Alcohol Preference Impacts Multi-Organ Transcriptome in MetALD
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study
2.2. Glucose Tolerance Testing
2.3. Plasma Analyte Analyses
2.4. Histological Analysis
2.5. Liver Tissue Analyte Analyses
2.6. 16S Sample Preparation and Sequencing Analysis
2.7. Whole-Organ mRNA Sequencing Analysis
2.8. Statistical Analyses
3. Results
3.1. Alcohol-Preferring and Non-Preferring Mice Have Similar Liver Lipid Changes
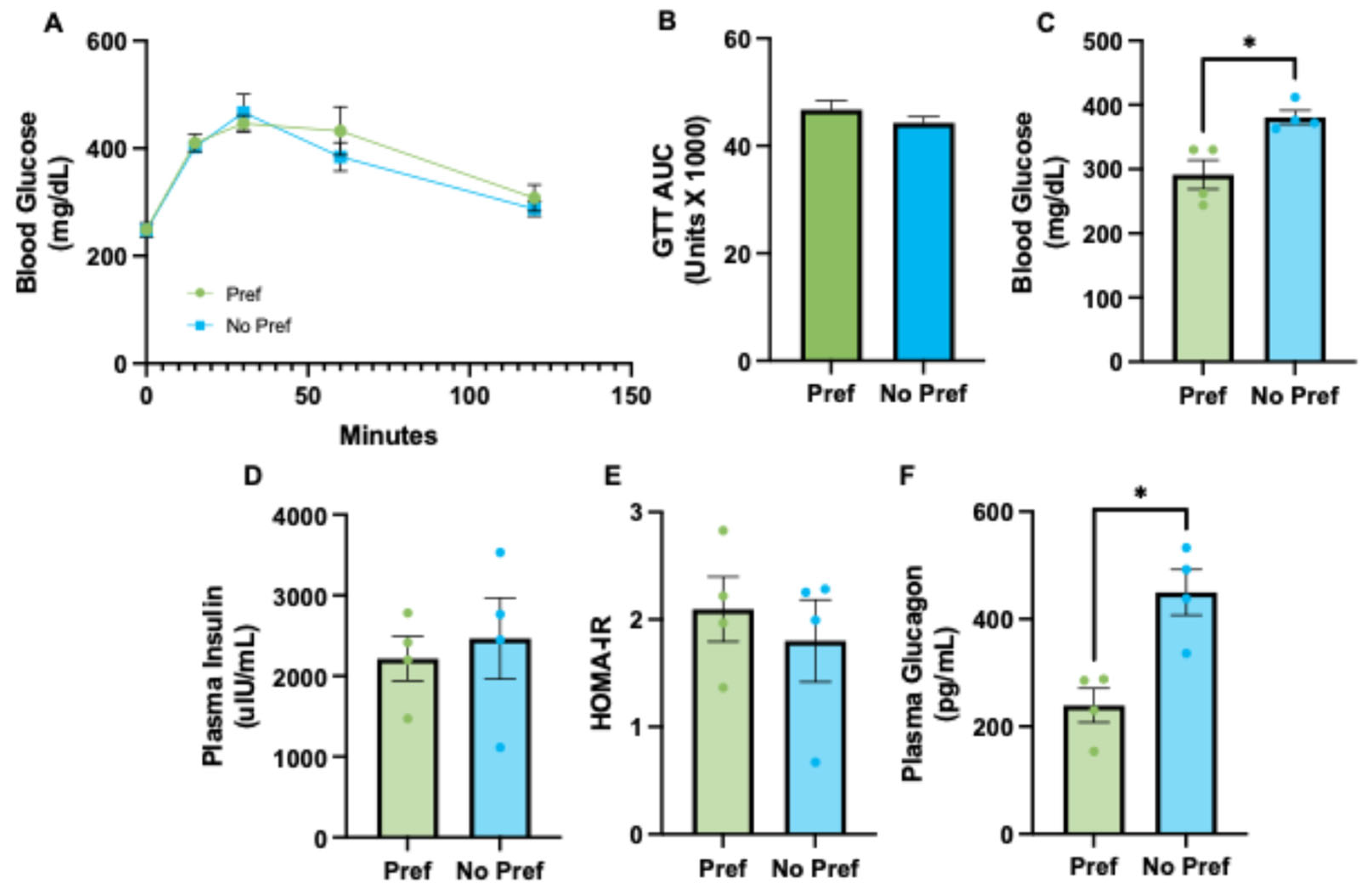
