Abstract
Background: The ubiquitin–proteasome system plays a critical role in plant antiviral defense, with HECT-type E3 ubiquitin ligases serving as key regulators of protein turnover. To explore the potential involvement of the HECT gene family in host resistance against tomato yellow leaf curl virus (TYLCV), a comprehensive analysis was conducted in Nicotiana benthamiana. Methods: In this study, the HECT gene family in N. benthamiana was systematically investigated using a genome-wide bioinformatic analysis. The potential roles of these genes in the response to TYLCV infection were further examined using a virus-induced gene silencing (VIGS) technique. Results: Using a Hidden Markov Model approach, 18 NbHECT genes were identified that phylogenetically clustered into four subfamilies with distinct structural features. Chromosomal location and synteny analyses indicated that these genes were unevenly distributed across 11 chromosomes, with 10 instances of segmental duplication identified. Tissue-specific expression profiling demonstrated that 17 NbHECTs were constitutively expressed, with Group III members showing the highest expression in reproductive tissues. Following TYLCV infection, NbHECT6 was significantly downregulated while NbHECT13 was upregulated in both inoculated and systemic leaves. Functional validation through the VIGS approach revealed that suppression of NbHECT6 and NbHECT13 increased host susceptibility, as evidenced by exacerbated symptom severity and enhanced viral DNA accumulation compared to controls. Conclusions: These findings establish NbHECT6 and NbHECT13 as critical components of the plant antiviral response, providing new insights into ubiquitin-mediated defense mechanisms against geminiviruses.
1. Introduction
In higher plants, the life cycle—from seed germination and subsequent growth and development to environmental adaptation—is orchestrated by a series of highly coordinated biological processes [1]. In plant cells, proteins function as central molecular executors that regulate a complex network of biochemical pathways, maintaining core physiological functions and facilitating adaptive responses [2]. However, the functions of proteins are not constant; they must be precisely activated, regulated, and degraded at specific times to accommodate the dynamic growth demands and environmental stresses faced by plants [3,4]. Central to this intricate regulatory network is the ubiquitin–proteasome system (UPS), which has been demonstrated to play a crucial role in plant growth, development, and stress responses [5,6,7,8]. The core mechanism underlying the UPS is ubiquitination, a biochemical process mediated by an enzymatic cascade in which ubiquitin (Ub) molecules are covalently linked through their C-terminus to lysine residues of target proteins [9,10]. Ub-tagged proteins are recognized explicitly by various intracellular enzyme complexes and organelles, such as the 26S proteasome, thereby determining the fate of the target proteins, including their degradation, transport, functional modification, or participation in signaling cascades [6,7,8,9,10].
The ubiquitination process is highly organized and sequentially mediated by three classes of Ub-related enzymes (E1, E2, and E3) [9,10,11,12]. Among these, E3 ligases serve as the primary determinants of substrate specificity in the ubiquitination process. Based on their domain architecture and mechanisms of catalyzing Ub chain formation, E3 ligases are broadly classified into four major categories: HECT (Homologous to E6-AP Carboxyl Terminus) type, RING (Really Interesting New Gene) type, CRL (Cullin-RING Ligase) type, and U-box type [13,14], which facilitate Ub transfer through two basic mechanisms: direct transfer and indirect transfer. HECT-type E3 ligases (HECTs) are mainly involved in the indirect Ub transfer [14,15,16]. These E3 ligases possess a conserved HECT domain, which is typically situated at the C-terminus and consists of approximately 350 amino acid residues [15,16]. This domain can be further divided into two functional subdomains: an N-terminal subdomain responsible for facilitating E2 binding, and a C-terminal subdomain that contains a critical catalytic cysteine residue [14,17,18]. This catalytic cysteine residue is crucial for the formation of the Ub-thioester intermediate, which ultimately enables the transfer of Ub to target proteins [17,19,20]. The distinct catalytic mechanism of HECTs and their pivotal role in mediating Ub chain formation constitute key molecular foundations for the identification, functional characterization, and evolutionary analysis of members within this gene family.
Tomato yellow leaf curl virus (TYLCV), which is a monopartite DNA virus within the family Geminiviridae, has spread worldwide rapidly since its initial identification in tomato plants in 1983 [21]. It has caused significant losses in tomato cultivation and has become the predominant viral pathogen, posing a threat to global Solanaceae crop production [22,23,24,25]. The TYLCV genome comprises eight genes that encode eight corresponding viral proteins (V1–V3 and C1–C5), which coordinately regulate key stages of viral infection [23,24,25,26]. During the invasion and immune evasion stage, the V1 protein (also known as coat protein, CP) assembles into the viral capsid structure, and V2 facilitates nuclear export of CP and functions as a suppressor to counteract both host antiviral gene silencing [27,28]. Furthermore, the C2 protein can interfere with protein ubiquitination and antagonize the jasmonic acid signaling pathway, therefore promoting viral infection [29,30]. During the replication and proliferation stage, the C1/Rep protein plays a central role in viral DNA replication, recruiting the host replication machinery [31]. Meanwhile, the C3 protein significantly enhances viral replication efficiency through its interactions with host factors [32,33]. During the symptom development process, the V3 protein localizes to the endoplasmic reticulum and the Golgi apparatus, inhibiting cell-to-cell spread of RNA silencing and restricting intercellular movement of the virus [34]. The C4 and C5 proteins serve as the primary determinant of symptom development, suppressing the intercellular spread of RNA silencing and interfering with host defense mechanisms and developmental processes [35,36,37]. Thus, it appears that each protein encoded by TYLCV plays a crucial role in its viral pathogenicity.
N. benthamiana, a member of the Solanaceae family, is extensively utilized as a model organism in plant virology due to its high susceptibility to a wide range of viruses [38]. To explore the role of HECT E3 ligases in the plant response to TYLCV infection, we conducted a comprehensive analysis of the HECT gene family in N. benthamiana. In this study, the HECT gene family in N. benthamiana was systematically investigated, and their potential roles in response to TYLCV infection were examined using a virus-induced gene silencing (VIGS) technique. A total of 18 members of the HECT gene family were identified in N. benthamiana, and two of them were found to be differentially changed during TYLCV infection. Further VIGS assay revealed that suppressing the expression of NbHECT6 and NbHECT13 in N. benthamiana enhances host susceptibility to TYLCV. Our findings provide new evidence that NbHECTs play a significant role in improving plant resistance to TYLCV infection.
2. Materials and Methods
2.1. Identification of the HECT Genes in N. benthamiana
The HECT protein sequences of Arabidopsis thaliana [39], Oryza sativa [40], Zea mays [41], Solanum lycopersicum [14], Glycine max [42], and Triticum aestivum [43] were obtained from the Ensembl Plants database (https://plants.ensembl.org/ (accessed on 2 December 2023)) (Table S1) and used to generate an HECT Hidden Markov Model (HMM) using HMMER software (v3.3.2) [44]. The genome sequence of N. benthamiana (v2.6.1) was downloaded from the Solanaceae Genomics Network database (https://solgenomics.net/ (accessed on 2 December 2023)) [45] and subjected to HMMER search as described previously [26]. The obtained protein sequences were further analyzed using the online InterPro database (https://www.ebi.ac.uk/interpro/ (accessed on 14 December 2023)) [46] to verify the presence of the conserved HECT domain (IPR000569). The candidate NbHECT genes identified in N. benthamiana were renamed according to their chromosomal distribution order. The physicochemical properties of NbHECT proteins were analyzed using the Compute pI/MW tool available on the Expasy website (https://www.expasy.org/ (accessed on 16 December 2023)) [47].
