread wheat landraces and cultivars. Crop Pasture Sci. 2021, 72, 173–182. [Google Scholar] [CrossRef]
- Li, P.; Ma, B.; Palta, J.; Ding, T.; Cheng, Z.; Lv, G.; Xiong, Y. Wheat breeding highlights drought tolerance while ignores the advantages of drought avoidance: A meta-analysis. Eur. J. Agron. 2021, 122, 126196. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Nat. Acad. Sci. 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Gul, A.; Ahmad, I.; Farooq, M.; Rauf, Y.; -ur Rahman, A.; Riaz, H. Genetic resources for some wheat abiotic stress tolerances. In Salinity and Water Stress: Improving Crop Efficiency; Springer: Dordrecht, The Netherlands, 2009; pp. 149–163. [Google Scholar]
- Xi, Y.; Du, Y.L.; Wang, D.; Ren, J.Y.; Luo, W.Y.; Peng, Q.; Wang-Ying, F.; Feng-Ming, L. Wheat genetic progress in biomass allocation and yield components: A global perspective. Field Crops Res. 2024, 318, 109617. [Google Scholar] [CrossRef]
- Ramya, P.; Chaubal, A.; Kulkarni, K.; Gupta, L.; Kadoo, N.; Dhaliwal, H.S.; Chhuneja, P.; Lagu, M.; Gupt, V. QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J. Appl. Genet. 2010, 51, 421–429. [Google Scholar] [CrossRef]
- Fan, M.; Zhang, X.; Nagarajan, R.; Zhai, W.; Rauf, Y.; Jia, H.; Yan, L. Natural variants and editing events provide insights into routes for spike architecture modification in common wheat. Crop J. 2023, 11, 148–156. [Google Scholar] [CrossRef]
- Zhang, D.; Fan, M.; Li, T.; Rauf, Y.; Liu, Y.; Zhu, X.; Jia, H.; Zhai, W.; Luzuriaga, J.C.; Carver, B.F.; et al. A natural allele of the transcription factor gene TaMYB-D7b is a genetic signature for phosphorus deficiency in wheat. Plant Physiol. 2025, kiaf224. [Google Scholar] [CrossRef]
- Wu, J.Z.; Qiao, L.Y.; Liu, Y.; Fu, B.S.; Ragupathi, N.; Rauf, Y.; Jia, H.Y.; Yan, L. Rapid identification and deployment of major genes for flowering time and awn traits in common wheat. Front. Plant Sci. 2022, 13, 992811. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, F.; Yan, X.; Zhang, X.; Dong, Z.; Cui, D.; Chen, F. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol. J. 2017, 15, 953–969. [Google Scholar] [CrossRef]
- Zhang, J.; Gill, H.S.; Halder, J.; Brar, N.K.; Ali, S.; Bernardo, A.; Amand, P.S.; Bai, G.; Turnipseed, B.; Sehgal, S.K. Multi-locus genome-wide association studies to characterize Fusarium head blight (FHB) resistance in hard winter wheat. Front. Plant Sci. 2022, 13, 946700. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, F.; Zhao, C.; Lv, G.; Sun, C.; Pan, Y.; Guo, X.; Chen, F. Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol. J. 2019, 17, 2106–2122. [Google Scholar] [CrossRef]
- Singh, K.; Saini, D.K.; Saripalli, G.; Batra, R.; Gautam, T.; Singh, R. WheatQTLdb V2.0: A supplement to the database for wheat QTL. Mol. Breeding 2022, 42, 56. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, H.; Deng, Z.; Wu, R.; Li, D.; Wang, M.; Tian, J. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica 2016, 212, 173–185. [Google Scholar] [CrossRef]
- Jaiswal, V.; Gahlaut, V.; Mathur, S.; Agarwal, P.; Khandelwal, M.K.; Khurana, J.P.; Tyagi, A.K.; Balyan, H.S.; Gupta, P.K. Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0129400. [Google Scholar] [CrossRef]
- Bednarek, J.; Boulaflous, A.; Girousse, C.; Ravel, C.; Tassy, C.; Barret, P.; Bouzidi, M.F.; Mouzeyar, S. Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 2012, 63, 5945–5955. [Google Scholar] [CrossRef]
- Hou, J.; Jiang, Q.; Hao, C.; Wang, Y.; Zhang, H.; Zhang, X. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 2014, 164, 1918–1929. [Google Scholar] [CrossRef]
- Ma, L.; Li, T.; Hao, C.; Wang, Y.; Chen, X.; Zhang, X. TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 2016, 14, 1269–1280. [Google Scholar] [CrossRef]
