Somatic TEK Mutation Identified in a Patient with Calvarial Venous Malformations
Abstract
1. Introduction
2. Materials and Methods
2.1. Exome Sequencing and Data Analysis
2.2. Single-Cell RNA-Seq and Protein–Protein Interaction Analysis
3. Results
3.1. Clinical Presentation
3.2. Integrative Genomic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Adj. P | Adjusted P-value |
| bAVM | Brain arteriovenous malformation |
| CADD | Combined annotation-dependent depletion |
| gnomAD | The genome aggregation database |
| INDELs | Insertion–deletions |
| MAF | Minor allele frequency |
| MCC | Maximum clique centrality |
| Mis-Z | Z-scores of the observed missense counts compared to expected |
| MRI | Magnetic resonance imaging |
| pLI | Probability of loss of function intolerance |
| scRNA-seq | Single-cell RNA sequencing |
| SNVs | Single-nucleotide variants |
| SP | Sinus pericrania |
| TEK | TEK receptor tyrosine kinase |
| VAF | Variant allele frequency |
| VM | Venous malformation |
| WES | Whole-exome sequencing |
References
- Legiehn, G.M.; Heran, M.K.S. Venous malformations: Classification, development, diagnosis, and interventional radiologic management. Radiol. Clin. N. Am. 2008, 46, 545–597. [Google Scholar] [CrossRef]
- Hassanein, A.H.; Mulliken, J.B.; Fishman, S.J.; Alomari, A.I.; Zurakowski, D.; Greene, A.K. Venous malformation: Risk of progression during childhood and adolescence. Ann. Plast. Surg. 2012, 68, 198–201. [Google Scholar] [CrossRef]