2.2. Phylogenetic Tree Construction, Exon–Intron Structure, and Promoter Analyses
The full-length sequences of HECT proteins from N. benthamiana, A. thaliana, O. sativa, Z. mays, S. lycopersicum, G. max, and T. aestivum (Table S1) were aligned using the MUSCLE algorithm implemented in MEGA 11.0 [48]. A phylogenetic tree was generated using MEGA 11.0, employing the maximum-likelihood (ML) method with the JTT + G model with 1000 bootstrap replicates, as previously described [26]. The exon–intron structural information of NbHECT genes was obtained from the N. benthamiana genome sequence database (v2.6.1) [45] and subsequently visualized using the online tool Gene Structure Display Server 2.0 (GSDS 2.0) [49]. The 2000 bp promoter sequences of NbHECT genes were retrieved from the N. benthamiana genome and analyzed using the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 18 April 2024)) [50]. The putative cis-acting elements were classified and summarized according to their functional roles and visualized using TBtools-II software (v2.148) [51].
2.3. Chromosomal Localization and Collinearity Analyses
The chromosomal locations of NbHECT genes were obtained from the GFF3 annotation file of the N. benthamiana genome. Collinearity analysis was performed using the MCScanX toolkit (v1.0.0) as described previously [52]. Tandem duplications were identified as genes with no more than five adjacent duplicates located within a 100-kb positional interval on the same chromosome as described previously [53,54]. The non-synonymous (Ka) and synonymous (Ks) substitution rates, along with the corresponding Ka/Ks ratio for each gene pair, were calculated using TBtools-II software (v2.148) [51]. For the evolutionary analysis, the divergence times (T) of duplication events were estimated using the formula T = Ks/2λ, where λ represents a substitution rate, which was set to 1.5 × 10−8 for N. benthamiana [55,56].
2.4. Tissue-Specific Expression Pattern Analysis
To explore the expression profiles of NbHECT genes in different tissues of N. benthamiana, we performed an integrative comparative analysis using publicly available RNA-sequencing datasets. Eight different tissues, including the roots, stems, leaves, flowers, capsules, apices, calli, and seedlings, under accession numbers, SRR696961, SRR696992, SRR696940, SRR696938, SRR696884, SRR685298, SRR697013, and SRR696988 of NCBI BioProject PRJNA188486 [57], were selected to analyze the expression patterns of NbHECTs in N. benthamiana. Sequence assembly was conducted using HISAT2 (v.2.1.0) [58], and the transcripts per million (TPM) values were computed using StringTie2 (v.2.1.5) [59]. The results were visualized using TBtools-II software (v2.148) [51].
2.5. Plant Materials and TYLCV Inoculation
In this study, wild-type N. benthamiana plants were used and were cultivated under controlled conditions in a greenhouse at Huzhou University as previously described [26]. The infectious clone of TYLCV was constructed as described by Zhang et al. [60]. Viral inoculation of N. benthamiana with TYLCV was performed as described previously [26,61]. At 6 and 14 days post-infiltration (dpi), inoculated and systemic leaves of N. benthamiana plants were sampled, respectively, from three independent biological replicates as described previously [62].
2.6. RNA Extraction and Quantitative PCR (qPCR) Analysis
Total RNA was isolated from leaf samples using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China), and residual genomic DNA was subsequently removed by DNase I following established protocols [63]. The concentration and integrity of RNA were assessed using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). cDNA synthesis and qPCR analysis were conducted as described previously [64]. Briefly, the cDNA was synthesized from 1 μg of total RNA using the Prime ScriptTM RT Reagent Kit (TaKaRa, Dalian, China), followed by qPCR conducted on a CFX96 Touch Deep Well Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). For each candidate gene, PCR reactions were conducted in duplicate, and relative mRNA expression levels were quantified using the 2−ΔΔCt method [65], with data derived from three independent biological replicates. N. benthamiana ACTIN2 gene (NbACTIN2) was used as an internal control [23,66]. The primers used for qPCR analysis were designed using TBtools-II software (v2.148) [51], and they are listed in Table S2. The results represent the averages of three biological replicates, with raw data provided in Table S3.
2.7. VIGS Experiment and Silencing Efficiency Assay
To suppress the expression of the NbHECT6 and NbHECT13 genes in N. benthamiana, a tobacco rattle virus (TRV)-based VIGS system was employed [67]. To construct the VIGS plasmids, a 300 bp fragment of the NbHECT6 and NbHECT13 genes was amplified and subsequently cloned into the EcoRI–BamHI restriction sites of the pTRV2 vector as described by Zhong et al. [68]. The primers used for VIGS plasmid construction are listed in Table S2. Agrobacterium-mediated infiltration of N. benthamiana was conducted as previously described [64]. N. benthamiana plants agroinfiltrated with TRV1 and TRV2:GFP served as the control. At 10 dpi, the silencing efficiency was determined by qPCR analysis, as previously described [26,68]. The primers used are listed in Table S2.
2.8. Genomic DNA Extraction and Viral Accumulation Determination
Total genomic DNA was extracted from systemically infected leaves using a cetyltrimethylammonium bromide (CTAB)-based method [66]. The accumulation of viral genomic DNA was assessed by qPCR, and the relative levels were determined using the 2−ΔΔCt method [65]. The N. benthamiana 25S nuclear rRNA gene (Nb25SrRNA) served as an internal reference [23]. The primers used are listed in Table S2.
2.9. Statistical Analysis
All experiments were conducted with three independent replicates, and data are presented as mean ± standard deviation (SD). Statistical significance was assessed using Student’s t-test, with a p-value <0.05 considered significant as previously described [26,69].
3. Results
3.1. Identification and Physicochemical Properties of the HECT Gene Family in N. benthamiana
In this study, an HMM was constructed based on the HECT protein sequences of six plant species: A. thaliana, O. sativa, Z. mays, S. lycopersicum, G. max, and T. aestivum (Table S1), and subsequently used to search against the whole-genome protein sequences of N. benthamiana. A total of 18 members of the HECT gene family were identified in N. benthamiana after the validation of conserved domains and the removal of redundant sequences. These genes were systematically designated as NbHECT1 to NbHECT18 according to their chromosomal locations (Table 1). An analysis of their physicochemical properties revealed that the genomic DNA sequences of the identified NbHECT genes ranged from 4235 to 56,645 bp in length, while the putative NbHECT proteins consisted of 588 to 3479 amino acid residues, with predicted molecular weights (MWs) ranging from 67.63 to 383.67 kDa. Notably, the isoelectric points (pIs) of the NbHECT proteins exhibited a slightly acidic characteristic, with 14 members having pI values below 7 (4.97–6.89), whereas only four members displayed weakly basic characteristics (7.21–8.56) (Table 1).

Table 1.
Structural and physicochemical characterization of the HECT-type E3 ubiquitin ligase (HECT) gene family in N. benthamiana.