3.2. Alcohol-Preferring Mice Have Reduced Plasma Glucagon and Lower Fasted Blood Glucose Levels
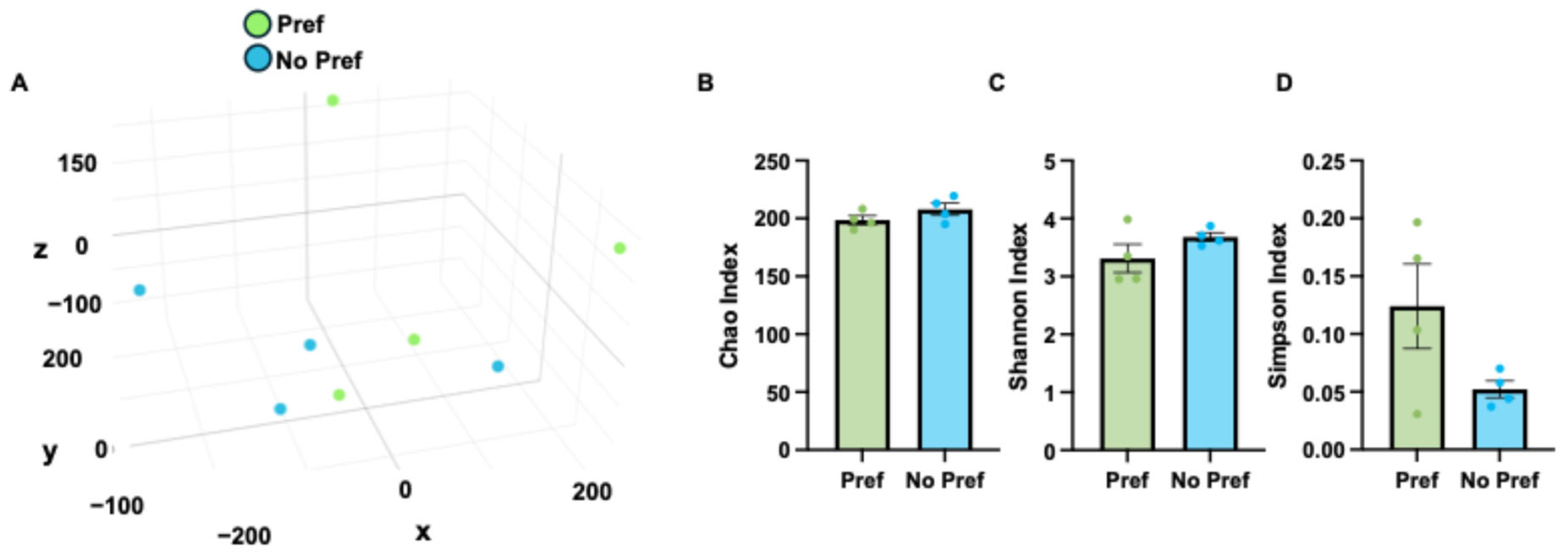
3.3. Microbiome Changes Associated with Alcohol Preference
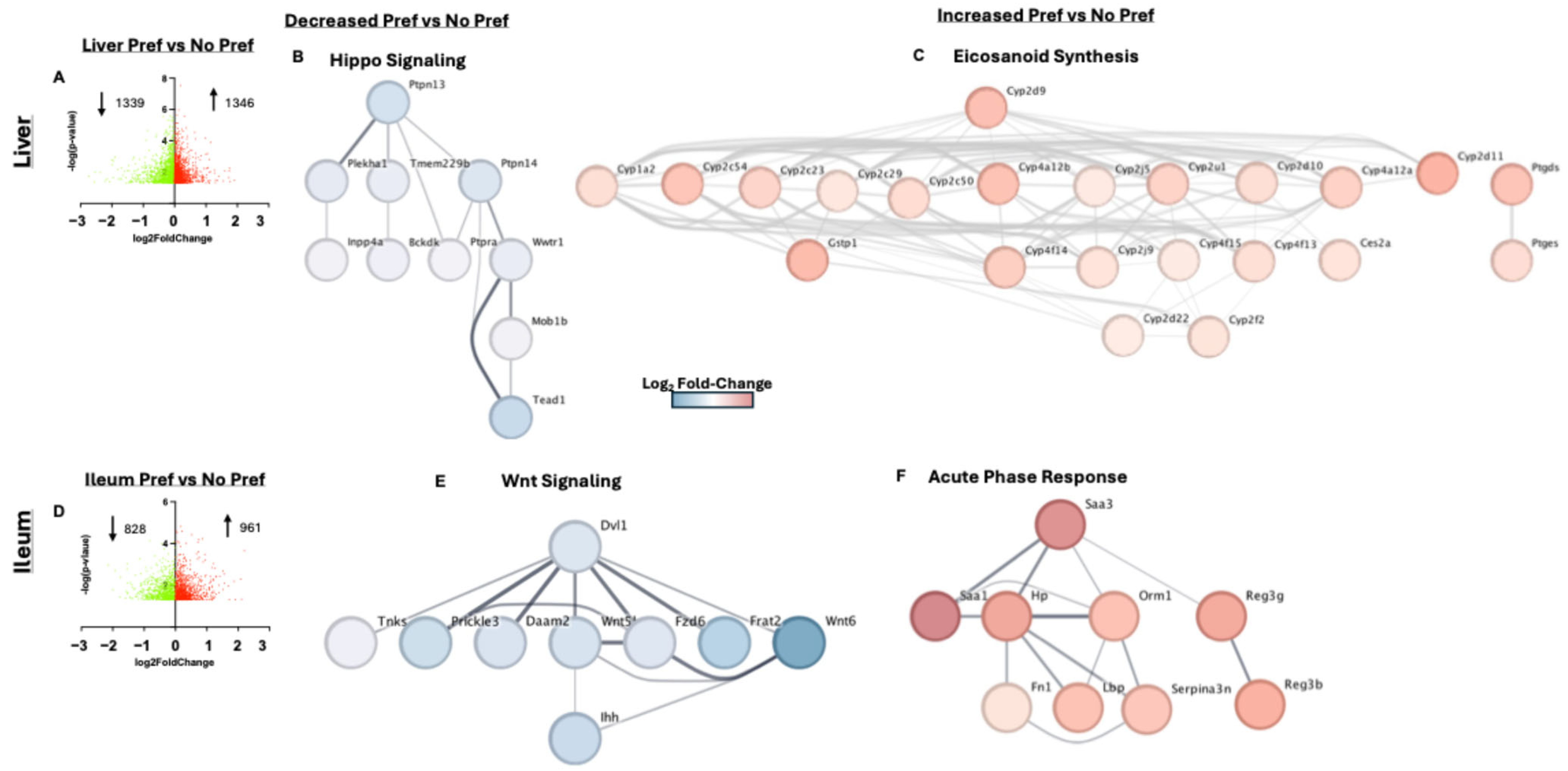
3.4. Liver and Ileum Tissue Had the Greatest Number of Differentially Expressed Genes Associated with Alcohol Preference
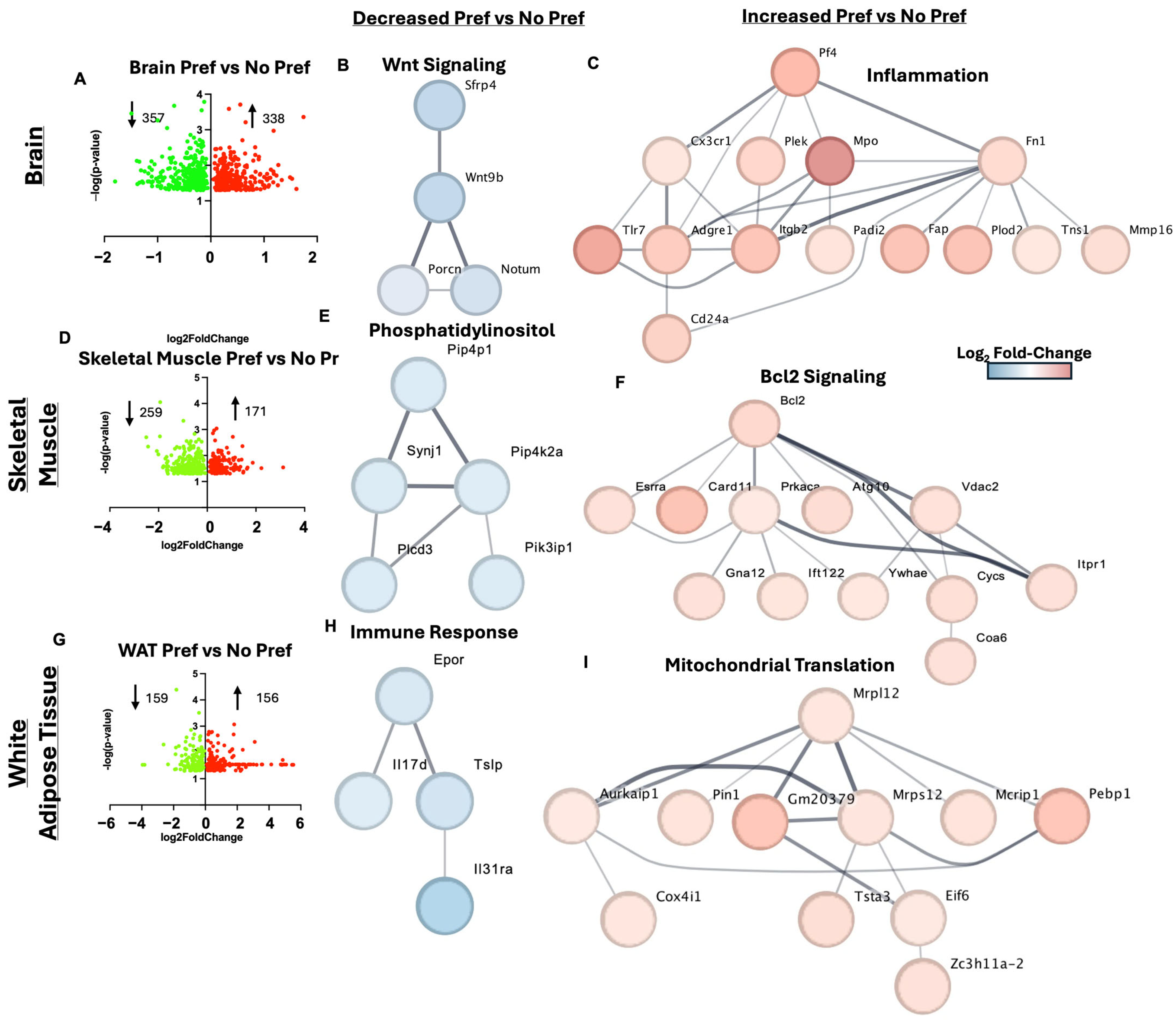
3.5. Brain, Skeletal Muscle, and White Adipose Tissue Gene Markers of Alcohol Preference
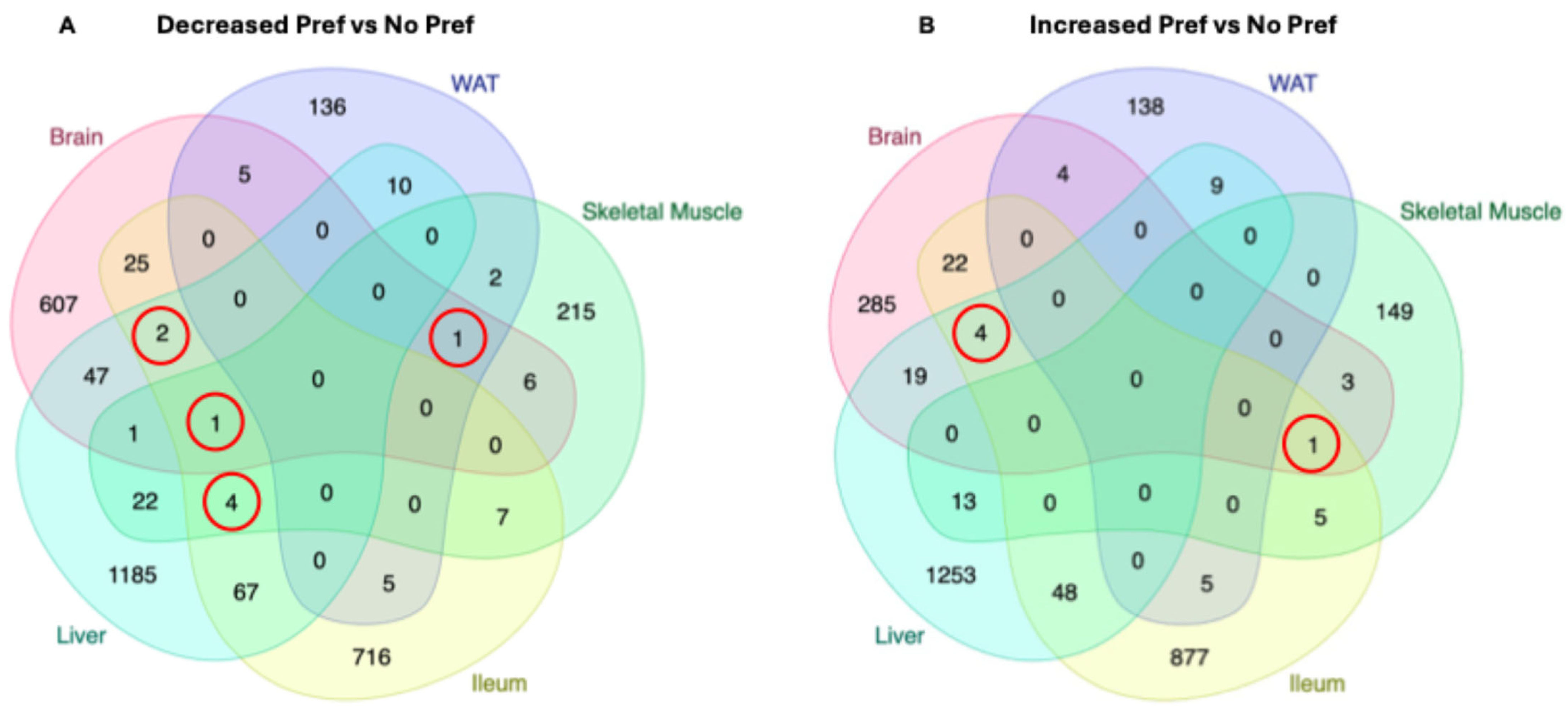
3.6. Shared DEGs Between Multiple Organs in Alcohol-Preferring Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ferreira, M.P.; Willoughby, D. Alcohol consumption: The good, the bad, and the indifferent. Appl. Physiol. Nutr. Metab. 2008, 33, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Cusack, S.E.; Wright, A.W.; Amstadter, A.B. Resilience and alcohol use in adulthood in the United States: A scoping review. Prev. Med. 2023, 168, 107442. [Google Scholar] [CrossRef] [PubMed]
- Boschuetz, N.; German, M.N. Alcohol use disorder: Recognition, testing, and initial management strategies. Clin. Liver Dis. 2023, 22, 18–22. [Google Scholar] [CrossRef]
- SAMHSA. Alcohol Use Disorder in Past Year: Among People Aged 12 or Older; by Age Group and Demographic Characteristics, Numbers in Thousands, 2023 and 2024; National Survey on Drug Use and Health. 2024. Available online: https://www.niaaa.nih.gov/alcohols-effects-health/alcohol-topics/alcohol-facts-and-statistics/alcohol-use-disorder-aud-united-states-age-groups-and-demographic-characteristics (accessed on 9 January 2025).
