Comparative Analysis of the Chloroplast Genomes of Grewia tembensis Fresen and Closely Related Species of Grewioideae Hochr: A Phylogenetic and Conservation Perspective
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Acquisition
2.2. DNA Extraction
2.3. Genomic Library Development
2.4. Genome Sequencing
2.5. Genome Composition and Annotation
2.6. Sequence Evaluation
2.7. Chloroplast Genome Repeat Analysis
2.8. Comparative Genome Examination
2.9. Nucleotide Variability Values
2.10. Phylogenetic Assessment
3. Results
3.1. Grewia Tembensis Chloroplast Genome Features
3.2. Relative Synonymous Codon Usage (RSCU)
3.3. RNA Editing Sites
3.4. Repeat Analysis
3.4.1. Prolonged Repetitions
3.4.2. Simple Sequence Repeats (SSRs)
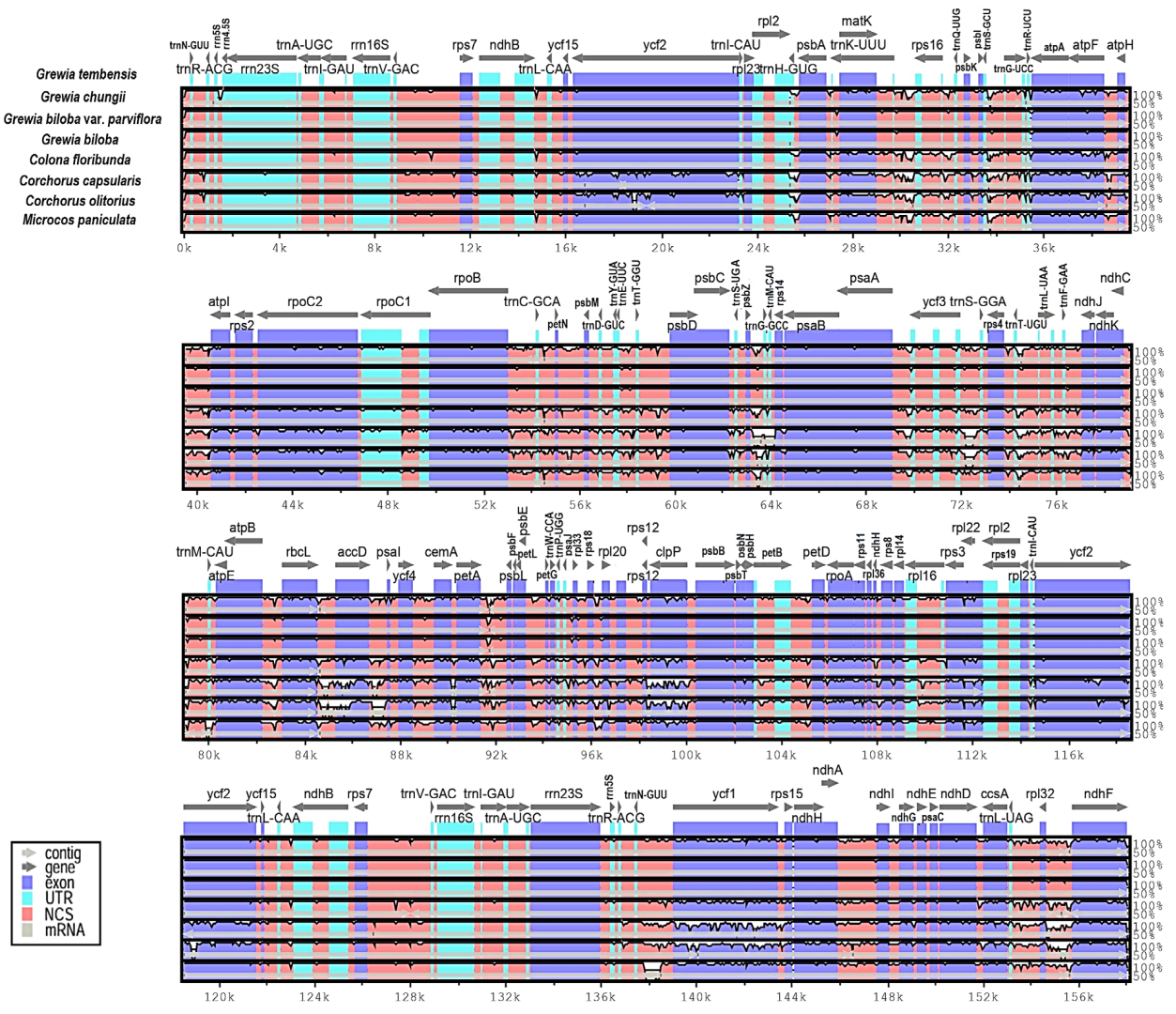
3.5. Comparison of Genomes
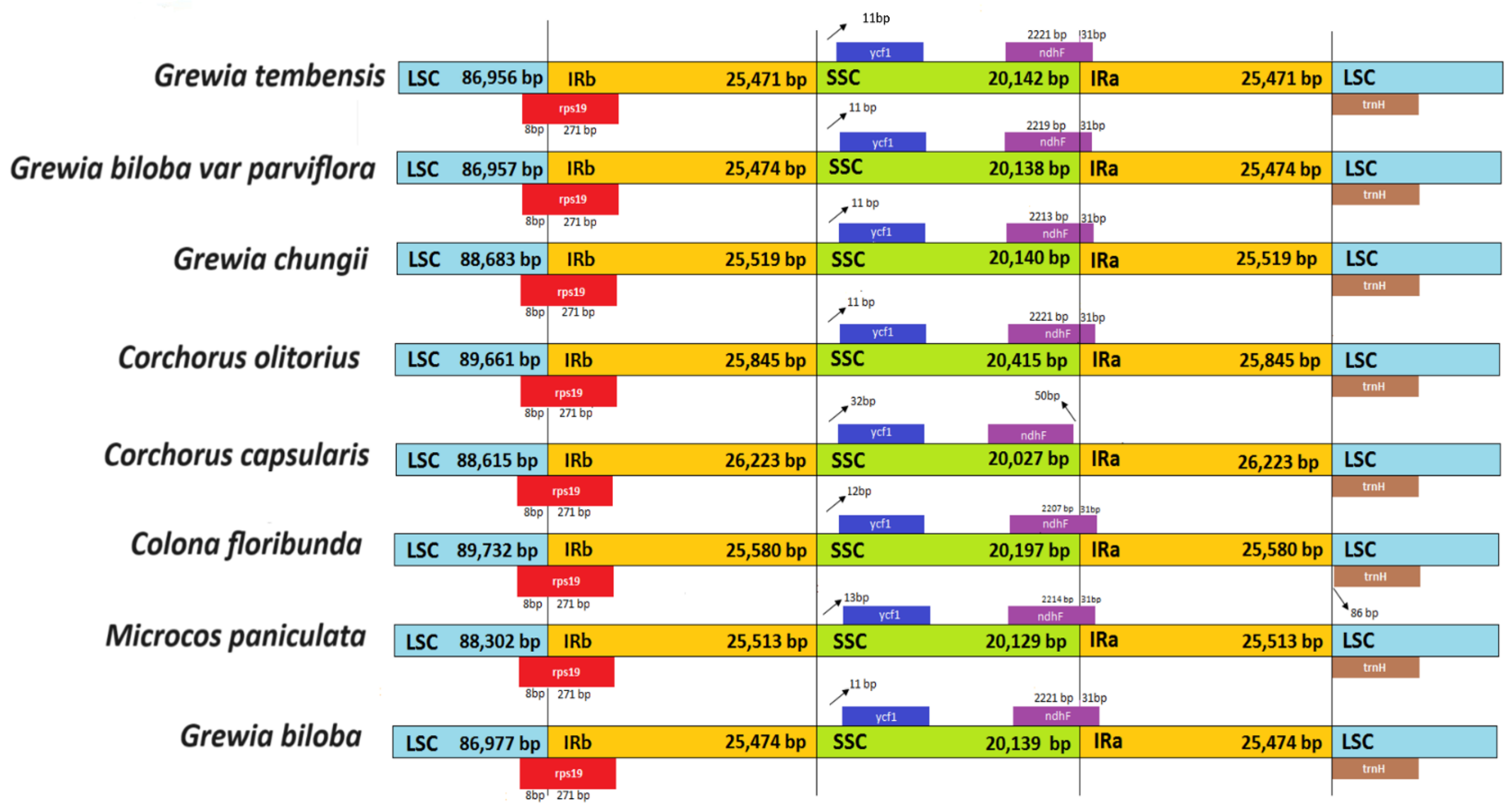
3.6. LSC/SSC and IR Boundaries
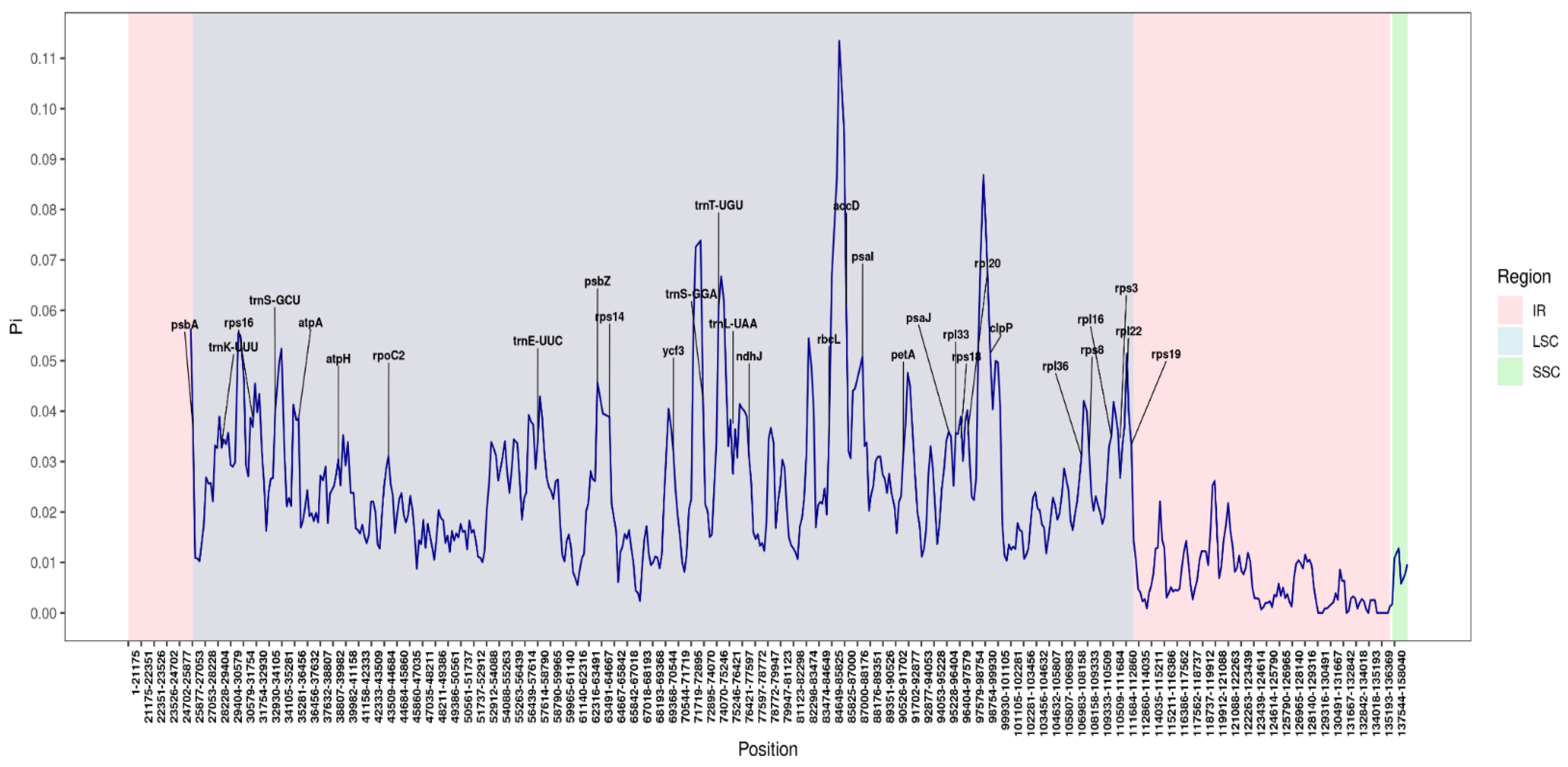
3.7. Nucleotide Variability
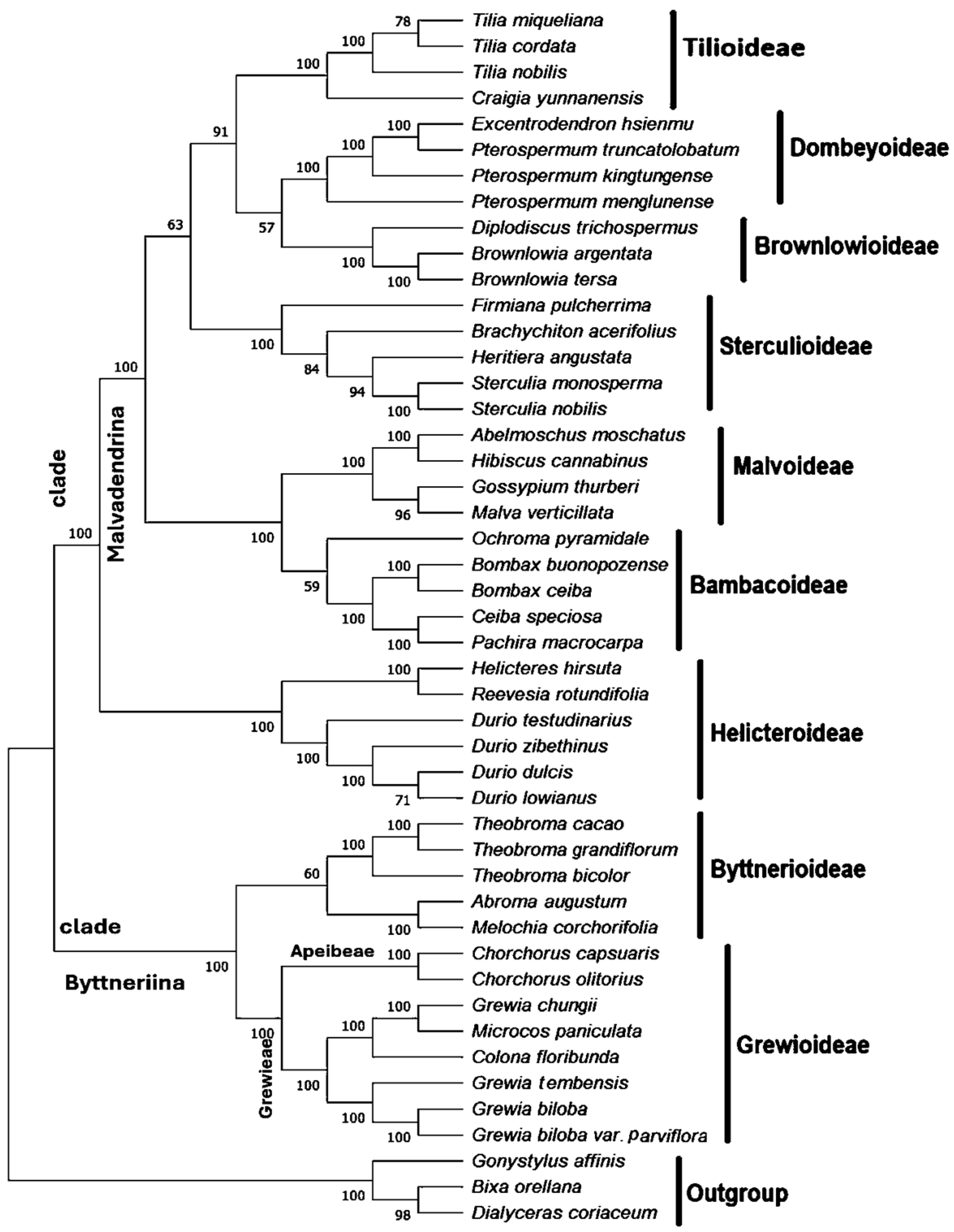
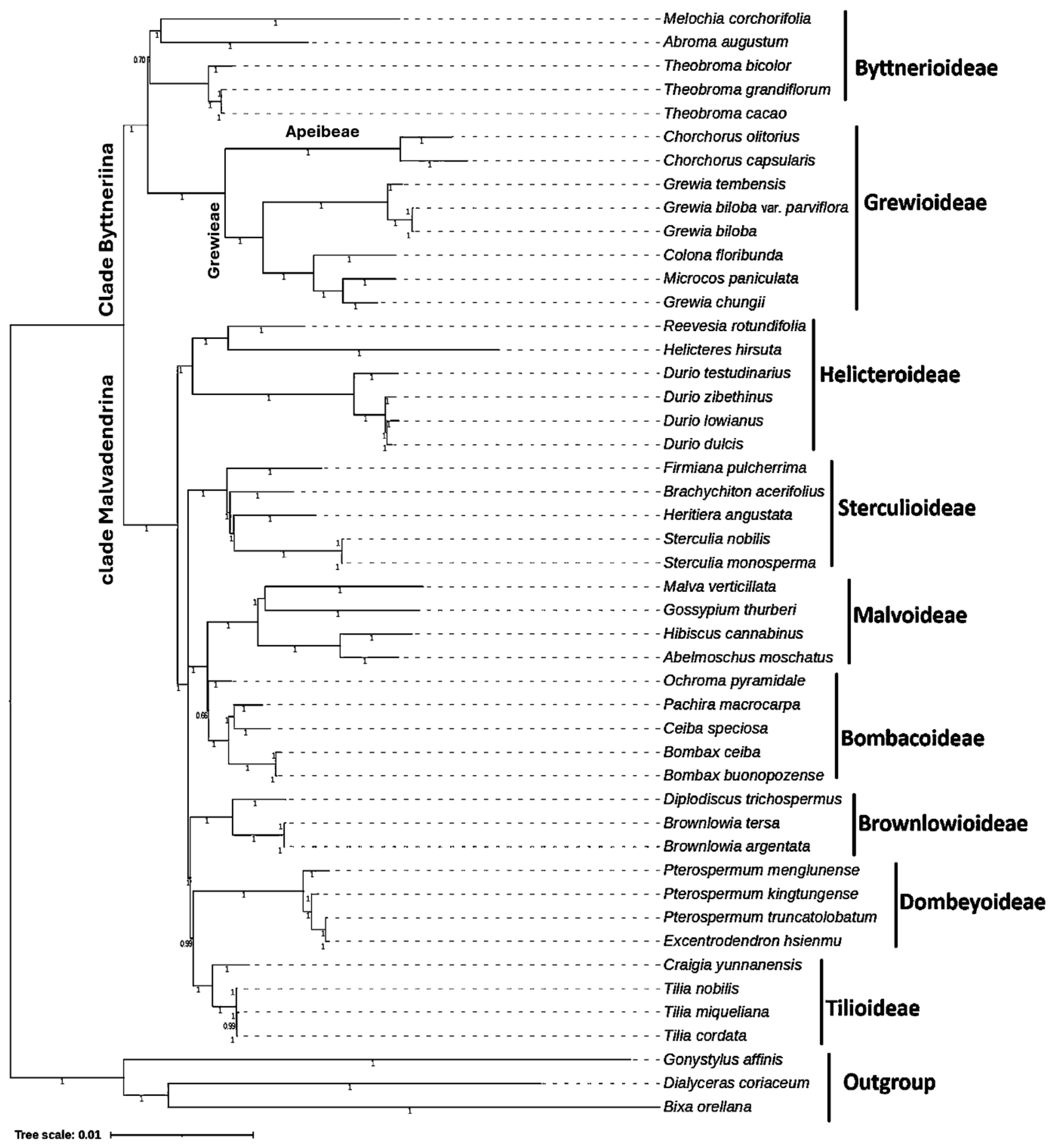
3.8. Phylogenetic Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CP | Chloroplast |
| IR | Inverted repeat |
| SSC | Small single copy |
| LSC | Large single copy |
| SC | Single copy |
| SSR | Simple sequence repeat |
| PI | Nucleotide diversity |
| ML | Maximum likelihood |
| BI | Bayesian inference analysis |
References
- Dorr, L.; Wurdack, K. Re-evaluating monotypic Eleutherostylis from New Guinea and the Moluccas and its inclusion in Grewia (Malvaceae, Grewioideae). PhytoKeys 2024, 237, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Bayer, C.; Fay, M.F.; De Bruijn, A.Y.; Savolainen, V.; Morton, C.M.; Kubitizki, K.; Alverson, W.S.; Chase, M.W. Support for an expanded family concept of Malvaceae within a Recircumscribed order Malvales: A combined analysis of plastid atpB and rbcL DNA sequences. Bot. J. Linn. 1999, 129, 267–303. [Google Scholar] [CrossRef]
- Royal Botanic Gardens, (Kew); Plants of the World Online. Grewia L. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:30123138-2 (accessed on 11 February 2025).