- Pavanello, M.; Melloni, I.; Antichi, E.; Severino, M.; Ravegnani, M.; Piatelli, G.; Cama, A.; Rossi, A.; Gandolfo, C. Sinus pericranii: Diagnosis and management in 21 pediatric patients. J. Neurosurg. Pediatr. 2015, 15, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Strauss, S.B.; Steinklein, J.M.; Phillips, C.D.; Shatzkes, D.R. Intraosseous venous malformations of the head and neck. Am. J. Neuroradiol. 2022, 43, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Brouillard, P.; Vikkula, M. Genetic causes of vascular malformations. Hum. Mol. Genet. 2007, 16, R140–R149. [Google Scholar] [CrossRef]
- Du, Z.; Zheng, J.; Zhang, Z.; Wang, Y. Review of the endothelial pathogenic mechanism of TIE2-related venous malformation. J. Vasc. Surg. Venous Lymphat. Disord. 2017, 5, 740–748. [Google Scholar] [CrossRef]
- Wouters, V.; Limaye, N.; Uebelhoer, M.; Irrthum, A.; Boon, L.M.; Mulliken, J.B.; Enjolras, O.; Baselga, E.; Berg, J.; Dompmartin, A.; et al. Hereditary cutaneomucosal venous malformations are caused by TIE2 mutations with widely variable hyper-phosphorylating effects. Eur. J. Hum. Genet. 2010, 18, 414–420. [Google Scholar] [CrossRef]
- Limaye, N.; Wouters, V.; Uebelhoer, M.; Tuominen, M.; Wirkkala, R.; Mulliken, J.B.; Eklund, L.; Boon, L.M.; Vikkula, M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2009, 41, 118–124. [Google Scholar] [CrossRef]
- Grima, N.; Henden, L.; Watson, O.; Blair, I.P.; Williams, K.L. Simultaneous Isolation of High-Quality RNA and DNA From Postmortem Human Central Nervous System Tissues for Omics Studies. J. Neuropathol. Exp. Neurol. 2022, 81, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.C.; Dong, W.; Kundishora, A.J.; Panchagnula, S.; Moreno-De-Luca, A.; Furey, C.G.; Allocco, A.A.; Walker, R.L.; Nelson-Williams, C.; Smith, H.; et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat. Med. 2020, 26, 1754–1765. [Google Scholar] [CrossRef]