- Esser, M.B.; Sherk, A.; Liu, Y.; Naimi, T.S. Deaths from Excessive Alcohol Use—United States, 2016–2021. Morb. Mortal. Wkly. Rep. 2024, 73, 154–161. [Google Scholar] [CrossRef]
- Shuey, B.; Halbisen, A.; Lakoma, M.; Zhang, F.; Argetsinger, S.; Williams, E.C.; Druss, B.G.; Wen, H.; Wharam, J.F. High-Acuity Alcohol-Related Complications During the COVID-19 Pandemic. JAMA Health Forum 2024, 5, e240501. [Google Scholar] [CrossRef] [PubMed]
- Kalligeros, M.; Vassilopoulos, A.; Vassilopoulos, S.; Victor, D.W.; Mylonakis, E.; Noureddin, M. Prevalence of Steatotic Liver Disease (MASLD, MetALD, and ALD) in the United States: NHANES 2017–2020. Clin. Gastroenterol. Hepatol. 2024, 22, 1330–1332.e4. [Google Scholar] [CrossRef]
- Gripshover, T.C.; Treves, R.S.; Hardesty, J.E. Identification of Preclinical Biomarkers of Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD) versus MASLD and Increased Alcohol Intake and the Impact of Diet. Am. J. Pathol. 2025. [Google Scholar] [CrossRef]
- Arab, J.P.; Díaz, L.A.; Rehm, J.; Im, G.; Arrese, M.; Kamath, P.S.; Lucey, M.R.; Mellinger, J.; Thiele, M.; Thursz, M.; et al. Metabolic dysfunction and alcohol-related liver disease (MetALD): Position statement by an expert panel on alcohol-related liver disease. J. Hepatol. 2025, 82, 744–756. [Google Scholar] [CrossRef]
- Perilli, M.; Toselli, F.; Franceschetto, L.; Cinquetti, A.; Ceretta, A.; Cecchetto, G.; Viel, G. Phosphatidylethanol (PEth) in Blood as a Marker of Unhealthy Alcohol Use: A Systematic Review with Novel Molecular Insights. Int. J. Mol. Sci. 2023, 24, 12175. [Google Scholar] [CrossRef]
- Treves, R.S.; Gripshover, T.C.; Hardesty, J.E. STATom@ic: R Package for Automated Statistical Analysis of Omic Datasets. Stats 2025, 8, 18. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Luginbühl, M.; Weinmann, W.; Butzke, I.; Pfeifer, P. Monitoring of direct alcohol markers in alcohol use disorder patients during withdrawal treatment and successive rehabilitation. Drug Test. Anal. 2019, 11, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Bishehsari, F.; Magno, E.; Swanson, G.; Desai, V.; Voigt, R.M.; Forsyth, C.B.; Keshavarzian, A. Alcohol and Gut-Derived Inflammation. Alcohol. Res. 2017, 38, 163–171. [Google Scholar]
- Ahmad, M.K.; Abdollah, N.A.; Shafie, N.H.; Yusof, N.M.; Razak, S.R.A. Dual-specificity phosphatase 6 (DUSP6): A review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 2018, 15, 14–28. [Google Scholar] [CrossRef]
- Russell, J.O.; Camargo, F.D. Hippo signalling in the liver: Role in development, regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.Y.; Blutt, S.E.; Zeng, X.L.; Chen, M.S.; Lo, Y.H.; Castillo-Azofeifa, D.; Klein, O.D.; Shroyer, N.F.; Donowitz, M.; Estes, M.K. Epithelial WNT Ligands Are Essential Drivers of Intestinal Stem Cell Activation. Cell Rep. 2018, 22, 1003–1015. [Google Scholar] [CrossRef]
- Górnicki, T.; Lambrinow, J.; Mrozowska, M.; Podhorska-Okołów, M.; Dzięgiel, P.; Grzegrzółka, J. Role of RBMS3 Novel Potential Regulator of the EMT Phenomenon in Physiological and Pathological Processes. Int. J. Mol. Sci. 2022, 23, 10875. [Google Scholar] [CrossRef]
- Beermann, M.L.; Ardelt, M.; Girgenrath, M.; Miller, J.B. Prdm1 (Blimp-1) and the expression of fast and slow myosin heavy chain isoforms during avian myogenesis in vitro. PLoS ONE 2010, 5, e9951. [Google Scholar] [CrossRef]
- Karpyak, V.M.; Winham, S.J.; Biernacka, J.M.; Cunningham, J.M.; Lewis, K.A.; Geske, J.R.; Colby, C.L.; Abulseoud, O.A.; Hall-Flavin, D.K.; Loukianova, L.L.; et al. Association of GATA4 sequence variation with alcohol dependence. Addict. Biol. 2014, 19, 312–315. [Google Scholar] [CrossRef]
- Hahn, J.A.; Fatch, R.; Barnett, N.P.; Marcus, G.M. Phosphatidylethanol vs Transdermal Alcohol Monitoring for Detecting Alcohol Consumption Among Adults. JAMA Netw. Open 2023, 6, e2333182. [Google Scholar] [CrossRef]