3.2. Phylogenetic Analysis of the NbHECT Gene Family
To elucidate the evolutionary relationships among members of the NbHECT gene family, a phylogenetic tree was constructed using 18 NbHECTs in combination with 7 A. thaliana ubiquitin-protein ligases (AtUPLs)/AtHECTs, 7 OsUPLs/OsHECTs, 12 ZmUPLs/ZmHECTs, 14 SlHECTs, 19 GmHECTs, and 25 TaHECTs (Table S1). As shown in Figure 1, these 102 HECTs were classified into four subfamilies: Groups I, II, III, and IV. Group I, the largest subgroup, comprised 6 NbHECT members, 2 AtHECT members, 3 OsHECT members, 4 ZmHECT members, 3 SlHECT members, 6 GmHECT members, and 9 TaHECT members. Group II consisted of 3 NbHECT members, 2 AtHECT members, 2 OsHECT members, 3 ZmHECT members, 2 SlHECT members, 5 GmHECT members, and 8 TaHECT members. Group III possessed 4 NbHECT members, 2 AtHECT members, 1 OsHECT member, 3 ZmHECT members, 2 SlHECT members, 6 GmHECT members, and 3 TaHECT members, and therefore was the smallest subfamily. Group IV contained 5 NbHECT members, 1 AtHECT member, 1 OsHECT member, 2 ZmHECT members, 7 SlHECT members, 2 GmHECT members, and 5 TaHECT members (Figure 1). Notably, Groups I and IV were the subfamilies with the highest number of NbHECTs (Figure 1). These results indicate a more closely related evolutionary relationship between the HECT genes of N. benthamiana and S. lycopersicum compared to other analyzed taxa.
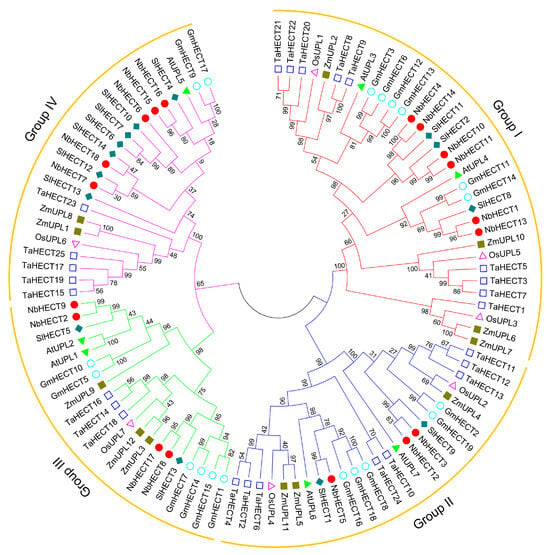
Figure 1.
Phylogenetic analysis of the HECT-type E3 ubiquitin ligase (HECT) gene family from N. benthamiana, A. thaliana, O. sativa, Z. mays, S. lycopersicum, G. max, and T. aestivum. A phylogenetic tree was generated in MEGA 11.0 using the maximum-likelihood (ML) method, with branch support assessed by 1000 bootstrap replicates. HECTs from different plant species are represented by distinct colors, with red, blue, green, and pink clusters denoting Groups I, II, III, and IV, respectively.
3.3. Gene Structure Analysis of the NbHECT Gene Family
To gain deeper insights into the structural diversity of NbHECT genes, we generated an ML phylogenetic tree and analyzed the exon–intron organization using the online tool GSDS 2.0 [49]. As shown in Figure 2, the number of exons exhibited considerable variation across different HECT genes. Among them, NbHECT5 showed the most complex exon organization, comprising 24 exons, whereas NbHECT6 and NbHECT7 displayed the simplest architecture, with only three exons each (Figure 2). Interestingly, all NbHECT genes belonging to Groups I, II, and III exhibited gene architectures comprising more than 10 exons, whereas those in Group IV contained fewer than 10 exons (Figure 2). It is noteworthy that the majority of NbHECT genes in the same subfamily possessed the same number of exons, such as NbHECT1, NbHECT10, NbHECT11, and NbHECT14 in Group I and NbHECT2, NbHECT8, and NbHECT9 in Group III (Figure 2). These results indicate that phylogenetic clustering is correlated with structural conservation among NbHECT genes, suggesting shared functional characteristics and evolutionary trajectories within each clade.
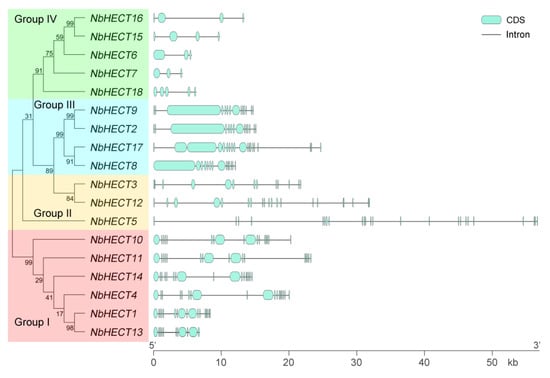
Figure 2.
Schematic representation of the molecular phylogenetic relationships and gene structures of the HECT-type E3 ubiquitin ligase (HECT) genes in N. benthamiana. The unrooted phylogenetic tree was generated using the full-length NbHECT protein sequences with 1000 bootstrap replicates. The exon–intron architecture of NbHECT genes was analyzed using the Gene Structure Display Server 2.0 (GSDS 2.0). The exon and intron lengths for each NbHECT gene were proportionally represented to illustrate structural features.
3.4. Chromosomal Location and Synteny Analyses of the NbHECT Genes
To determine the chromosomal distribution of NbHECT genes in the N. benthamiana genome, we analyzed their chromosomal positions using the locus data obtained from the N. benthamiana Genome Project. As shown in Figure 3, a total of 18 NbHECT genes were unevenly distributed across 19 chromosomes. A single locus of genes was identified on chromosomes 02, 09, 10, 13, 14, and 15; chromosomes 04, 12, and 19 each harbored two genes; chromosomes 01 and 06 each contained three genes; while chromosomes 03, 05, 07, 08, 11, 16, 17, and 18 did not contain any NbHECT genes (Figure 3). To obtain a more comprehensive understanding of the evolutionary duplication patterns of NbHECT genes, a collinearity analysis was conducted using their chromosomal positions. As a result, a total of 10 segmental duplication events and no tandem duplication events were identified in the N. benthamiana genome (Figure 3 and Table 2). This indicates that segmental duplication may act as a primary driving force in the evolution of NbHECT genes in N. benthamiana. To assess evolutionary divergence and selection pressures among these duplicated gene pairs, we calculated Ka and Ks substitution rates using TBtools-II (v2.148) [51]. As shown in Table 2, the Ka/ Ks ratios of 10 duplicated gene pairs were all below 1, indicating that the HECT gene family in N. benthamiana has predominantly undergone purifying selection. Furthermore, the evolutionary dates of these duplication events were estimated using the formula T = Ks/2λ, which has been previously established for N. benthamiana [56]. These results suggest that 10 segmental duplications likely occurred between 1.75 and 21.38 million years ago.
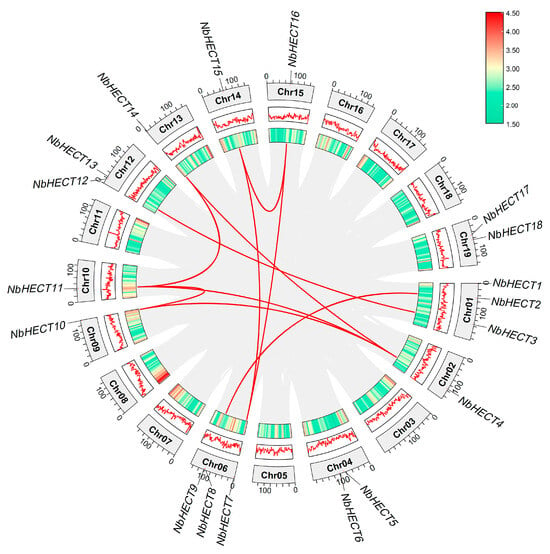
Figure 3.