- Pei, S.; Liu, T.; Ren, X.; Li, W.; Chen, C.; Xie, Z. Benchmarking variant callers in next-generation and third-generation sequencing analysis. Brief. Bioinform. 2021, 22, bbaa148. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Wu, Y.; Choi, J.; Allington, G.; Zhao, S.; Khanfar, M.; Yang, K.; Fu, P.-Y.; Wrubel, M.; Yu, X.; et al. Computational genomics in the era of precision medicine: Applications to variant analysis and gene therapy. J. Pers. Med. 2022, 12, 175. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Wälchli, T.; Ghobrial, M.; Schwab, M.; Takada, S.; Zhong, H.; Suntharalingham, S.; Vetiska, S.; Gonzalez, D.R.; Wu, R.; Rehrauer, H.; et al. Single-cell atlas of the human brain vasculature across development, adulthood and disease. Nature 2024, 632, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Speir, M.L.; Bhaduri, A.; Markov, N.S.; Moreno, P.; Nowakowski, T.J.; Papatheodorou, I.; A Pollen, A.; Raney, B.J.; Seninge, L.; Kent, W.J.; et al. UCSC Cell Browser: Visualize your single-cell data. Bioinformatics 2021, 37, 4578–4580. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Vrinceanu, D.; Dumitru, M.; Marinescu, A.; Dorobat, B.; Palade, O.D.; Manole, F.; Muresian, H.; Popa-Cherecheanu, M.; Ciornei, C.M. New Insights into Cervicofacial Vascular Anomalies. J. Clin. Med. 2024, 13, 3515. [Google Scholar] [CrossRef]
- Vollherbst, D.F.; Bendszus, M.; Möhlenbruch, M.A. Vascular malformations of the brain and its coverings. J. Neuroendovasc. Ther. 2020, 14, 285–294. [Google Scholar] [CrossRef]