- Al-Hawshabi, O.S.S. Taxonomic Study on Grewia (Grewioideae) of Malvaceae sensu lato in Toor Al-Baha District Lahej Governorate Yemen. J. Fac. Educ. 2023, 17, 93–116. [Google Scholar]
- Bhandari, K.B.; Subedi, S.; Kunwar, R.M.; Bussmann, R.W.; Paniagua-Zambrana, N.Y. Kunwar, R.M., Sher, H., Bussmann, R.W., Eds.; Grewia disperma Rottler ex Spreng. Malvaceae. In Ethnobotany of the Himalayas. Ethnobotany of Mountain Regions; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Nyalo, P. In Vitro Antibacterial and Antioxidant Activities of Ethyl Acetate Extracts of Xerophyta spekei (Baker), Senna singueana (Delile) and Grewia tembensis (Fresen). BSc. Dissertation, Medical Laboratory Sciences, Kenyatta University, Nairobi, Kenya, 2022. [Google Scholar]
- Aljuhani, W.S. Grewia tembensis Fresen and Grewia trichocarpa Hochst. ex A.Rich. (Grewioideae Hochr; Malvaceae Juss.) Micromorphological Study and Comparison via Electron Microscopy. Diversity 2025, 17, 340. [Google Scholar] [CrossRef]
- Jung, J.; Deng, T.; Kim, G.; Kim, C.; Sun, H.; Kim, J.H. Comparative phylogenomic study of East Asian endemic genus, Corchoropsis Siebold & Zucc. (Malvaceae s.l.), based on complete plastome sequences. BMC Genom. 2024, 25, 854. [Google Scholar] [CrossRef]
- Kan, J.; Nie, L.; Wang, M.; Tiwari, R.; Tembrock, L. The Mendelian Pea pan-plastome: Insights into genomic structure, evolutionary history, and genetic diversity of an essential food crop. Genomics Commun. 2024, 1, e004. [Google Scholar] [CrossRef]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef]
- Song, W.; Shi, W.; Wang, H.; Zhang, Z.; Tao, R.; Liu, J.; Wang, S.; Engel, M.S.; Shi, C. Comparative analysis of 12 water lily plastid genomes reveals genomic divergence and evolutionary relationships in early flowering plants. Mar. Life Sci. Tech. 2024, 6, 425–441. [Google Scholar] [CrossRef]
- Yan, X.; Kan, S.; Wang, M.; Li, Y.; Tembrock, L. Genetic diversity and evolution of the plastome in allotetraploid cotton (Gossypium spp.). J. Syst. Evol. 2024, 62, 1118–11136. [Google Scholar] [CrossRef]
- Kan, S.; Liao, X.; Lan, L.; Kong, J.; Wang, J. Cytonuclear interactions and subgenome dominance shape the evolution of organelle-targeted genes in the Brassica triangle of U. Mol. Biol. Evol. 2024, 41, msae043. [Google Scholar] [CrossRef]
- Wang, J.; Kan, J.; Wang, J.; Yan, X.; Li, Y. The pan-plastome of Prunus mume: Insights into Prunus diversity, phylogeny, and domestication history. Front. Plant Sci. 2024, 15, 1404071. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-R.; Zhou, S.-D.; Shi, X.-Q. The complete chloroplast genome of Hibiscus taiwanensis (Malvaceae). Mitochondrial DNA B Resour. 2019, 4, 2533–2534. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, J.; Xu, L.; Duan, B.; Qian, J. The complete chloroplast genome of Malva verticillata (Malvaceae). Mitochondrial DNA B Resour. 2020, 5, 1609–1610. [Google Scholar] [CrossRef]