Schematic representation of the chromosomal distribution and duplication patterns of 18 HECT-type E3 ubiquitin ligase (HECT) genes in N. benthamiana. Red dots and black lines indicate the chromosomal locations of NbHECT genes, and the red lines represent duplicated gene pairs. Chromosome numbers are labeled within each respective chromosome. The scale bar indicates the gene density across the chromosomes of N. benthamiana.

Table 2.
Divergence analysis of duplicated HECT-type E3 ubiquitin ligase (HECT) gene pairs in N. benthamiana.
3.5. Promoter Analysis of the NbHECT Genes
To further elucidate the potential functions and regulatory mechanisms of NbHECTs, we performed a prediction and analysis of cis-acting elements in the promoter regions of NbHECT genes using the PlantCARE database [50]. The results revealed a large number of phytohormone- and growth-related regulatory elements in the regulatory regions of the NbHECT genes. These included the MeJA-responsive element (MeJARE), gibberellin-responsive element (GARE), salicylic acid (SA)-responsive element (SARE), abscisic acid (ABA)-responsive element (ABARE), meristem expression element (CAT-box), and auxin-responsive element (AuxRE) (Figure 4). Among the 18 NbHECT genes, 15 harbored the MeJARE element, 15 contained the GARE element, 12 possessed the SARE element, 10 included the ABARE element, 6 possessed the CAT-box element, and 5 contained the AuxRE element (Figure 4 and Table S4). In addition to the phytohormone- and growth-related regulatory elements, a considerable number of stress-responsive regulatory elements were also identified within the promoter regions of NbHECT genes (Figure 4). These included the anaerobic response element (ARE), defense- and stress-responsive element (DSRE), low-temperature-responsive element (LTRE), and MYB binding site (MBS). Of the 18 NbHECT genes, 16 contained the ARE element, 8 harbored the DSRE element, 7 contained the LTRE element, and 6 possessed the MBS element (Figure 4 and Table S4). These findings indicate that the promoters of NbHECT genes contain diverse cis-acting elements related to phytohormone signaling, growth processes, and stress responses, suggesting that NbHECTs may play crucial roles in modulating multiple phytohormone pathways, developmental programs, and plant responses to environmental stresses.
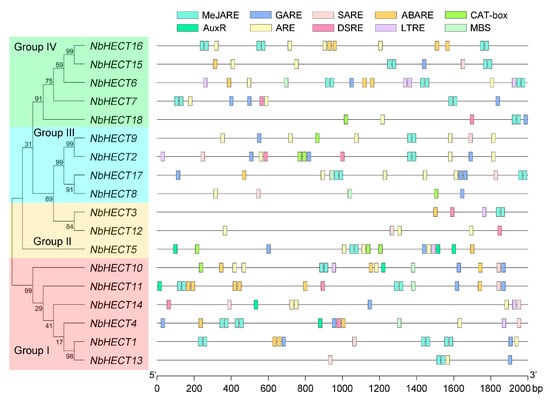
Figure 4.
Schematic representation of the molecular phylogenetic relationships and 2000 bp promoters of the HECT-type E3 ubiquitin ligase (HECT) genes in N. benthamiana. The unrooted phylogenetic tree was generated using the full-length NbHECT protein sequences with 1000 bootstrap replicates. The 2000 bp promoter sequences of HECT genes were analyzed using PlantCARE and visualized using TBtools-II (v2.148). Different colored boxes represent different cis-acting elements.
3.6. Tissue-Specific Expression Patterns of the NbHECT Genes
To elucidate the spatiotemporal expression patterns of NbHECT genes, we analyzed their transcript abundance across eight distinct tissue types of N. benthamiana using published RNA-sequencing datasets under the BioProject accession number PRJNA188486 [57]. The results indicated that all 18 NbHECT genes exhibited expression in at least one tissue (Figure 5). Strikingly, 17 of them (NbHECT1–NbHECT17) showed constitutive expression patterns across all examined tissues, with TPM values greater than 1.0 (TPM > 1.0) (Figure 5 and Table S5). Interestingly, all NbHECT genes (NbHECT2, NbHECT8, NbHECT9, and NbHECT17) in Group III exhibited high expression levels (TPM ≥ 5.0), suggesting that this subfamily may play a crucial role in regulating the growth and development of N. benthamiana (Figure 5 and Table S5). Furthermore, 11 genes, including NbHECT1, NbHECT2, NbHECT4, NbHECT6, NbHECT8–NbHECT11, NbHECT14, NbHECT15, and NbHECT17, displayed high expression levels in the stems and capsules of N. benthamiana (TPM ≥ 15.0). Among them, NbHECT11 showed the highest expression in the stems (TPM ≥ 42.0), while NbHECT17 exhibited the highest expression in the capsules (TPM ≥ 48.0) (Figure 5 and Table S5). Seven genes, namely, NbHECT2, NbHECT8, NbHECT10, NbHECT11, NbHECT14, NbHECT15, and NbHECT17, displayed high expression levels in the roots, leaves, flowers, and calli of N. benthamiana (TPM ≥ 15.0), with the highest expression levels observed for NbHECT17 in the roots, capsules, and calli (TPM ≥ 20.0), and NbHECT11 in the stems, leaves, and flowers (TPM ≥ 20.0), respectively (Figure 5 and Table S5). These results indicate that NbHECT genes exhibit distinct tissue-specific expression patterns, suggesting their potential roles in modulating diverse biological processes in N. benthamiana.
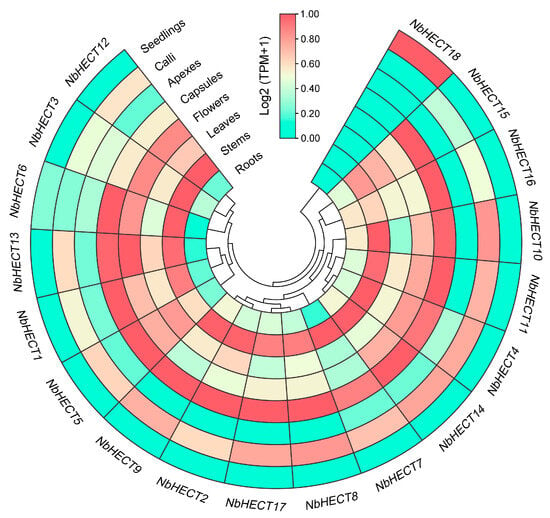
Figure 5.
Expression profiles of the HECT-type E3 ubiquitin ligase (HECT) genes in N. benthamiana. The data used for tissue expression were obtained from the NCBI sequence read archive (SRA) database under the accession number PRJNA188486 [57], and the expression level of each gene is colored based on its Log2 (TPM + 1) values calculated from eight tissues: roots, stems, leaves, flowers, capsules, apices, calli, and seedlings. The heatmap was produced using TBtools-II (v2.148).