- Mahajan, P.; Bergstrom, K.L.; Phung, T.L.; Metry, D.W. The genetics of vascular birthmarks. Clin. Dermatol. 2022, 40, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Daido, W.; Nakashima, T.; Masuda, T.; Sakamoto, S.; Yamaguchi, K.; Horimasu, Y.; Miyamoto, S.; Iwamoto, H.; Fujitaka, K.; Hamada, H.; et al. Nestin and Notch3 collaboratively regulate angiogenesis, collagen production, and endothelial-mesenchymal transition in lung endothelial cells. Cell Commun. Signal. 2023, 21, 247. [Google Scholar] [CrossRef]
- Thompson, E.J.; Dorward, E.L.; Jurrius, K.; Nataren, N.; Tondl, M.; Min, K.K.M.; Cockshell, M.P.; Fouladzadeh, A.; Toubia, J.; DeNichilo, M.; et al. Inhibitor of DNA binding-1 is a key regulator of cancer cell vasculogenic mimicry. Mol. Oncol. 2025, 19, 2537–2556. [Google Scholar] [CrossRef]
- Dong, Z.-X.; Chan, S.-H.; Chen, S.-N.; Li, M.; Zhang, X.-D.; Liu, X.-Q. TJP1 promotes vascular mimicry in bladder cancer by facilitating VEGFA expression and transcriptional activity through TWIST1. Transl. Oncol. 2023, 32, 101666. [Google Scholar] [CrossRef]
- Bell, L.M.; Holm, A.; Matysiak, U.; Driever, W.; Rößler, J.; Schanze, D.; Wieland, I.; Niemeyer, C.M.; Zenker, M.; Kapp, F.G. Functional assessment of two variants of unknown significance in TEK by endothelium-specific expression in zebrafish embryos. Hum. Mol. Genet. 2021, 31, 10–17. [Google Scholar] [CrossRef]
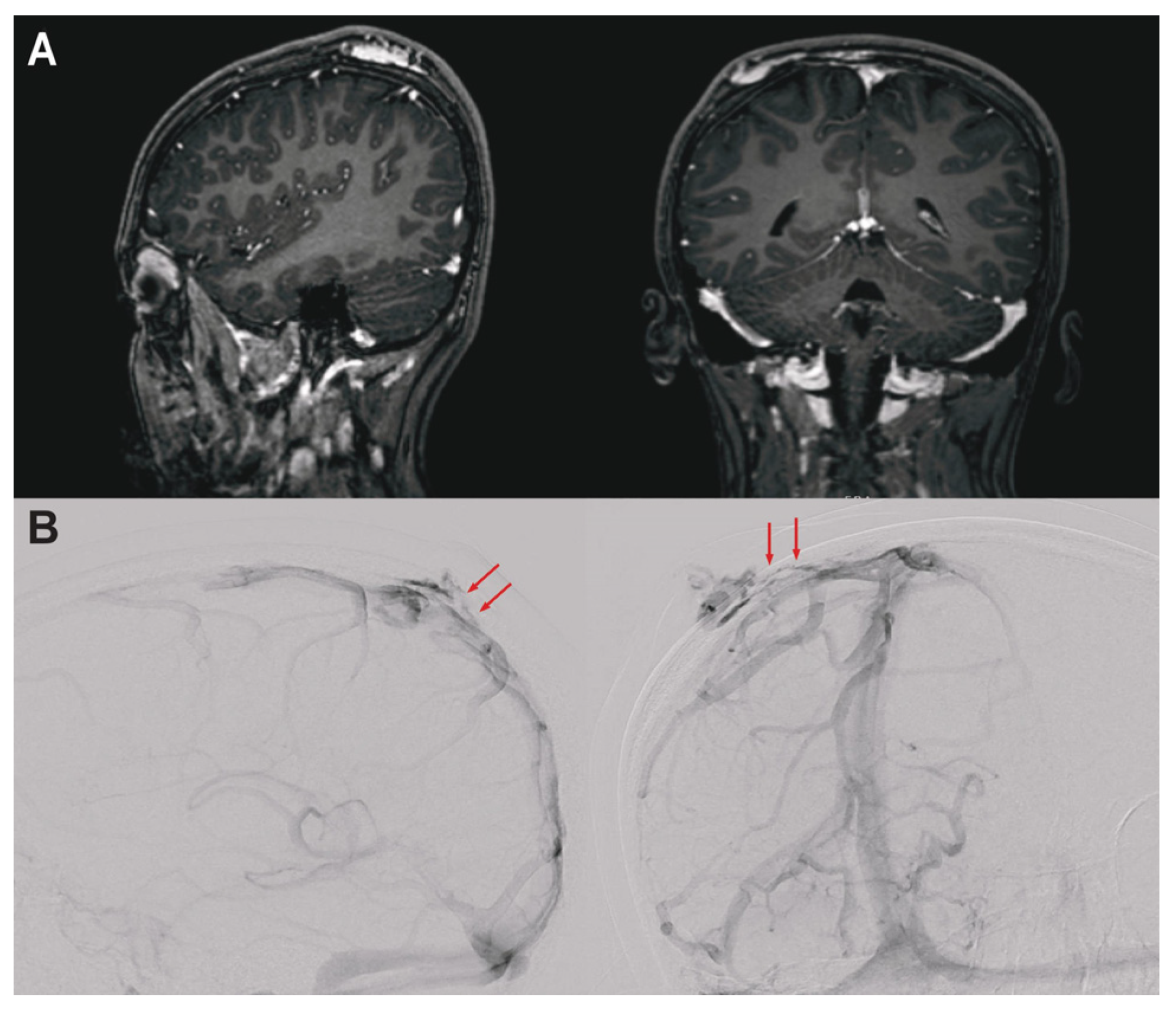
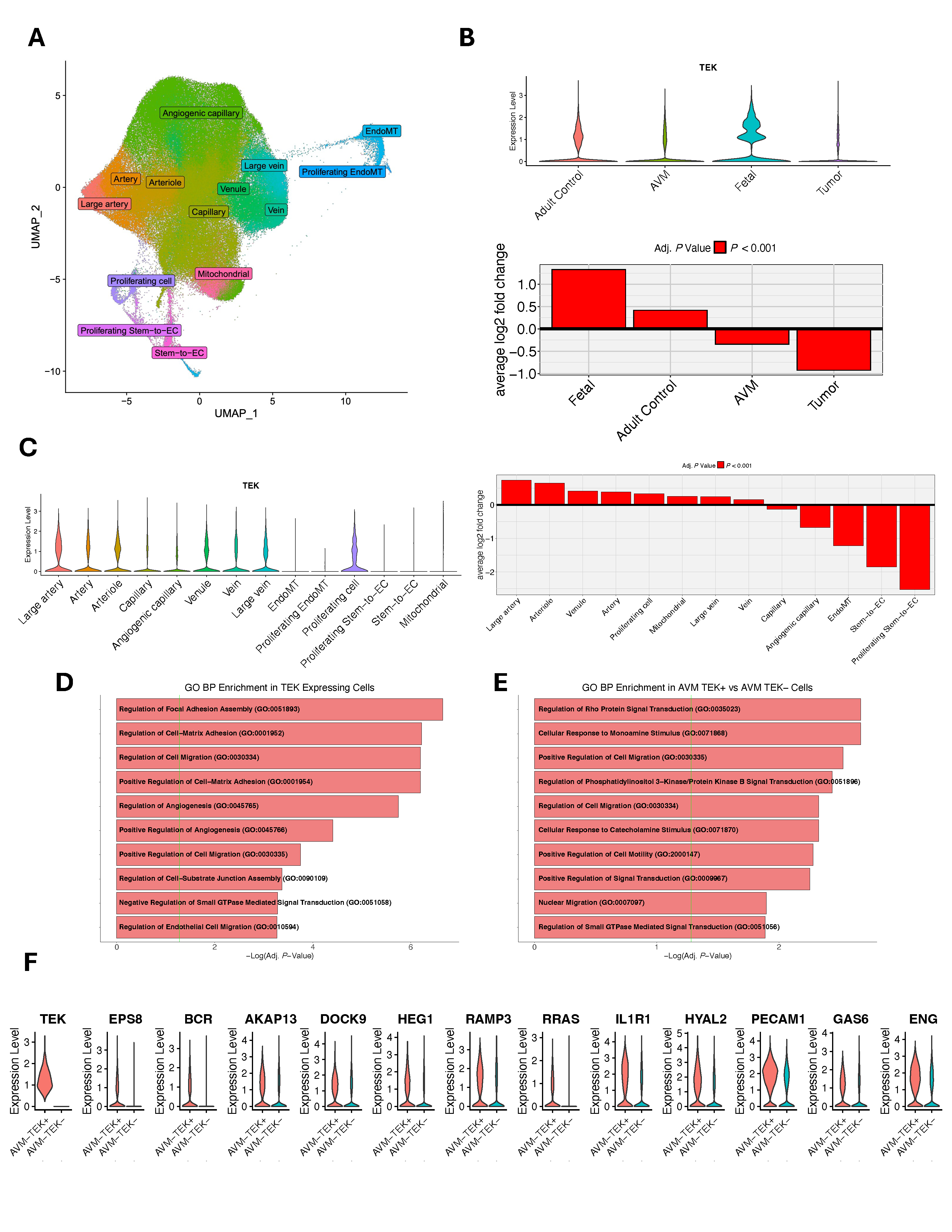
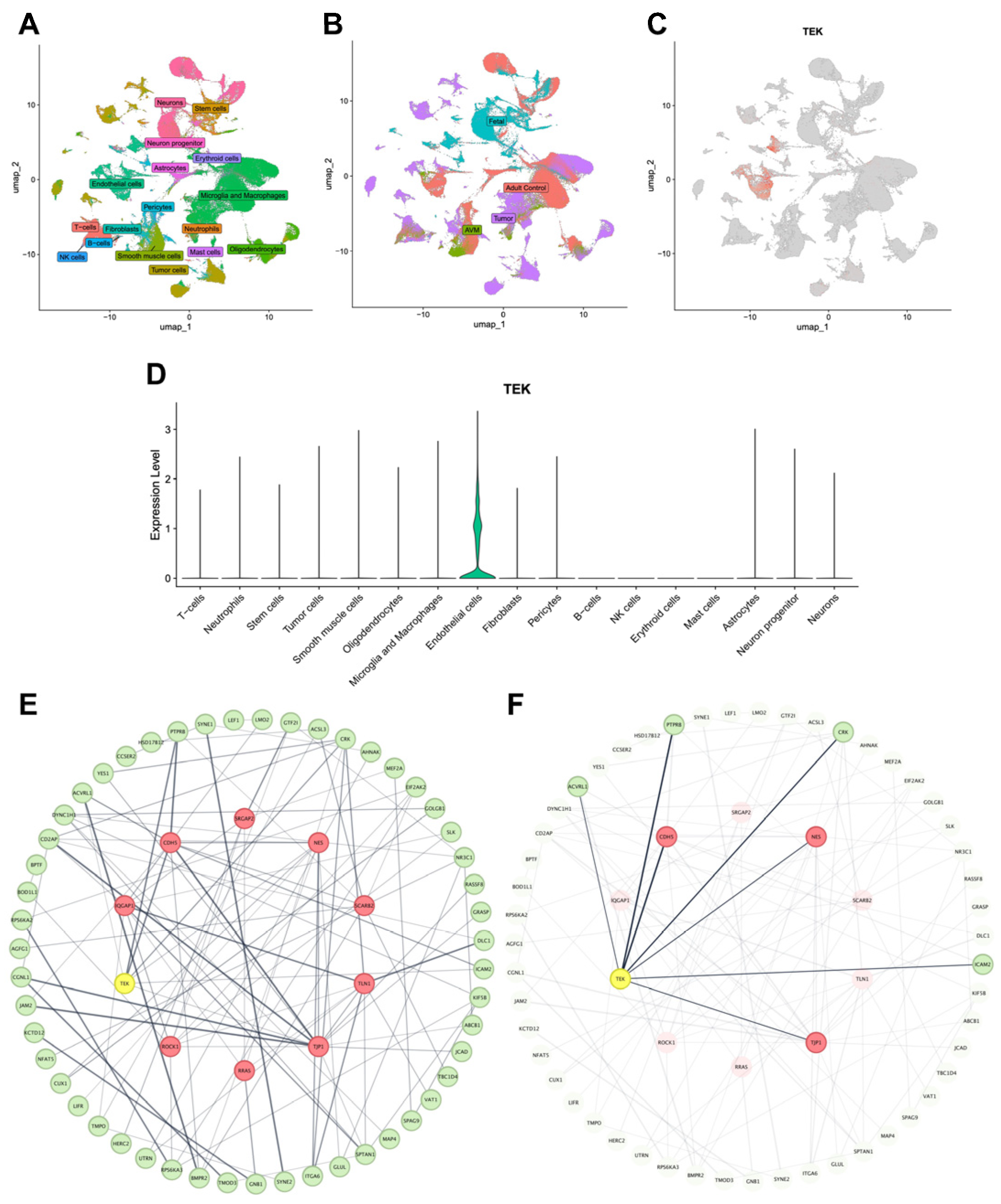