- Kim, Y.; Oh, Y.J.; Han, K.Y.; Kim, G.H.; Ko, J.; Park, J. The complete chloroplast genome sequence of Hibiscus syriacus L. Mamonde’ (Malvaceae). Mitochondrial DNA B Resour. 2019, 4, 558–559. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, L.; Qi, J.; Zhang, L. Complete Chloroplast Genome Sequence of Hibiscus cannabinus and Comparative Analysis of the Malvaceae Family. Front. Plant Sci. 2020, 11, 227. [Google Scholar] [CrossRef]
- Wu, M.; He, L.; Ma, G.; Zhang, K.; Yang, H.; Yang, X. The complete chloroplast genome of Diplodiscus trichospermus and phylogenetic position of Brownlowioideae within Malvaceae. BMC Genom. 2023, 24, 571. [Google Scholar] [CrossRef]
- Zhong, Y.; Bai, B.; Sun, Y.; Wen, K.; Qiao, Y.; Guo, L.; Deng, H.; Ye, Y.; Feng, L.; Feng, X. Comparative genomics and phylogenetic analysis of six Malvaceae species based on chloroplast genomes. BMC Plant Biol. 2024, 24, 1245. [Google Scholar] [CrossRef]
- Tineo, D.M.; Calderon, S.; Maicelo, J.L.; Oliva, M.; Huamán-Pilco, Á.F.; Ananco, O.; Bustamante, D.E. Characterization and phylogenetic analysis of the complete plastid genome of Theobroma bicolor (Malvaceae) from Peru. Mitochondrial DNA B Resour. 2024, 9, 227–232. [Google Scholar] [CrossRef]
- Huy, T.G.; Thi, N.P.A.; Do, H.D.K.; Khang, D.T. The complete chloroplast genome of Durio zibethinus L. cultivar Ri6 (Helicteroideae, Malvaceae). Mitochondrial DNA B Resour. 2024, 9, 625–630. [Google Scholar]
- Xu, D.; Liu, M.; Ren, H.; Zhang, B.; Liu, Z.; Wang, H. Chloroplast genome and phylogenetic analysis of Grewia biloba. Mitochondrial DNA B Resour. 2021, 6, 6. [Google Scholar]
- Hou, W.J.; Men, W.X.; Bian, C.; Song, Y.Y.; Yang, Y.; Xu, L.; Kang, T.G. The complete chloroplast genome of Grewia biloba var. parviflora (Bunge) Hand.-Mazz. (Malvaceae). Mitochondrial DNA B Resour. 2023, 8, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Moore, M.J.; Wang, H.; Zhu, Z.X.; Wang, H.F. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene 2021, 765, 145103. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.M.; Al-Kahtani, M.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Badry, M.O. Floristic Diversity and Phytogeography of JABAL Fayfa: A Subtropical Dry Zone, South-West Saudi Arabia. Diversity 2020, 12, 345. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom. Bioinform. 2020, 2, lqz011. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Chan, P.; Lowe, T. Kollmar, M., Ed.; Sequences. In Gene Prediction: Methods and Protocols; Springer: New York, NY, USA, 2019. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. Organellar Genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- McInerney, J.O. GCUA: General codon usage analysis. Bioinformatics 1998, 14, 372–373. [Google Scholar] [CrossRef]