3.7. Expression Profiles of NbHECT Genes in Response to TYLCV Infection
To elucidate the potential roles of NbHECT genes in TYLCV defense responses, we selected NbHECTs with basal expression levels in the leaves of N. benthamiana (TPM ≥1.0) (Table S5) and examined their expression profiles following TYLCV infection using qPCR. The results showed that the expression of NbHECT1–NbHECT5, NbHECT7, NbHECT9–NbHECT11, and NbHECT13–NbHECT17 was significantly upregulated in the inoculated leaves of N. benthamiana following TYLCV infection, while NbHECT6 exhibited a pronounced downregulation pattern (Figure 6). In the systemically infected leaves, only two genes, NbHECT6 and NbHECT13, displayed downregulated and upregulated patterns, respectively, following TYLCV infection (Figure 6). However, due to the exceptionally low basal expression level of NbHECT18 in all examined tissues (Figure 5 and Table S5), its expression was not detected in either the inoculated leaves or the systemically infected leaves. These results suggest that NbHECTs are likely to play a key role in the early defense response to TYLCV infection, particularly NbHECT6 and NbHECT13, which exhibit a consistent expression pattern in both inoculated and systemic leaves.
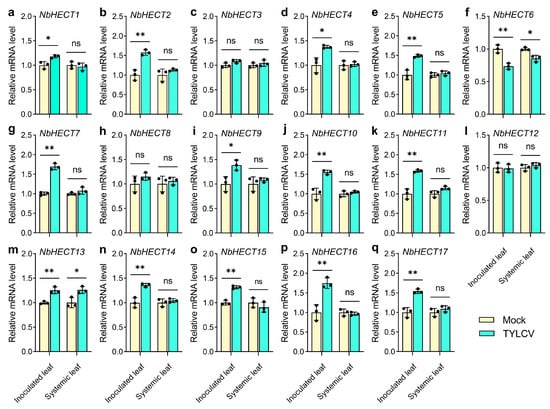
Figure 6.
Expression levels of the HECT-type E3 ubiquitin ligase (HECT) genes in N. benthamiana in response to tomato yellow leaf curl virus (TYLCV) infection. (a–q) Relative expression levels of NbHECT1–NbHECT17 in inoculated and systemically infected leaves of N. benthamiana following TYLCV infection. The data represent relative mRNA expression levels in leaves infected with Agrobacterium tumefaciens carrying an empty vector (Mock), which were normalized to a reference value of 1.0. Results are presented as the mean ± standard deviation from three independent biological replicates. Statistically significant differences are indicated by asterisks: * p < 0.05 or ** p < 0.01; ns, not significant (Student’s t-test).
3.8. Disruption of the Expression of NbHECTs Increases Host Susceptibility to TYLCV
To further clarify the functional roles of NbHECTs in the response to TYLCV infection, we employed the VIGS system to investigate their specific involvement during viral infection. According to the tissue-specific expression patterns and gene expression profiles of NbHECTs following TYLCV infection (Figure 5 and Figure 6), two genes, NbHECT6 and NbHECT13, were selected for individual or together silencing using a TRV-mediated VIGS approach [67]. Compared with the vector control (N. benthamiana agroinfiltrated with TRV:GFP), the mRNA levels of NbHECT6 and NbHECT13 in plants infiltrated with the silencing constructs were reduced by about 60–80% at 10 dpi (Figure 7a), confirming successful silencing of the target genes through VIGS. Following VIGS treatment, both control and gene-silenced plants were inoculated with an infectious clone of TYLCV and systematically monitored over time. At 21 dpi, compared to the TRV:GFP control plants, systemic leaves of NbHECT6- and NbHECT13-silenced plants exhibited severe curling and wrinkling symptoms induced by TYLCV (Figure 7b). These findings indicate that the suppression of NbHECT6 and NbHECT13 enhances the susceptibility of N. benthamiana to TYLCV infection. To further confirm these observations, the accumulation of TYLCV genomic DNA was assessed by determining the expression levels of TYLCV AC1 and CP genes via qPCR, following established protocols [26,70]. The results revealed that silencing of NbHECT6 and NbHECT13 in N. benthamiana plants led to an approximately three-fold increase in TYLCV genomic DNA accumulation relative to control plants (Figure 7c). Interestingly, viral DNA accumulation in the double gene-silenced plants was found to be intermediate between that observed in the two single gene-silenced groups (Figure 7c). Collectively, these results suggest that the suppression of NbHECT6 and NbHECT13 significantly compromises the resistance of N. benthamiana to TYLCV, as indicated by a marked increase in viral genomic DNA accumulation in host tissues.
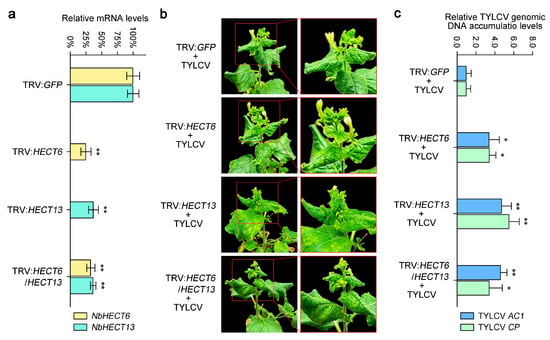
Figure 7.
Silencing the NbHECT6 and NbHECT13 genes in N. benthamiana increases its susceptibility to tomato yellow leaf curl virus (TYLCV) infection. (a) Silencing efficiency was evaluated by quantitative PCR (qPCR) at 10 days post-inoculation (dpi). The data are expressed as relative mRNA levels, with values for the control group (N. benthamiana plants agroinfiltrated with TRV:GFP) set at 100%. (b) Disease symptoms induced by TYLCV in NbHECT6 and NbHECT13 silenced N. benthamiana plants at 21 dpi. TRV:GFP agroinfiltrated N. benthamiana seedlings infected with TYLCV served as the control. (c) Relative accumulation of TYLCV genomic DNA was assessed in the NbHECT6 and NbHECT13 silenced N. benthamiana plants. Viral accumulation was quantified by qPCR, as depicted in (a). Data are presented as relative viral DNA accumulation, with values for the TRV:GFP control plants set to 1.0. For panels (a,c), data are presented as means ± standard deviation of three independent biological replicates. Statistically significant differences are indicated by asterisks: * p < 0.05 or ** p < 0.01, as determined by Student’s t-test.
4. Discussion
Previous studies have shown that the UPS plays a central role in regulating plant growth, development, and stress responses by controlling protein turnover and signaling cascades [5,6,7,8]. In this study, we systematically identified and characterized 18 NbHECTs in N. benthamiana, providing new insights into their structural diversity, evolutionary conservation, and functional roles in antiviral defense. Phylogenetic analysis demonstrated that NbHECTs clustered into four distinct subfamilies (Groups I–IV) (Figure 1), consistent with the classification observed in S. lycopersicum and A. thaliana [14,39], suggesting an ancient evolutionary origin of this gene family in plants. Notably, the closer evolutionary relationship between HECT genes in N. benthamiana and S. lycopersicum, as compared to those in other species (Figure 1), may reflect their shared susceptibility to viral pathogens such as TYLCV [22,23,24,25]. This phylogenetic conservation implies that NbHECTs may have retained critical regulatory functions in Solanaceae-specific stress responses.
Structural analysis revealed that NbHECT genes exhibit significant variation in exon–intron organization, with exon numbers ranging from 3 (NbHECT6 and NbHECT7) to 24 (NbHECT5) (Figure 2 and Table 1). This structural diversity likely reflects functional specialization, as genes within the same phylogenetic group maintained conserved exon numbers (e.g., Group I and III members). Such conservation suggests strong selective pressures to maintain gene architecture, possibly due to constraints on protein domain organization or regulatory elements [16,17,20]. The predominance of segmental duplication events (10 detected) over tandem duplications (Figure 3 and Table 2) suggests that whole-genome duplications rather than localized gene amplifications drove NbHECT family expansion. This finding is consistent with the gene duplication patterns observed in HECT gene families across other plant species [42,43]. The estimated divergence times (1.75–21.38 million years ago) coincide with periods of Solanaceae diversification [53,54,71], further supporting their role in adaptive evolution. Furthermore, the Ka/Ks ratios (<1) across all duplicated pairs (Table 2) suggest purifying selection has maintained critical functional divergence, particularly the catalytic HECT domain required for Ub transfer [15,16,17].