| Gene | Variant (GRCh38) 1 | Amino Acid Change | VAF | MAF (gnomAD) | CADD | Mis-Z | pLI |
|---|---|---|---|---|---|---|---|
| EIF3E | 8-108217434-C-T | p.C250Y | 0.0138 | 0 | 31.0 | 3.10 | 0 |
| NEMF | 14-49785100-T-C | p.K1022R | 0.0231 | 6.2 × 10−7 | 23.8 | 1.29 | 0 |
| RALY | 20-34077060-G-A | p.G231S | 0.0115 | 0.0065 | 0.035 | 1.45 | 0.03 |
| TEK 2 | 9-27212760-C-T | p.L914F | 0.0339 | 0 | 24.0 | 2.89 | 1 |
| TTC28 | 22-28679648-G-T | p.P26T | 0.0600 | 0.033 | 14.3 | 3.47 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, B.; Dennis, E.; Mehta, N.H.; Davalan, W.; Fortes, C.; Swamy, A.; Muñoz, W.; Jaimes, C.; Hale, A.T.; Kahle, K.T. Somatic TEK Mutation Identified in a Patient with Calvarial Venous Malformations. Genes 2025, 16, 1123. https://doi.org/10.3390/genes16101123
Fan B, Dennis E, Mehta NH, Davalan W, Fortes C, Swamy A, Muñoz W, Jaimes C, Hale AT, Kahle KT. Somatic TEK Mutation Identified in a Patient with Calvarial Venous Malformations. Genes. 2025; 16(10):1123. https://doi.org/10.3390/genes16101123
Chicago/Turabian StyleFan, Baojian, Evan Dennis, Neel H. Mehta, William Davalan, Carla Fortes, Aditi Swamy, William Muñoz, Camilo Jaimes, Andrew T. Hale, and Kristopher T. Kahle. 2025. "Somatic TEK Mutation Identified in a Patient with Calvarial Venous Malformations" Genes 16, no. 10: 1123. https://doi.org/10.3390/genes16101123
APA StyleFan, B., Dennis, E., Mehta, N. H., Davalan, W., Fortes, C., Swamy, A., Muñoz, W., Jaimes, C., Hale, A. T., & Kahle, K. T. (2025). Somatic TEK Mutation Identified in a Patient with Calvarial Venous Malformations. Genes, 16(10), 1123. https://doi.org/10.3390/genes16101123