- Lenz, H.; Hein, A.; Knoop, V. Plant organelle RNA editing and its specificity factors: Enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinform. 2018, 19, 255. [Google Scholar] [CrossRef]
- Thie, T.; Michalek, W.; Varshney, R. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. LAGAN and multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003, 13, 721–731. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.; Poliakov, A.; Rubin, E.; Frazer, K.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Delbarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Emms, D.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Yang, J.X.; Li, H.K.; Zhao, H.S. Chloroplast Genomes of Two Species of Cypripedium: Expanded Genome Size and Proliferation of AT-Biased Repeat Sequences. Front. Plant Sci. 2021, 12, 609729. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.; Depamphilis, C.; Müller, K.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Duan, H.; Tao, K.; Luo, Y.; Li, Q.; Li, L. Complete chloroplast genome structural characterization of two Phalaenopsis (Orchidaceae) species and comparative analysis with their alliance. BMC Genom. 2023, 24, 359. [Google Scholar] [CrossRef]
- Li, B.; Huang, P.; Guo, W.; Zheng, Y. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Sci. Rep. 2017, 7, 10073. [Google Scholar] [CrossRef]
- Zong, D.; Zhang, Y.; Zou, X.; Li, D.; Duan, A.; He, C. Characterization of the complete chloroplast genomes of five Populus species from the western Sichuan plateau, southwest China: Comparative and phylogenetic analyses. PeerJ 2019, 7, e6386. [Google Scholar] [CrossRef]
- Liu, X.; Chang, E.M.; Liu, J.F.; Huang, Y.N.; Wang, Y.; Yao, N.; Jiang, Z.P. Complete chloroplast genome sequence and phylogenetic analysis of Quercus bawanglingensis Huang, Li et Xing, a vulnerable oak tree in China. Forests 2019, 10, 587. [Google Scholar] [CrossRef]
- Fan, R.; Ma, W.; Liu, S.; Huang, Q. Integrated analysis of three newly sequenced fern chloroplast genomes: Genome structure and comparative analysis. Ecol. Evol. 2021, 11, 4550–4563. [Google Scholar] [CrossRef]
- Aljuhania, W.S.; Al Thagafi, N.T.S. Micromorphological characterization and electron microscopic study of parasitic species of the family Loranthaceae Juss. Braz. J. Biol. 2023, 83, e278994. [Google Scholar] [CrossRef]