Promoter analysis revealed the presence of numerous cis-acting regulatory elements associated with phytohormone signaling and stress responses (Figure 4 and Table S4), indicating that NbHECTs may serve as integrators of multiple signaling pathways. The high prevalence of MeJAREs and GAREs (15 out of 18 genes) is particularly notable, given the critical role of jasmonate and gibberellin signaling pathways in antiviral defense [72,73,74]. Additionally, the presence of stress-responsive elements (AREs, DSREs, and LTREs) in most NbHECT promoters further supports their involvement in abiotic and biotic stress responses (Figure 4 and Table S4). These findings align with studies in A. thaliana, Malus domestica, Brassica rapa, B. oleracea, and Phyllostachys edulis, where HECTs modulate hormone crosstalk and stress responses [39,75,76,77], suggesting a conserved regulatory network across plant species. Tissue-specific expression analysis demonstrated that 17 out of 18 NbHECTs are constitutively expressed (TPM > 1.0) across all examined tissues (Figure 5 and Table S5), consistent with their putative roles in fundamental cellular functions, including protein homeostasis and signal transduction pathways [6,9]. Notably, Group III members (NbHECT2, NbHECT8, NbHECT9, and NbHECT17) exhibited the highest expression levels in reproductive tissues (TPM ≥ 5.0), suggesting their specialized functions in reproductive development or stress responses [20,78,79]. Furthermore, the strong expression of NbHECT6 and NbHECT13 in leaves (Figure 5) and their differential regulation during TYLCV infection (Figure 6) further highlights their potential role in leaf-specific antiviral responses.
It has been shown that an HECT domain-containing protein, Rsp5p, can bind to the p33 and p92 proteins encoded by tomato bushy stunt virus and inhibit its replication in yeast [80]. In S. lycopersicum and N. benthamiana, plant HECT proteins have also been reported to participate in antiviral responses [14,81]. In this study, it was found that NbHECTs showed a differential expression pattern during TYLCV infection (Figure 6), providing direct evidence for their involvement in antiviral responses. Although most NbHECTs were upregulated in inoculated leaves (Figure 6), the consistent downregulation of NbHECT6 in both local and systemic infections suggests TYLCV may actively suppress this gene to evade host defenses, similar to how geminiviral C2 protein interferes with ubiquitination [82,83]. Conversely, the upregulation of NbHECT13 suggests a potentially positive role in antiviral signaling, which may be mediated through the ubiquitination and subsequent degradation of viral proteins or via the modulation of defense-related hormonal pathways [84,85]. Functional validation through VIGS revealed that the silencing of NbHECT6 and NbHECT13 aggravates TYLCV symptoms (Figure 7b) and leads to approximately a threefold increase in viral DNA accumulation (Figure 7c). These phenotypes are consistent with the effects observed upon the disruption of UPS components in other plant–virus systems [86,87], supporting an evolutionarily conserved role for E3 ligases in antiviral defense. These findings align with emerging models of Ub-mediated antiviral defense in higher plants [88,89]. Although the exact roles of NbHECTs in the response of N. benthamiana to TYLCV infection remain to be fully elucidated, our findings enhance the understanding of the expansion of the NbHECT gene family and offer valuable insights into the functional characterization of these genes in mediating resistance against TYLCV.
5. Conclusions
In this study, we conducted the first comprehensive analysis of the HECT gene family in N. benthamiana through gene identification, phylogenetic analysis, gene structure characterization, promoter analysis, and tissue-specific and TYLCV-responsive expression profiling. A total of 18 NbHECT genes were identified across the entire genome, providing valuable insights for the functional characterization of the HECT gene family in N. benthamiana. Furthermore, our VIGS and qPCR data revealed that TYLCV genomic DNA accumulation was significantly enhanced upon silencing of the two NbHECTs (NbHECT6 and NbHECT13) in N. benthamiana. These findings contribute to a better understanding of the molecular processes mediated by the NbHECT genes in response to TYLCV infection and lay the groundwork for a more systematic exploration of the functional mechanisms of the HECT gene family in N. benthamiana.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16101150/s1, Table S1: Protein sequences of HECT E3 ligases of Arabidopsis thaliana, Oryza sativa, Zea mays, Solanum lycopersicum, Glycine max, and Triticum aestivum, and Nicotiana benthamiana; Table S2: List of primers used in this study; Table S3: Raw data used for qPCR analysis of NbHECT genes in response to TYLCV infection; Table S4: Cis-acting elements in the promoter regions of NbHECT genes in Nicotiana benthamiana; Table S5: Tissue-specific expression profiles of NbHECT genes in Nicotiana benthamiana.
Author Contributions
Conceptualization, X.Z. and Z.W.; methodology, J.S., S.Y. and F.Y.; software, J.S., S.Y. and Y.Z.; validation, J.S., S.Y., X.W. and M.S.; formal analysis, Y.S. and Z.L.; investigation, G.Z., Z.Y. and Z.W.; resources, G.Z., X.L. and X.Z.; data curation, J.S. and S.Y.; writing—original draft preparation, J.S., S.Y., X.Z. and Z.W.; writing—review and editing, X.Z. and Z.W.; visualization, J.S., S.Y., X.Z. and Z.W.; supervision, X.Z. and Z.W.; project administration, X.Z. and Z.W.; funding acquisition, J.S., X.Z. and Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant numbers: 32102162 and 31701759), the Huzhou Public Welfare Application Research Project of China (grant number: 2024GZ34), and the Innovation and Entrepreneurship Training Program for College Students of China (grant number: 202410347025).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study have been included in this article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Niklas, K.J.; Kutschera, U. The evolution of the land plant life cycle. New Phytol. 2010, 185, 27–41. [Google Scholar] [CrossRef]
- Morris, R.; Black, K.A.; Stollar, E.J. Uncovering protein function: From classification to complexes. Essays Biochem. 2022, 66, 255–285. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under stress: How plants balance growth and the stress response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Bartel, B.; Citovsky, V. Focus on ubiquitin in plant biology. Plant Physiol. 2012, 160, 1. [Google Scholar] [CrossRef][Green Version]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef]