- Johnson, E.B.; Nalam, V.J.; Zemetra, R.S.; Riera-Lizarazu, O. Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica 2008, 163, 193–201. [Google Scholar] [CrossRef]
- Xie, Q.; Li, N.; Yang, Y.; Lv, Y.; Yao, H.; Wei, R.; Sparkes, D.L.; Ma, Z. Pleiotropic effects of the wheat domestication gene Q on yield and grain morphology. Planta 2018, 247, 1089–1098. [Google Scholar] [CrossRef]
- Niroula, R.K.; Bimb, H.P. Overview of wheat x maize system of crosses for dihaploid induction in wheat. World Appl. Sci. J. 2009, 7, 1037–1045. [Google Scholar]
- Salembier Clairon, P.J.; Wilkinson, M. Connected operators: A review of region-based morphological image processing techniques. IEEE Signal Process. Mag. 2009, 26, 136–157. [Google Scholar] [CrossRef]
- Sintorn, I.-M.; Bischof, L.; Jackway, P.; Haggarty, S.; Buckley, M. Gradient based intensity normalization. J. Microsc. 2010, 240, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Whan, A.P.; Smith, A.B.; Cavanagh, C.R.; Ral, J.P.; Shaw, L.M.; Howitt, C.A.; Bischof, L. GrainScan: A low cost, fast method for grain size and colour measurements. Plant Methods 2014, 10, 23. [Google Scholar] [CrossRef]
- Rauf, Y.; Bajgain, P.; Rouse, M.N.; Khanzada, K.A.; Bhavani, S.; Huerta-Espino, J.; Singh, R.P.; Imtiaz, M.; Anderson, J.A. Molecular characterization of genomic regions for adult plant resistance to stem rust in a spring wheat mapping population. Plant Dis. 2022, 106, 439–450. [Google Scholar] [CrossRef]
- Rauf, Y.; Lan, C.; Randhawa, M.; Singh, R.P.; Huerta-Espino, J.; Anderson, J.A. Quantitative trait loci mapping reveals the complexity of adult plant resistance to leaf rust in spring wheat ‘Copio’. Crop Sci. 2022, 62, 1037–1050. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Wang, Z.; Dhakal, S.; Cerit, M.; Wang, S.; Rauf, Y.; Yu, S.; Maulana, F.; Huang, W.; Anderson, J.D.; Ma, X.F.; et al. QTL mapping of yield components and kernel traits in wheat cultivars TAM 112 and Duster. Front. Plant Sci. 2022, 13, 1057701. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A one-penny imputed genome from next generation reference panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Li, T.; Deng, G.; Su, Y.; Yang, Z.; Tang, Y.; Wang, J. Genetic dissection of quantitative trait loci for grain size and weight by high-resolution genetic mapping in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 257–271. [Google Scholar] [CrossRef]
- Kumari, S.; Jaiswal, V.; Mishra, V.K.; Paliwal, R.; Balyan, H.S.; Gupta, P.K. QTL mapping for some grain traits in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 2018, 24, 909–920. [Google Scholar] [CrossRef]
- Duan, X.; Yu, H.; Ma, W.; Sun, J.; Zhao, Y.; Yang, R. A major and stable QTL controlling wheat thousand grain weight: Identification, characterization, and CAPS marker development. Mol. Breed. 2020, 40, 68. [Google Scholar] [CrossRef]
- Ji, G.; Xu, Z.; Fan, X.; Zhou, Q.; Chen, L.; Yu, Q. Identification and validation of major QTL for grain size and weight in bread wheat (Triticum aestivum L.). Crop J. 2022, 11, 564–572. [Google Scholar] [CrossRef]
- Rasheed, A.; Xia, X.; Ogbonnaya, F.; Mahmood, T.; Zhang, Z.; Kazi, A.M. Genome-wide association for grain morphology in synthetic hexaploid wheats using digital imaging analysis. BMC Plant Biol. 2018, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del Blanco, A. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Halder, J.; Gill, H.S.; Zhang, J.; Altameemi, R.; Olson, E.; Turnipseed, B.; Sehgal, S.K. Genome-wide association analysis of spike and kernel traits in the U.S. hard winter wheat. Plant Genome 2023, 16, e20300. [Google Scholar] [CrossRef] [PubMed]




| Ch | Start Position | End Position | Length (Mb) | No. of Markers | Marker Density |