- Aljuhani, W.S.; Aljohani, A.Y. Complete chloroplast genome of the medicinal plant Cleome paradoxa R.Br. Ex DC: Comparative analysis, and phylogenetic relationships among the members of Cleomaceae. Gene 2022, 845, 146851. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, G.; Zhang, C.; Zhang, X.; Jiang, X. Comparative analysis of chloroplast genomes of endangered heterostylous species Primula wilsonii and its closely related species. Ecol. Evol. 2023, 13, e9730. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Deng, P.; Zhang, S.; Zhao, Z. Genetic Diversity and Population Genetic Structure of Ancient Platycladus orientalis L. (Cupressaceae) in the Middle Reaches of the Yellow River by Chloroplast Microsatellite Markers. Forests 2021, 12, 592. [Google Scholar] [CrossRef]
- Moosavi, S.J.; Mueller, M.; Gailing, O. Development of New Chloroplast Microsatellites for Pinus gerardiana and their Application in Genetic Diversity Analyses. Ecol. Evol. 2025, 15, e71185. [Google Scholar] [CrossRef]
- Guo, Q.; Xue, X.; Wang, D.; Zhang, L.; Liu, W.; Wang, E.; Cui, X.; Hou, X. Genetic diversity and population genetic structure of Paeonia suffruticosa by chloroplast DNA simple sequence repeats (cpSSRs). Hortic. Plant J. 2025, 11, 367–376. [Google Scholar] [CrossRef]
- Li, J.; Quan, C.; Wei, R.; Wei, F.; Ma, Q.; Huang, Y.; Xu, M.; Tang, D. Genome-wide identification and development of SSR molecular markers for genetic diversity studies in Ilex asprella. Front. Plant Sci. 2025, 16, 1582154. [Google Scholar] [CrossRef]
- Yang, J.B.; Yang, S.X.; Li, H.T.; Yang, J.; Li, D.Z. Comparative chloroplast genomes of Camellia species. PLoS ONE 2013, 8, e73053. [Google Scholar] [CrossRef]
- Ren, T.; Yang, Y.C.; Zhou, T.; Liu, Z.L. Comparative plastid genomes of Primula species: Sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 2018, 19, 1050. [Google Scholar] [CrossRef]
- Lin, N.; Zhangab, X.; Dengc, T.; Zhang, J.; Meng, A.; Wang, H.; Sun, H.; Sun, Y. Plastome sequencing of Myripnois dioica and comparison within Asteraceae. Plant Divers. 2019, 41, 315–322. [Google Scholar] [CrossRef]
- Ni, Z.; Ye, Y.; Bai, T.; Xu, M.; Xu, L.-A. Complete Chloroplast Genome of Pinus massoniana (Pinaceae): Gene Rearrangements, Loss of ndh Genes, and Short Inverted Repeats Contraction, Expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef]
- Mehmood, F.; Li, M.; Bertolli, A.; Prosser, F.; Varotto, C. Comparative Plastomics of Plantains (Plantago, Plantaginaceae) as a Tool for the Development of Species-Specific DNA Barcodes. Plants 2024, 13, 2691. [Google Scholar] [CrossRef]
- Cauz-Santos, L.A.; da Costa, Z.P.; Sader, M.A.; den Berg, C.V.; Vieira, M.L.C. Chloroplast genomic insights into adaptive evolution and rapid radiation in the genus Passiflora (Passifloraceae). BMC Plant Biol. 2025, 25, 192. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Li, H.T.; Luo, Y.; Gan, L.; Ma, P.F.; Gao, L.M.; Yang, J.B.; Cai, J.; Gitzendanner, M.A.; Fritsch, P.W.; Zhang, T.; et al. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 2021, 19, 232. [Google Scholar] [CrossRef]