- Lobaina, D.P.; Tarazi, R.; Castorino, T.; Vaslin, M.F.S. The ubiquitin-proteasome system (UPS) and viral infection in plants. Plants 2022, 11, 2476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Isono, E.; Li, J.; Pulido, P.; Siao, W.; Spoel, S.H.; Wang, Z.; Zhuang, X.; Trujillo, M. Protein degrons and degradation: Exploring substrate recognition and pathway selection in plants. Plant Cell 2024, 36, 3074–3098. [Google Scholar] [CrossRef]
- Mattern, M.; Sutherland, J.; Kadimisetty, K.; Barrio, R.; Rodriguez, M.S. Using ubiquitin binders to decipher the ubiquitin code. Trends Biochem. Sci. 2019, 44, 599–615. [Google Scholar] [CrossRef]
- Agrata, R.; Komander, D. Ubiquitin-A structural perspective. Mol. Cell 2025, 85, 323–346. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Saxena, H.; Negi, H. Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum. Sci. Rep. 2021, 11, 15891. [Google Scholar] [CrossRef]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef]
- Toma-Fukai, S.; Shimizu, T. Structural diversity of ubiquitin E3 Ligase. Molecules 2021, 26, 6682. [Google Scholar] [CrossRef]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Maspero, E.; Mari, S.; Valentini, E.; Musacchio, A.; Fish, A.; Pasqualato, S.; Polo, S. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011, 12, 342–349. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2021, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Miao, Y. New aspects of HECT-E3 ligases in cell senescence and cell death of plants. Plants 2019, 8, 483. [Google Scholar] [CrossRef]
- Makkouk, K.M.; Laterrot, H. Epidemiology and Control of Tomato Yellow Leaf Curl Virus. In Plant Virus Epidemiology; Plumb, R.T., Thresh, J.M., Eds.; Blackwell: Oxford, UK, 1983; pp. 315–321. [Google Scholar]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato yellow leaf curl virus: Impact, challenges, and management. Trends Plant Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Durán, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qiao, R.; Yang, X.; Gong, P.; Zhou, X. Occurrence, distribution, and management of tomato yellow leaf curl virus in China. Phytopathol. Res. 2022, 4, 28. [Google Scholar] [CrossRef]
- Cao, X.; Huang, M.; Wang, S.; Li, T.; Huang, Y. Tomato yellow leaf curl virus: Characteristics, influence, and regulation mechanism. Plant Physiol. Biochem. 2024, 213, 108812. [Google Scholar] [CrossRef]
- Zhong, X.; Li, J.; Yang, L.; Wu, X.; Xu, H.; Hu, T.; Wang, Y.; Wang, Y.; Wang, Z. Genome-wide identification and expression analysis of wall-associated kinase (WAK) and WAK-like kinase gene family in response to tomato yellow leaf curl virus infection in Nicotiana benthamiana. BMC Plant Biol. 2023, 23, 146. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161. [Google Scholar] [CrossRef]
- Zhao, W.; Wu, S.; Barton, E.; Fan, Y.; Ji, Y.; Wang, X.; Zhou, Y. Tomato yellow leaf curl virus V2 protein plays a critical role in the nuclear export of V1 protein and viral systemic infection. Front. Microbiol. 2020, 11, 1243. [Google Scholar] [CrossRef]
- Wang, L.; Tan, H.; Medina-Puche, L.; Wu, M.; Garnelo Gomez, B.; Gao, M.; Shi, C.; Jimenez-Gongora, T.; Fan, P.; Ding, X.; et al. Combinatorial interactions between viral proteins expand the potential functional landscape of the tomato yellow leaf curl virus proteome. PLoS Pathog. 2022, 18, e1010909. [Google Scholar] [CrossRef]
- Rosas-Diaz, T.; Cana-Quijada, P.; Wu, M.; Hui, D.; Fernandez-Barbero, G.; Macho, A.P.; Solano, R.; Castillo, A.G.; Wang, X.W.; Lozano-Duran, R.; et al. The transcriptional regulator JAZ8 interacts with the C2 protein from geminiviruses and limits the geminiviral infection in Arabidopsis. J. Integr. Plant Biol. 2023, 65, 1826–1840. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Wu, M.; Wei, H.; Tan, H.; Pan, S.; Liu, Q.; Bejarano, E.R.; Lozano-Durán, R. Plant DNA polymerases α and δ mediate replication of geminiviruses. Nat. Commun. 2021, 12, 2780. [Google Scholar] [CrossRef]
- Ghosh, D.; Chakraborty, S. Selective REcruitmeNt of plant DNA polymerases by geminivirus. Trends Genet. 2022, 38, 211–213. [Google Scholar] [CrossRef]
- Gong, P.; Zhao, S.; Liu, H.; Chang, Z.; Li, F.; Zhou, X. Tomato yellow leaf curl virus V3 protein traffics along microfilaments to plasmodesmata to promote virus cell-to-cell movement. Sci. China Life Sci. 2022, 65, 1046–1049. [Google Scholar] [CrossRef]
- Corrales-Gutierrez, M.; Medina-Puche, L.; Yu, Y.; Wang, L.; Ding, X.; Luna, A.P.; Bejarano, E.R.; Castillo, A.G.; Lozano-Duran, R. The C4 protein from the geminivirus Tomato yellow leaf curl virus confers drought tolerance in Arabidopsis through an ABA-independent mechanism. Plant Biotechnol. J. 2020, 18, 1121–1123. [Google Scholar] [CrossRef]
- Padmanabhan, C.; Zheng, Y.; Shamimuzzaman, M.; Wilson, J.R.; Gilliard, A.; Fei, Z.; Ling, K.S. The tomato yellow leaf curl virus C4 protein alters the expression of plant developmental genes correlating to leaf upward cupping phenotype in tomato. PLoS ONE 2022, 17, e0257936. [Google Scholar] [CrossRef]
- Zhao, S.; Gong, P.; Ren, Y.; Liu, H.; Li, H.; Li, F.; Zhou, X. The novel C5 protein from tomato yellow leaf curl virus is a virulence factor and suppressor of gene silencing. Stress Biol. 2022, 2, 19. [Google Scholar] [CrossRef]
- Bally, J.; Jung, H.; Mortimer, C.; Naim, F.; Philips, J.G.; Hellens, R.; Bombarely, A.; Goodin, M.M.; Waterhouse, P.M. The rise and rise of Nicotiana benthamiana: A Plant for all reasons. Annu. Rev. Phytopathol. 2018, 56, 405–426. [Google Scholar] [CrossRef]
- Downes, B.P.; Stupar, R.M.; Gingerich, D.J.; Vierstra, R.D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003, 35, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Hua, K.; Xu, R.; Zeng, D.; Wang, R.; Dong, G.; Zhang, G.; Lu, X.; Fang, N.; Wang, D.; et al. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 2021, 33, 1212–1228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, C.; Qin, A.; Deng, W.; Chen, R.; Yu, H.; Wang, Y.; Song, J.; Zeng, L. Genome-wide identification and transcriptome profiling expression analysis of the U-box E3 ubiquitin ligase gene family related to abiotic stress in maize (Zea mays L.). BMC Genom. 2024, 25, 132. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, C.; Rahman, S.U.; Wang, Y.; Wang, A.; Tao, S. Genome-wide identification and evolution of HECT genes in soybean. Int. J. Mol. Sci. 2015, 16, 8517–8535. [Google Scholar] [CrossRef]
- Meng, X.; Yang, T.; Liu, J.; Zhao, M.; Wang, J. Genome-wide identification and evolution of HECT genes in wheat. PeerJ 2020, 8, e10457. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yan, M.; Chen, S.; Sun, J.; Wang, J.; Meng, D.; Li, J.; Zhang, L.; Guo, L. The complete genome assembly of Nicotiana benthamiana reveals the genetic and epigenetic landscape of centromeres. Nat. Plants 2024, 10, 1928–1943. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Tung, J.; Zhang, X.; Liu, D.; Deng, Y.; Tian, Z.; Chen, H.; Wang, T.; Yin, W.; et al. High-quality assembled and annotated genomes of Nicotiana tabacum and Nicotiana benthamiana reveal chromosome evolution and changes in defense arsenals. Mol. Plant 2024, 17, 423–437. [Google Scholar] [CrossRef]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N.; et al. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef]