- Lanng, A.R.; Gasbjerg, L.S.; Bergmann, N.C.; Bergmann, S.; Helsted, M.M.; Gillum, M.P.; Hartmann, B.; Holst, J.J.; Vilsbøll, T.; Knop, F.K. Gluco-metabolic effects of oral and intravenous alcohol administration in men. Endocr. Connect. 2019, 8, 1372–1382. [Google Scholar] [CrossRef]
- Arky, R.A. Hypoglycemia associated with liver disease and ethanol. Endocrinol. Metab. Clin. N. Am. 1989, 18, 75–90. [Google Scholar] [CrossRef]
- Piacentino, D.; Grant-Beurmann, S.; Vizioli, C.; Li, X.; Moore, C.F.; Ruiz-Rodado, V.; Lee, M.R.; Joseph, P.V.; Fraser, C.M.; Weerts, E.M.; et al. Gut microbiome and metabolome in a non-human primate model of chronic excessive alcohol drinking. Transl. Psychiatry 2021, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, L.; Gong, C.; Li, T.; Xia, Y. The diversity of the intestinal microbiota in patients with alcohol use disorder and its relationship to alcohol consumption and cognition. Front. Psychiatry 2022, 13, 1054685. [Google Scholar] [CrossRef]
- Driskill, J.H.; Pan, D. The Hippo Pathway in Liver Homeostasis and Pathophysiology. Annu. Rev. Pathol. 2021, 16, 299–322. [Google Scholar] [CrossRef]
- Michaelis, U.R.; Falck, J.R.; Schmidt, R.; Busse, R.; Fleming, I. Cytochrome P4502C9-derived epoxyeicosatrienoic acids induce the expression of cyclooxygenase-2 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.B.; Hardesty, J.E.; Song, Y.L.; Floyd, A.T.; Deng, Z.; Jebet, A.; He, L.; Zhang, X.; McClain, C.J.; Hammock, B.D.; et al. Hepatic Transcriptome and Its Regulation Following Soluble Epoxide Hydrolase Inhibition in Alcohol-Associated Liver Disease. Am. J. Pathol. 2024, 194, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Zhang, G.; Wang, Y.; Xu, L.; Wu, M.; Zhou, Y.; Xu, T.; Meng, X.; Huang, C.; Zhang, L. β-catenin ISGylation promotes lipid deposition and apoptosis in ethanol-stimulated liver injury models. Redox Rep. 2022, 27, 239–248. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Austin, C.R.; Vincan, E.; Phesse, T.J. Wnt Signalling in Gastrointestinal Epithelial Stem Cells. Genes 2018, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Gregorieff, A.; Pinto, D.; Begthel, H.; Destrée, O.; Kielman, M.; Clevers, H. Expression Pattern of Wnt Signaling Components in the Adult Intestine. Gastroenterology 2005, 129, 626–638. [Google Scholar] [CrossRef]
- Yuan, G.; Regel, I.; Lian, F.; Friedrich, T.; Hitkova, I.; Hofheinz, R.D.; Ströbel, P.; Langer, R.; Keller, G.; Röcken, C.; et al. WNT6 is a novel target gene of caveolin-1 promoting chemoresistance to epirubicin in human gastric cancer cells. Oncogene 2013, 32, 375–387. [Google Scholar] [CrossRef]
- Gulhar, R.; Ashraf, M.A.; Jialal, I. Physiology, Acute Phase Reactants. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2025. [Google Scholar]
- Molmenti, E.P.; Ziambaras, T.; Perlmutter, D.H. Evidence for an acute phase response in human intestinal epithelial cells. J. Biol. Chem. 1993, 268, 14116–14124. [Google Scholar] [CrossRef]
- Stärkel, P.; Leclercq, S.; de Timary, P.; Schnabl, B. Intestinal dysbiosis and permeability: The yin and yang in alcohol dependence and alcoholic liver disease. Clin. Sci. 2018, 132, 199–212. [Google Scholar] [CrossRef]
- Alkailani, M.I.; Aittaleb, M.; Tissir, F. WNT signaling at the intersection between neurogenesis and brain tumorigenesis. Front. Mol. Neurosci. 2022, 15, 1017568. [Google Scholar] [CrossRef]
- Aghaizu, N.D.; Jin, H.; Whiting, P.J. Dysregulated Wnt Signalling in the Alzheimer’s Brain. Brain Sci. 2020, 10, 902. [Google Scholar] [CrossRef]
- Gastfriend, B.D.; Nishihara, H.; Canfield, S.G.; Foreman, K.L.; Engelhardt, B.; Palecek, S.P.; Shusta, E.V. Wnt signaling mediates acquisition of blood-brain barrier properties in naïve endothelium derived from human pluripotent stem cells. Elife 2021, 10, e70992. [Google Scholar] [CrossRef] [PubMed]
- Swoboda, J.; Mittelsdorf, P.; Chen, Y.; Weiskirchen, R.; Stallhofer, J.; Schüle, S.; Gassler, N. Intestinal Wnt in the transition from physiology to oncology. World J. Clin. Oncol. 2022, 13, 168–185. [Google Scholar] [CrossRef]