- Cvetković, T.; Areces-Berazain, F.; Hinsinger, D.D.; Thomas, D.C.; Wieringa, J.J.; Ganesan, S.K. Phylogenomics resolves deep subfamilial relationships in Malvaceae s. l. G3 Genes Genomes Genet. 2021, 11, jkab136. [Google Scholar] [CrossRef]

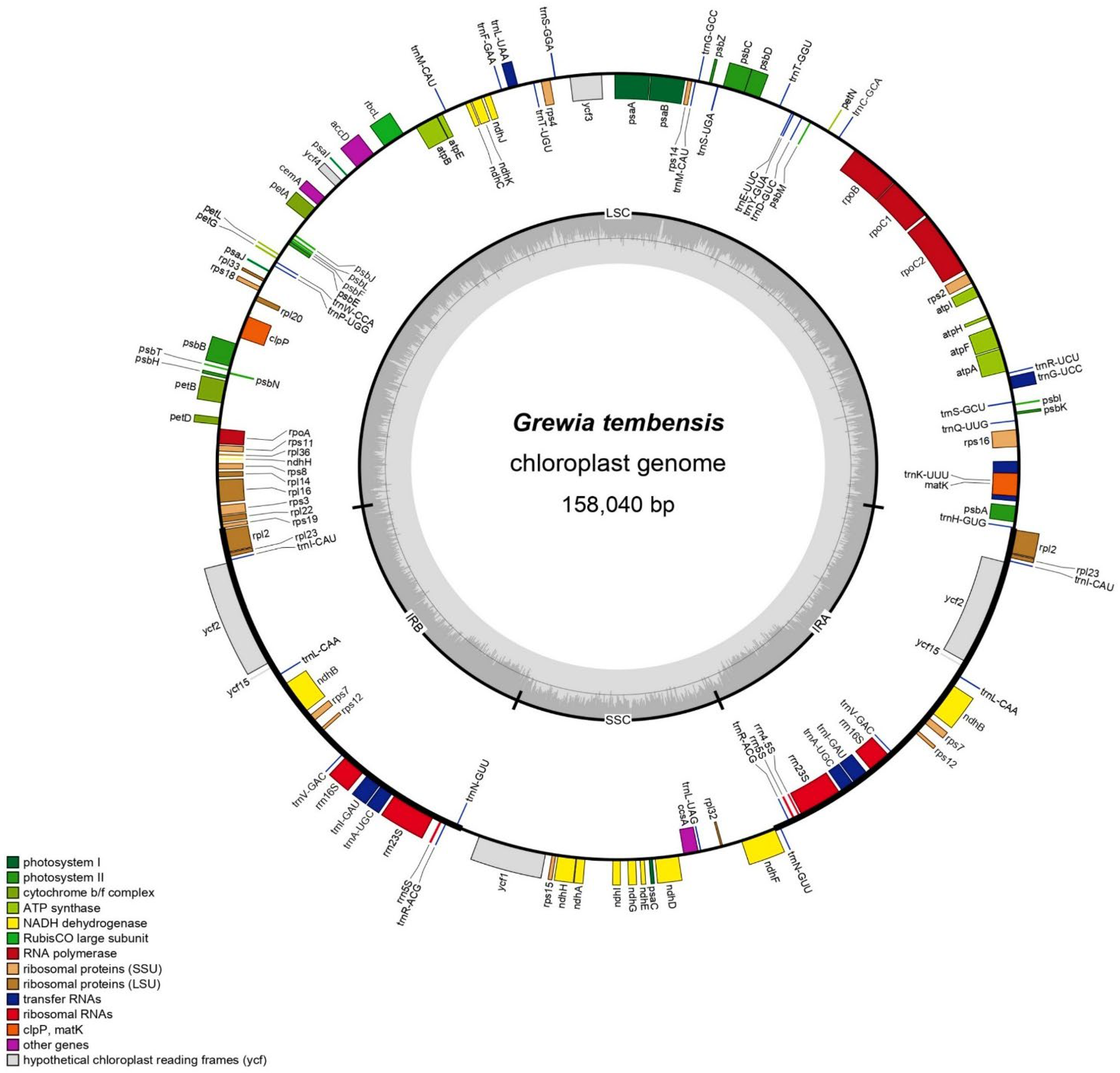
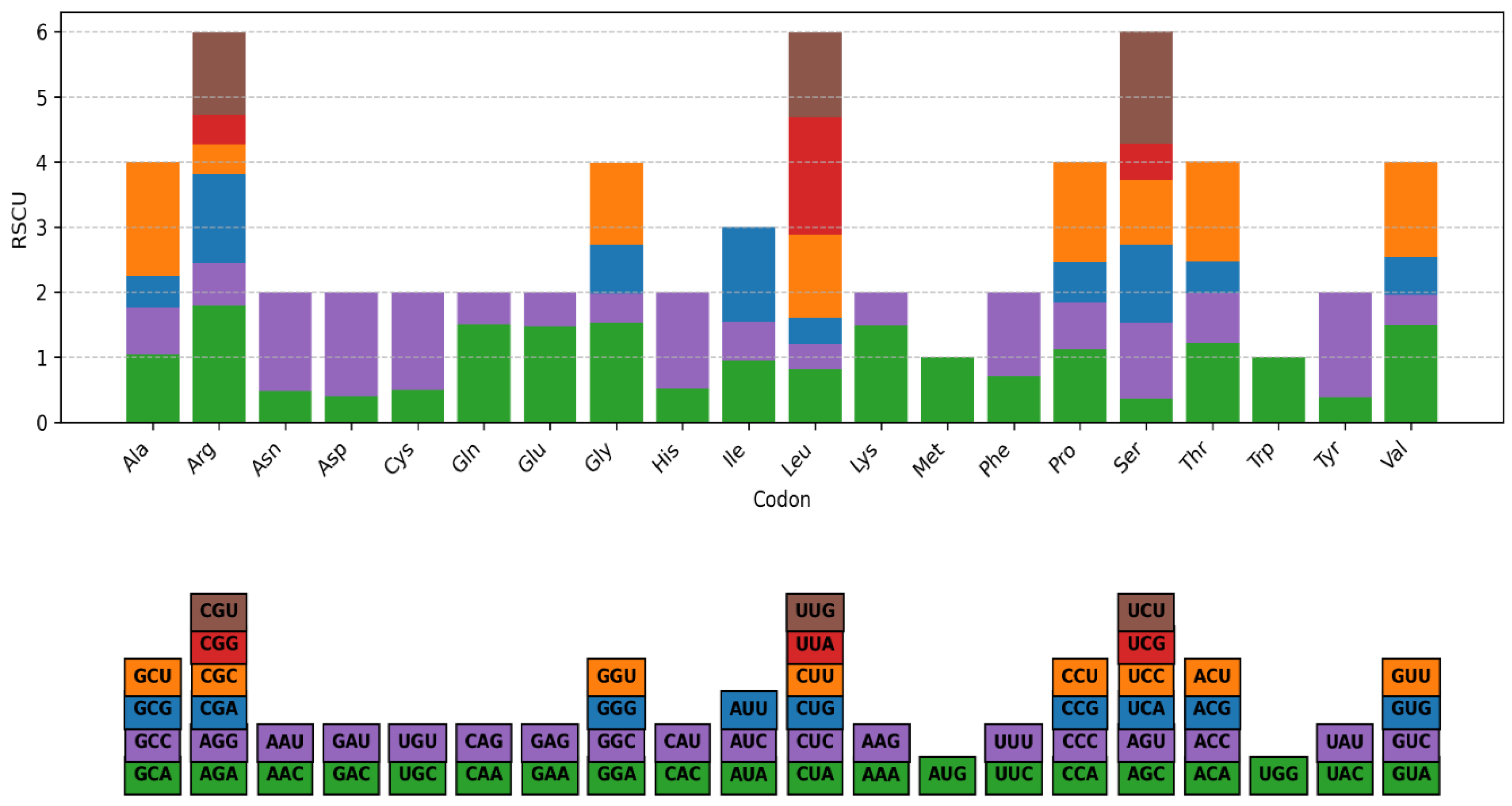
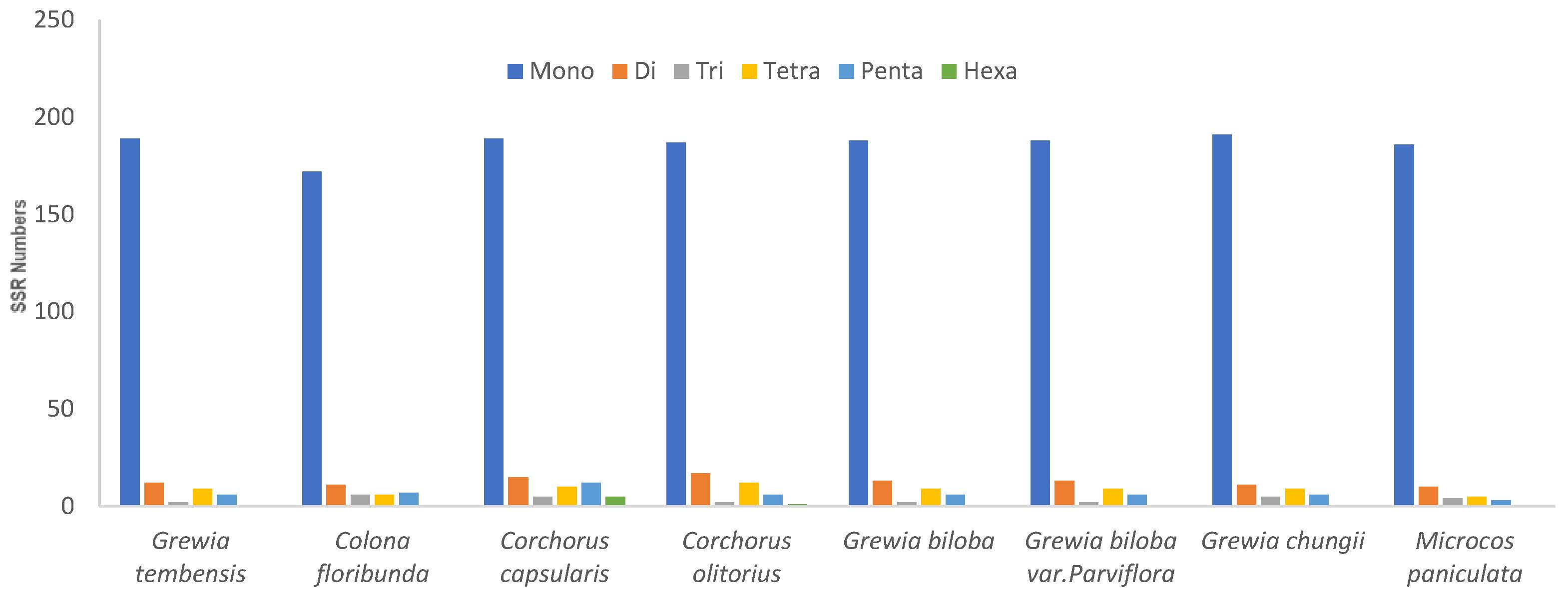
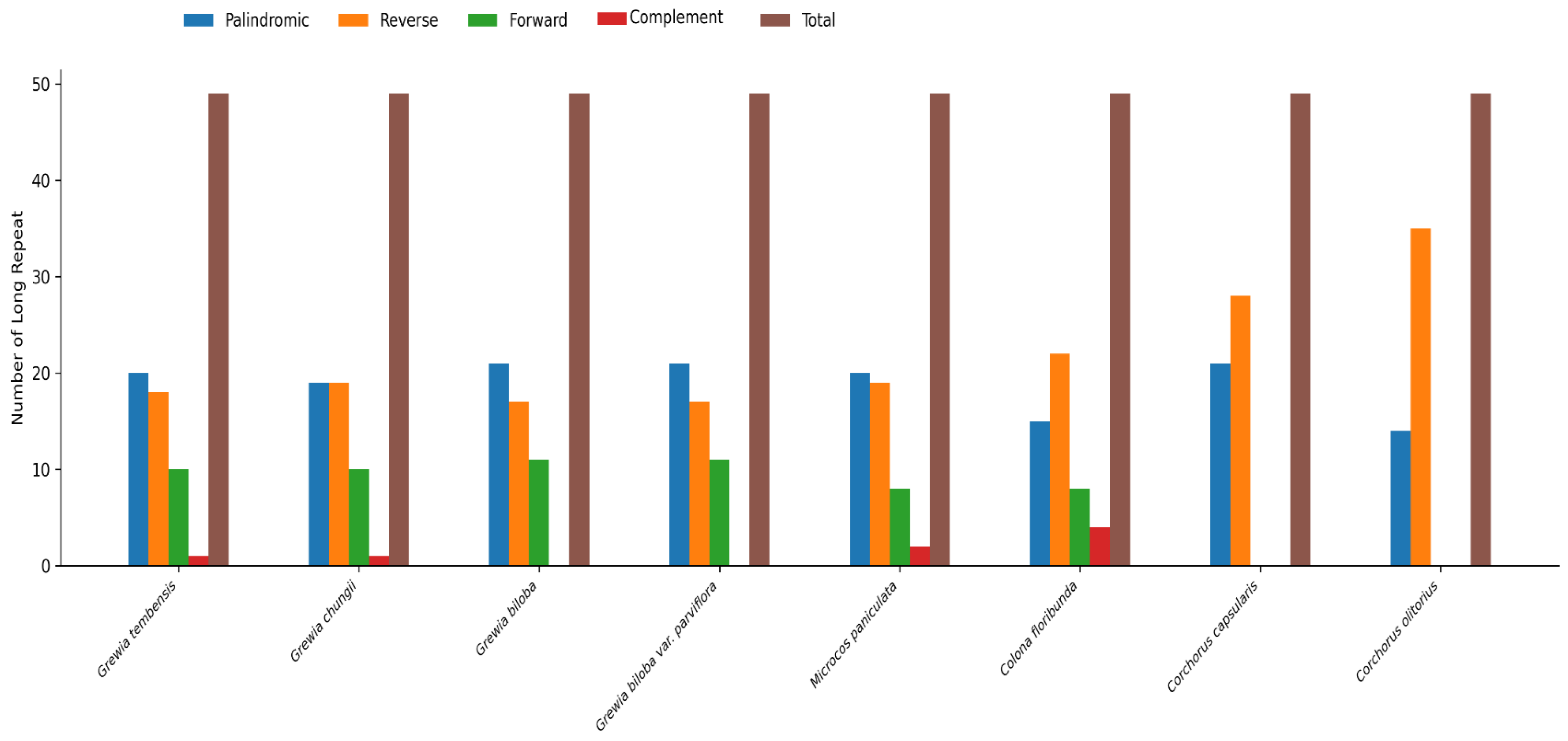
| Characteristics | Number |
|---|---|
| Genome size (bp) | 158,040 |
| IRA (bp) | 25,471 |
| IRB (bp) | 25,471 |
| LSC (bp) | 86,956 |
| SSC (bp) | 20,142 |
| Total no of genes | 130 |
| Total no of unique genes | 110 |
| rRNA | 3 |
| tRNA | 28 |
| Protein-coding genes | 79 |
| A% | 30.89 |
| T (U)% | 31.76 |
| G% | 18.51 |
| C% | 18.85 |
| GC% | 37.36 |
| Gene | Regions | Location Start | Location End | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|---|
| atpF | LSC | 37,103 | 38,520 | 145 | 863 | 410 | ||
| clpP | LSC | 98,503 | 99,987 | 71 | 894 | 520 | ||
| ndhB | IR | 12,388 | 14,613 | 775 | 693 | 758 | ||
| 123,142 | 125,367 | 775 | 693 | 758 | ||||
| rpl16 | LSC | 109,214 | 110,724 | 9 | 1103 | 399 | ||
| rpl2 | IR | 23,800 | 25,318 | 399 | 589 | 531 | ||
| 112,437 | 113,955 | 399 | 589 | 531 | ||||
| rpoC1 | LSC | 46,878 | 497,18 | 432 | 783 | 1626 | ||
| rps16 | LSC | 30,619 | 317,56 | 40 | 871 | 227 | ||
| trnA-UGC | IR | 4827 | 5700 | 37 | 801 | 36 | ||
| 132,055 | 132,928 | 37 | 801 | 36 | ||||
| trnK-UUU | LSC | 27,121 | 29,720 | 37 | 2528 | 35 | ||
| trnL-UAA | LSC | 75,203 | 75,845 | 35 | 558 | 50 | ||
| ycf3 | LSC | 69,898 | 71,931 | 124 | 744 | 230 | 783 | 153 |
| petB | LSC | 10,2866 | 104,325 | 6 | 812 | 642 | ||
| trnG-UCC | LSC | 34,345 | 35,148 | 31 | 713 | 62 | ||
| trnI-GAU | IR | 130,963 | 131,991 | 32 | 959 | 40 | ||
| 5764 | 6792 | 32 | 959 | 40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AL-Juhani, W.S. Comparative Analysis of the Chloroplast Genomes of Grewia tembensis Fresen and Closely Related Species of Grewioideae Hochr: A Phylogenetic and Conservation Perspective. Genes 2025, 16, 1124. https://doi.org/10.3390/genes16101124
AL-Juhani WS. Comparative Analysis of the Chloroplast Genomes of Grewia tembensis Fresen and Closely Related Species of Grewioideae Hochr: A Phylogenetic and Conservation Perspective. Genes. 2025; 16(10):1124. https://doi.org/10.3390/genes16101124
Chicago/Turabian StyleAL-Juhani, Widad S. 2025. "Comparative Analysis of the Chloroplast Genomes of Grewia tembensis Fresen and Closely Related Species of Grewioideae Hochr: A Phylogenetic and Conservation Perspective" Genes 16, no. 10: 1124. https://doi.org/10.3390/genes16101124
APA StyleAL-Juhani, W. S. (2025). Comparative Analysis of the Chloroplast Genomes of Grewia tembensis Fresen and Closely Related Species of Grewioideae Hochr: A Phylogenetic and Conservation Perspective. Genes, 16(10), 1124. https://doi.org/10.3390/genes16101124