- Ranawaka, B.; An, J.; Lorenc, M.T.; Jung, H.; Sulli, M.; Aprea, G.; Roden, S.; Llaca, V.; Hayashi, S.; Asadyar, L.; et al. A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology. Nat. Plants 2023, 9, 1558–1571. [Google Scholar] [CrossRef]
- Nakasugi, K.; Crowhurst, R.N.; Bally, J.; Wood, C.C.; Hellens, R.P.; Waterhouse, P.M. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS ONE 2013, 8, e59534. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gong, H.; Zhou, X. Molecular characterization and pathogenicity of tomato yellow leaf curl virus in China. Virus Genes 2009, 39, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Souvan, J.M.; Bally, J.; de Felippes, F.F.; Waterhouse, P.M. Exploring the source of TYLCV resistance in Nicotiana benthamiana. Front. Plant Sci. 2024, 15, 1404160. [Google Scholar] [CrossRef]
- Wu, M.; Ding, X.; Fu, X.; Lozano-Duran, R. Transcriptional reprogramming caused by the geminivirus Tomato yellow leaf curl virus in local or systemic infections in Nicotiana benthamiana. BMC Genom. 2019, 20, 542. [Google Scholar] [CrossRef]
- Luo, C.; Wang, Z.Q.; Liu, X.; Zhao, L.; Zhou, X.; Xie, Y. Identification and analysis of potential genes regulated by an alphasatellite (TYLCCNA) that contribute to host resistance against tomato yellow leaf curl China virus and its betasatellite (TYLCCNV/TYLCCNB) infection in Nicotiana benthamiana. Viruses 2019, 11, 442. [Google Scholar] [CrossRef]
- Zhong, X.; Yang, L.; Li, J.; Tang, Z.; Wu, C.; Zhang, L.; Zhou, X.; Wang, Y.; Wang, Z. Integrated next-generation sequencing and comparative transcriptomic analysis of leaves provides novel insights into the ethylene pathway of Chrysanthemum morifolium in response to a Chinese isolate of chrysanthemum virus B. Virol. J. 2022, 19, 182. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Cao, L.; Wang, Y.; Xie, Y.; Zhou, X. Mimic phosphorylation of a βC1 protein encoded by TYLCCNB impairs its functions as a viral suppressor of RNA silencing and a symptom determinant. J. Virol. 2017, 91, e00300-17. [Google Scholar] [CrossRef] [PubMed]
- Dinesh-Kumar, S.P.; Anandalakshmi, R.; Marathe, R.; Schiff, M.; Liu, Y. Virus-induced gene silencing. Methods Mol. Biol. 2003, 236, 287–294. [Google Scholar] [PubMed]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Wang, Y.; Xie, Y.; Zhou, X. iTRAQ analysis of the tobacco leaf proteome reveals that RNA-directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection. J. Proteom. 2017, 152, 88–101. [Google Scholar] [CrossRef]
- Cheng, R.; Mei, R.; Yan, R.; Chen, H.; Miao, D.; Cai, L.; Fan, J.; Li, G.; Xu, R.; Lu, W.; et al. A new distinct geminivirus causes soybean stay-green disease. Mol. Plant 2022, 15, 927–930. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef]
- Deanna, R.; Acosta, M.C.; Scaldaferro, M.; Chiarini, F. Chromosome Evolution in the Family Solanaceae. Front. Plant Sci. 2022, 12, 787590. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, Y.; Yang, J.; Yao, S.; Zhao, K.; Wang, D.; Qin, Q.; Bian, Z.; Li, Y.; Lan, Y.; et al. Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 2020, 28, 89–103.e8. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, D.; Li, Y.; Wang, Z.; Wu, Z.; Zhang, Q.; Jia, H.; Dong, X.; Qi, L.; Shi, J.; et al. Gibberellin positively regulates tomato resistance to tomato yellow leaf curl virus (TYLCV). Plants 2024, 13, 1277. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xing, S.; Cui, H.; Chen, X.; Wang, X. Genome-wide identification and characterization of the apple (Malus domestica) HECT ubiquitin-protein ligase family and expression analysis of their responsiveness to abiotic stresses. Mol. Genet. Genom. 2016, 291, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Cui, D.L.; Batool, K.; Yang, Y.Q.; Lu, Y.H. Comprehensive genomic survey, characterization and expression analysis of the hect gene family in Brassica rapa L. and Brassica oleracea L. Genes 2019, 10, 400. [Google Scholar] [PubMed]
- Xie, X.; Hu, S.; Liu, L.; Pan, H.; Huang, H.; Cao, X.; Qiao, G.; Han, X.; Qiu, W.; Lu, Z.; et al. Genome-wide analysis of HECT E3 ligases members in Phyllostachys edulis provides insights into the role of PeHECT1 in plant abiotic stress response. Int. J. Mol. Sci. 2024, 25, 11896. [Google Scholar] [CrossRef]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 ligases and their role in abiotic stress response. Cells 2022, 11, 890. [Google Scholar] [CrossRef]
- Wang, S.; Lv, X.; Zhang, J.; Chen, D.; Chen, S.; Fan, G.; Ma, C.; Wang, Y. Roles of E3 ubiquitin ligases in plant responses to abiotic stresses. Int. J. Mol. Sci. 2022, 23, 2308. [Google Scholar] [CrossRef]
- Barajas, D.; Li, Z.; Nagy, P.D. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 2009, 83, 11751–11764. [Google Scholar] [CrossRef]
- OuYang, X.; Wang, L.; Luo, X.; Li, C.; An, X.; Yao, L.; Huang, W.; Zhang, Z.; Zhang, S.; Liu, Y.; et al. Pepper vein yellow virus P0 protein triggers NbHERC3, NbBax, and NbCRR mediated hypersensitive response. J. Basic Microbiol. 2024, 64, e2400023. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Rosas-Díaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co-option of SCF-mediated ubiquitination. Plant Signal. Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef]
- Shen, Q.; Hu, T.; Bao, M.; Cao, L.; Zhang, H.; Song, F.; Xie, Q.; Zhou, X. Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef]
- Chen, I.H.; Chang, J.E.; Wu, C.Y.; Huang, Y.P.; Hsu, Y.H.; Tsai, C.H. An E3 ubiquitin ligase from Nicotiana benthamiana targets the replicase of Bamboo mosaic virus and restricts its replication. Mol. Plant Pathol. 2019, 20, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jin, L.; Huang, X.; Geng, Y.; Li, F.; Qin, Q.; Wang, R.; Ji, S.; Zhao, S.; Xie, Q.I.; et al. OsRFPH2-10, a ring-H2 finger E3 ubiquitin ligase, is involved in rice antiviral defense in the early stages of rice dwarf virus infection. Mol. Plant 2014, 7, 1057–1060. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.H.; Qin, X.Y.; Guo, H.F.; Zheng, C.; Zhang, Z.Y.; Chen, Q.; Wang, X.B.; Han, C.G.; Wang, Y. The E3 ligase HRD1 enhances plant antiviral immunity by targeting viral movement proteins. Cell Rep. 2025, 44, 115449. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, J.; Sun, X.; Li, J.; Cao, X.; Yao, S.; Han, Y.; Chen, C.; Du, L.; Li, S.; et al. Perception of viral infections and initiation of antiviral defence in rice. Nature 2025, 641, 173–181. [Google Scholar] [CrossRef]
- De Silva, A.; Kim, K.; Weiland, J.; Hwang, J.; Chung, J.; Pereira, H.S.; Patel, T.R.; Teyra, J.; Patel, A.; Mira, M.M.; et al. Suppressing Tymovirus replication in plants using a variant of ubiquitin. PLoS Pathog. 2025, 21, e1012899. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).