- Erickson, E.K.; Grantham, E.K.; Warden, A.S.; Harris, R.A. Neuroimmune signaling in alcohol use disorder. Pharmacol. Biochem. Behav. 2019, 177, 34–60. [Google Scholar] [CrossRef] [PubMed]
- Lanquetin, A.; Leclercq, S.; de Timary, P.; Segobin, S.; Naveau, M.; Coulbault, L.; Maccioni, P.; Lorrai, I.; Colombo, G.; Vivien, D.; et al. Role of inflammation in alcohol-related brain abnormalities: A translational study. Brain Commun. 2021, 3, fcab154. [Google Scholar] [CrossRef]
- Lékó, A.H.; Ray, L.A.; Leggio, L. The vicious cycle between (neuro)inflammation and alcohol use disorder: An opportunity to develop new medications? Alcohol. Clin. Exp. Res. 2023, 47, 843–847. [Google Scholar] [CrossRef]
- Adams, C.; Perry, N.; Conigrave, J.; Hurzeler, T.; Stevens, J.; Yacou Dunbar, K.P.; Sweeney, A.; Lee, K.; Sutherland, G.; Haber, P.; et al. Central markers of neuroinflammation in alcohol use disorder: A meta-analysis of neuroimaging, cerebral spinal fluid, and postmortem studies. Alcohol. Clin. Exp. Res. 2023, 47, 197–208. [Google Scholar] [CrossRef]
- Dasarathy, J.; McCullough, A.J.; Dasarathy, S. Sarcopenia in Alcoholic Liver Disease: Clinical and Molecular Advances. Alcohol. Clin. Exp. Res. 2017, 41, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, F.; Modrak, S.; Little, A.; Zhang, H. Chronic Alcohol Consumption Enhances Skeletal Muscle Wasting in Mice Bearing Cachectic Cancers: The Role of TNFα/Myostatin Axis. Alcohol. Clin. Exp. Res. 2020, 44, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Fedorenko, O.Y.; Mikhalitskaya, E.V.; Toshchakova, V.A.; Loonen, A.J.M.; Bokhan, N.A.; Ivanova, S.A. Association of PIP4K2A Polymorphisms with Alcohol Use Disorder. Genes 2021, 12, 1642. [Google Scholar] [CrossRef]
- Ivanova, H.; Vervliet, T.; Monaco, G.; Terry, L.E.; Rosa, N.; Baker, M.R.; Parys, J.B.; Serysheva, I.I.; Yule, D.I.; Bultynck, G. Bcl-2-Protein Family as Modulators of IP(3) Receptors and Other Organellar Ca(2+) Channels. Cold Spring Harb. Perspect. Biol. 2020, 12, a035089. [Google Scholar] [CrossRef] [PubMed]
- Chin, H.S.; Li, M.X.; Tan, I.K.L.; Ninnis, R.L.; Reljic, B.; Scicluna, K.; Dagley, L.F.; Sandow, J.J.; Kelly, G.L.; Samson, A.L.; et al. VDAC2 enables BAX to mediate apoptosis and limit tumor development. Nat. Commun. 2018, 9, 4976. [Google Scholar] [CrossRef]
- Maleki, B.; Modarres, P.; Salehi, P.; Vallian, S. Identification of ITPR1 gene as a novel target for hsa-miR-34b-5p in non-obstructive azoospermia: A Ca(2+)/apoptosis pathway cross-talk. Sci. Rep. 2023, 13, 21873. [Google Scholar] [CrossRef]
- Lee, Y.; Clinton, J.; Yao, C.; Chang, S.H. Interleukin-17D Promotes Pathogenicity During Infection by Suppressing CD8 T Cell Activity. Front. Immunol. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Starnes, T.; Broxmeyer, H.E.; Robertson, M.J.; Hromas, R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J. Immunol. 2002, 169, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Rajvanshi, P.K.; Rogers, H.M.; Yoshida, T.; Kopp, J.B.; An, X.; Gassmann, M.; Noguchi, C.T. Erythropoietin regulates energy metabolism through EPO-EpoR-RUNX1 axis. Nat. Commun. 2024, 15, 8114. [Google Scholar] [CrossRef]
- Hwang, S.; Gao, B. How does your fat affect your liver when you drink? J. Clin. Investig. 2019, 129, 2181–2183. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Baloh, R.H.; Milbrandt, J. The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J. Cell Sci. 2009, 122, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, R.; Yu, H.; Liu, J.; Zheng, S.; Li, Y.; Ye, L. NTF3 Correlates With Prognosis and Immune Infiltration in Hepatocellular Carcinoma. Front. Med. 2021, 8, 795849. [Google Scholar] [CrossRef]
- Malewska-Kasprzak, M.; Skibińska, M.; Dmitrzak-Węglarz, M. Alterations in Neurotrophins in Alcohol-Addicted Patients during Alcohol Withdrawal. Brain Sci. 2024, 14, 583. [Google Scholar] [CrossRef]