|---|---|---|---|---|---|
| 1A | 1.95 | 597.36 | 595.41 | 4465 | 0.133 |
| 1B | 1.42 | 700.37 | 698.95 | 3094 | 0.226 |
| 1D | 2.88 | 498.05 | 495.17 | 1168 | 0.424 |
| 2A | 2.4 | 787.72 | 785.32 | 2858 | 0.275 |
| 2B | 1.88 | 812.03 | 810.15 | 3581 | 0.226 |
| 2D | 2.22 | 565.44 | 563.22 | 2058 | 0.274 |
| 3A | 7.23 | 747.44 | 740.21 | 3912 | 0.189 |
| 3B | 0.07 | 848.26 | 848.19 | 8322 | 0.102 |
| 3D | 2.69 | 619.37 | 616.68 | 1869 | 0.330 |
| 4A | 4.02 | 754.04 | 750.02 | 3451 | 0.217 |
| 4B | 1.74 | 672.25 | 670.51 | 1567 | 0.428 |
| 4D | 0.46 | 514.92 | 514.46 | 1079 | 0.477 |
| 5A | 0.85 | 712.17 | 711.32 | 1902 | 0.374 |
| 5B | 0.08 | 714.55 | 714.47 | 3297 | 0.217 |
| 5D | 1.58 | 568.63 | 567.05 | 1487 | 0.381 |
| 6A | 1.54 | 621.88 | 620.34 | 2765 | 0.224 |
| 6B | 1.31 | 731.18 | 729.87 | 3980 | 0.183 |
| 6D | 0.06 | 494.6 | 494.54 | 1433 | 0.345 |
| 7A | 0.23 | 743.92 | 743.69 | 3357 | 0.222 |
| 7B | 1.04 | 763.61 | 762.57 | 2525 | 0.302 |
| 7D | 1.25 | 642.43 | 641.18 | 1312 | 0.489 |
| Whole genome | 14,073.32 | 59,482 | 0.236 | ||
| SNP | Position (bp) | Position (Mb) | Alleles | Chr | p Value | LOD | MAF | PVE | Environment/Trait |
|---|---|---|---|---|---|---|---|---|---|
| S1A_47840044 | 47,840,044 | 47.840044 | A/G | 1A | 3.09 × 10−8 | 7.51 | 0.05 | 10.6 | BI20. Length |
| S2A_498737202 | 498,737,202 | 498.737202 | C/T | 2A | 7.41 × 10−10 | 9.13 | 0.42 | 08.2 | BD20.1000 KW |
| S2A_211351736 | 211,351,736 | 211.351736 | C/T | 2A | 4.02 × 10−8 | 7.4 | 0.06 | 05.2 | BI20. Length |
| S2A_251496962 | 251,496,962 | 251.496962 | G/A | 2A | 6.49 × 10−8 | 7.19 | 0.45 | 05.7 | BD20. Peri |
| S2B_664436363 | 664,436,363 | 664.436363 | G/A | 2B | 1.86 × 10−10 | 9.73 | 0.09 | 37.1 | BD20. Length |
| S2B_702381983 | 702,381,983 | 702.381983 | G/C | 2B | 1.13 × 10−9 | 8.95 | 0.13 | 04.4 | BD20. Peri |
| S2B_740979562 | 740,979,562 | 740.979562 | G/A | 2B | 2.51 × 10−9 | 8.6 | 0.33 | 13.8 | BI20. H.Wt |
| S2B_792756832 | 792,756,832 | 792.756832 | G/A | 2B | 6.17 × 10−8 | 7.21 | 0.39 | 17.3 | BD20. Width |
| S4A_663097002 | 663,097,002 | 663.097002 | T/G | 4A | 3.96 × 10−11 | 10.4 | 0.23 | 31.8 | BD20.1000 KW |
| S4B_592421708 | 592,421,708 | 592.421708 | C/T | 4B | 1.83 × 10−13 | 12.74 | 0.25 | 11.7 | BD20. Area |
| S4B_592421708 | 592,421,708 | 592.421708 | C/T | 4B | 4.33 × 10−12 | 11.36 | 0.25 | 21.6 | BD20. Width |
| S6B_567706088 | 567,706,088 | 567.706088 | A/G | 6B | 7.69 × 10−9 | 8.11 | 0.25 | 11.2 | BD20.1000 KW |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rauf, Y.; Wang, Z.; Parker, K.; Baker, S.A.; Baker, J.A.; Rudd, J.C.; Xue, Q.; Ibrahim, A.; Liu, S. Association Mapping for Biomass and Kernel Traits in Doubled-Haploid Population Derived from Texas Wheat Cultivars. Genes 2025, 16, 1172. https://doi.org/10.3390/genes16101172
Rauf Y, Wang Z, Parker K, Baker SA, Baker JA, Rudd JC, Xue Q, Ibrahim A, Liu S. Association Mapping for Biomass and Kernel Traits in Doubled-Haploid Population Derived from Texas Wheat Cultivars. Genes. 2025; 16(10):1172. https://doi.org/10.3390/genes16101172
Chicago/Turabian StyleRauf, Yahya, Zhen Wang, Kyle Parker, Shannon A. Baker, Jason A. Baker, Jackie C. Rudd, Qingwu Xue, Amir Ibrahim, and Shuyu Liu. 2025. "Association Mapping for Biomass and Kernel Traits in Doubled-Haploid Population Derived from Texas Wheat Cultivars" Genes 16, no. 10: 1172. https://doi.org/10.3390/genes16101172
APA StyleRauf, Y., Wang, Z., Parker, K., Baker, S. A., Baker, J. A., Rudd, J. C., Xue, Q., Ibrahim, A., & Liu, S. (2025). Association Mapping for Biomass and Kernel Traits in Doubled-Haploid Population Derived from Texas Wheat Cultivars. Genes, 16(10), 1172. https://doi.org/10.3390/genes16101172