- Hulea, L.; Nepveu, A. CUX1 transcription factors: From biochemical activities and cell-based assays to mouse models and human diseases. Gene 2012, 497, 18–26. [Google Scholar] [CrossRef]
- Feng, F.; Zhao, Z.; Zhou, Y.; Cheng, Y.; Wu, X.; Heng, X. CUX1 Facilitates the Development of Oncogenic Properties Via Activating Wnt/β-Catenin Signaling Pathway in Glioma. Front. Mol. Biosci. 2021, 8, 705008. [Google Scholar] [CrossRef]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Alcohol consumption and site-specific cancer risk: A comprehensive dose-response meta-analysis. Br. J. Cancer 2015, 112, 580–593. [Google Scholar] [CrossRef]
- Tong, J.; Han, C.J.; Zhang, J.Z.; He, W.Z.; Zhao, G.J.; Cheng, X.; Zhang, L.; Deng, K.Q.; Liu, Y.; Fan, H.F.; et al. Hepatic Interferon Regulatory Factor 6 Alleviates Liver Steatosis and Metabolic Disorder by Transcriptionally Suppressing Peroxisome Proliferator-Activated Receptor γ in Mice. Hepatology 2019, 69, 2471–2488. [Google Scholar] [CrossRef]

| Gene | p-Value | Log2 Fold-Change (Pref/No Pref) |
|---|---|---|
| Cyp2a4 | 0.0401689 | −2.9088035 |
| Gm20274 | 0.01709653 | −2.7401112 |
| Cyp2c69 | 0.04493801 | −2.3655704 |
| Elf3 | 0.02857143 | −2.148375 |
| Ccnb2 | 0.04900357 | −2.0679971 |
| Col6a6 | 0.03273774 | 1.66892196 |
| 4931413K12Rik | 0.00385552 | 1.73018036 |
| Gm44620 | 0.00917213 | 1.74749083 |
| Mup19 | 0.00916766 | 2.05966658 |
| Gm14201 | 0.03302515 | 2.1428187 |
| Gene | p-Value | Log2 Fold-Change (Pref/No Pref) |
|---|---|---|
| Hspe1-ps5 | 0.00010337 | −2.1375433 |
| Klhdc7b | 0.02068844 | −1.9994922 |
| Cdh9 | 0.0092713 | −1.9256851 |
| Gm23123 | 0.03234469 | −1.8374187 |
| Gm43378 | 0.0009537 | −1.7702924 |
| Pcolce2 | 0.02857143 | 1.51095014 |
| Sprr2a3 | 0.02857143 | 1.71544386 |
| Saa3 | 0.02162142 | 1.87438718 |
| Gm28875 | 0.00886639 | 1.90822057 |
| Mrap | 0.00166207 | 2.1954666 |
| Gene | p-Value | Log2 Fold-Change (Pref/No Pref) |
|---|---|---|
| FUT4 | 0.00173059 | −1.857677 |
| Gm23119 | 0.02857143 | −1.791453 |
| Gm42471 | 0.00113454 | −1.76197 |
| Fam24b | 0.01099322 | −1.6992738 |
| Hsd3b7 | 0.00034175 | −1.4872414 |
| Kcng3 | 0.04148866 | 1.70387877 |
| Tex15 | 0.00042448 | 1.74945007 |
| Acsm3 | 0.01424366 | 1.78181541 |
| Gm37212 | 0.01554785 | 1.90608096 |
| H2ac6 | 0.00935297 | 1.91243398 |
| Gene | p-Value | Log2 Fold-Change (Pref/No Pref) |
|---|---|---|
| Meis2 | 0.00197744 | −2.502308894 |
| Gm26642 | 0.00453452 | −2.430666649 |
| A430035B10Rik | 0.00668697 | −2.068963567 |
| 5430416N02Rik | 0.00906632 | −2.008322262 |
| Tmem255a | 0.00183859 | −1.943820645 |
| Zfp773 | 0.0324688 | 1.62777072 |
| Gm45844 | 0.02882195 | 1.655560946 |
| Adamts8 | 0.01884012 | 1.86746702 |
| Gm43307 | 0.03048931 | 2.225161348 |
| Slc4a1 | 0.02857143 | 3.116616888 |
| Gene | p-Value | Log2 Fold-Change (Pref/No Pref) |
|---|---|---|
| Gata4 | 0.02940105 | −3.9475123 |
| Gm17455 | 0.02940105 | −3.8202673 |
| Gm25492 | 0.00499012 | −2.628718 |
| Aqp5 | 0.0283722 | −2.3473238 |
| Il31ra | 0.04392121 | −1.9193447 |
| Samd5 | 0.02940105 | 5.52984238 |
| Cdh17 | 0.02107057 | 1.00 × 1099 |
| Clec18a | 0.02107057 | 1.00 × 1099 |
| Ttc39d | 0.02107057 | 1.00 × 1099 |
| Kndc1 | 0.02107057 | 1.00 × 1099 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikhwal, S.; Gripshover, T.C.; Treves, R.S.; Hardesty, J.E. Alcohol Preference Impacts Multi-Organ Transcriptome in MetALD. Genes 2025, 16, 1121. https://doi.org/10.3390/genes16101121
Sikhwal S, Gripshover TC, Treves RS, Hardesty JE. Alcohol Preference Impacts Multi-Organ Transcriptome in MetALD. Genes. 2025; 16(10):1121. https://doi.org/10.3390/genes16101121
Chicago/Turabian StyleSikhwal, Saumya, Tyler C. Gripshover, Rui S. Treves, and Josiah E. Hardesty. 2025. "Alcohol Preference Impacts Multi-Organ Transcriptome in MetALD" Genes 16, no. 10: 1121. https://doi.org/10.3390/genes16101121
APA StyleSikhwal, S., Gripshover, T. C., Treves, R. S., & Hardesty, J. E. (2025). Alcohol Preference Impacts Multi-Organ Transcriptome in MetALD. Genes, 16(10), 1121. https://doi.org/10.3390/genes16101121





