Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and T. harzianum and F. solani Inoculation
2.2. RNA Sequencing and Functional Annotation
2.3. Differentially Expressed Gene Analysis
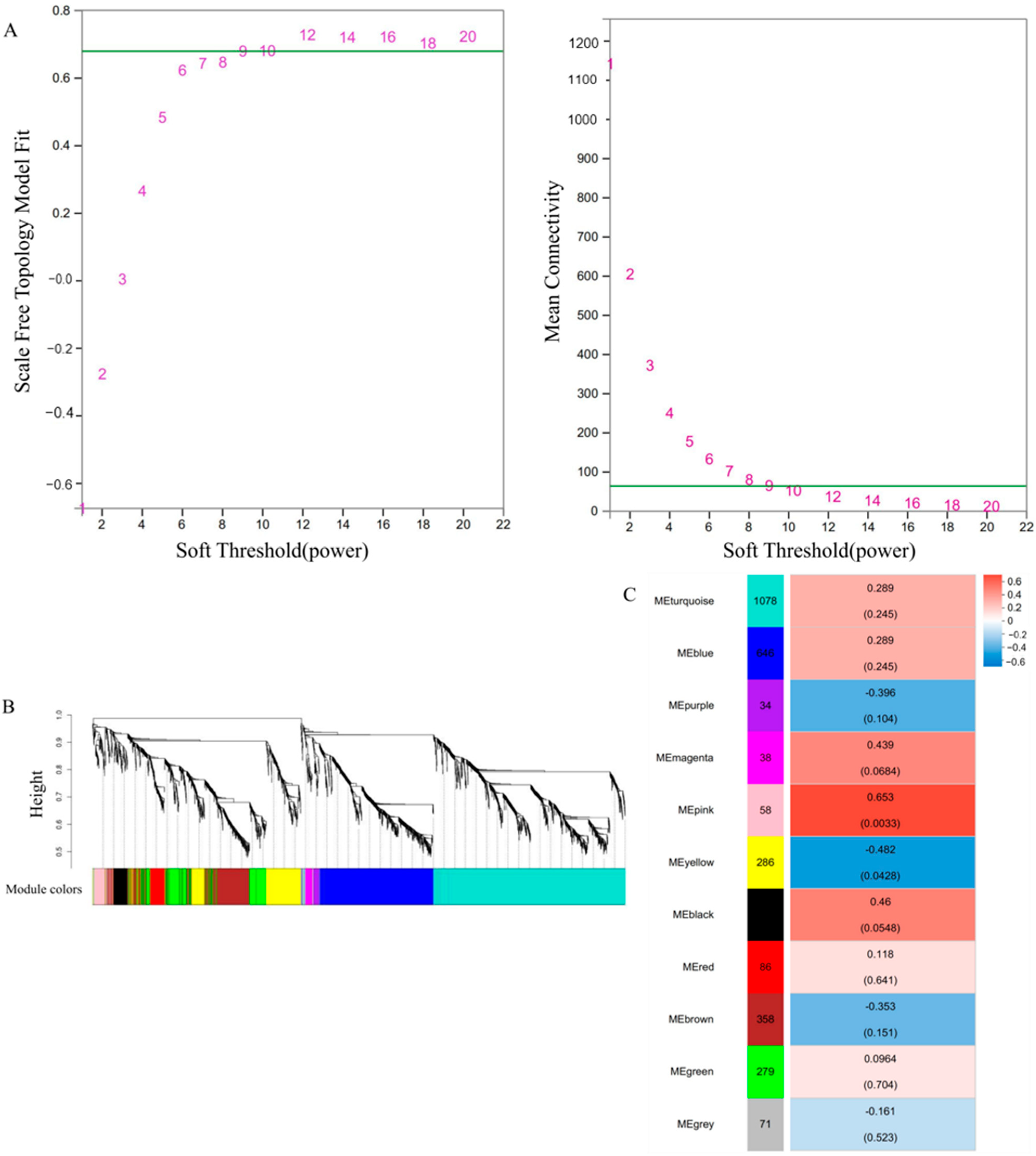
2.4. Weighted Gene Co-Expression Network Analysis
2.5. RT-qPCR Analysis
2.6. H2O2 Content Assays
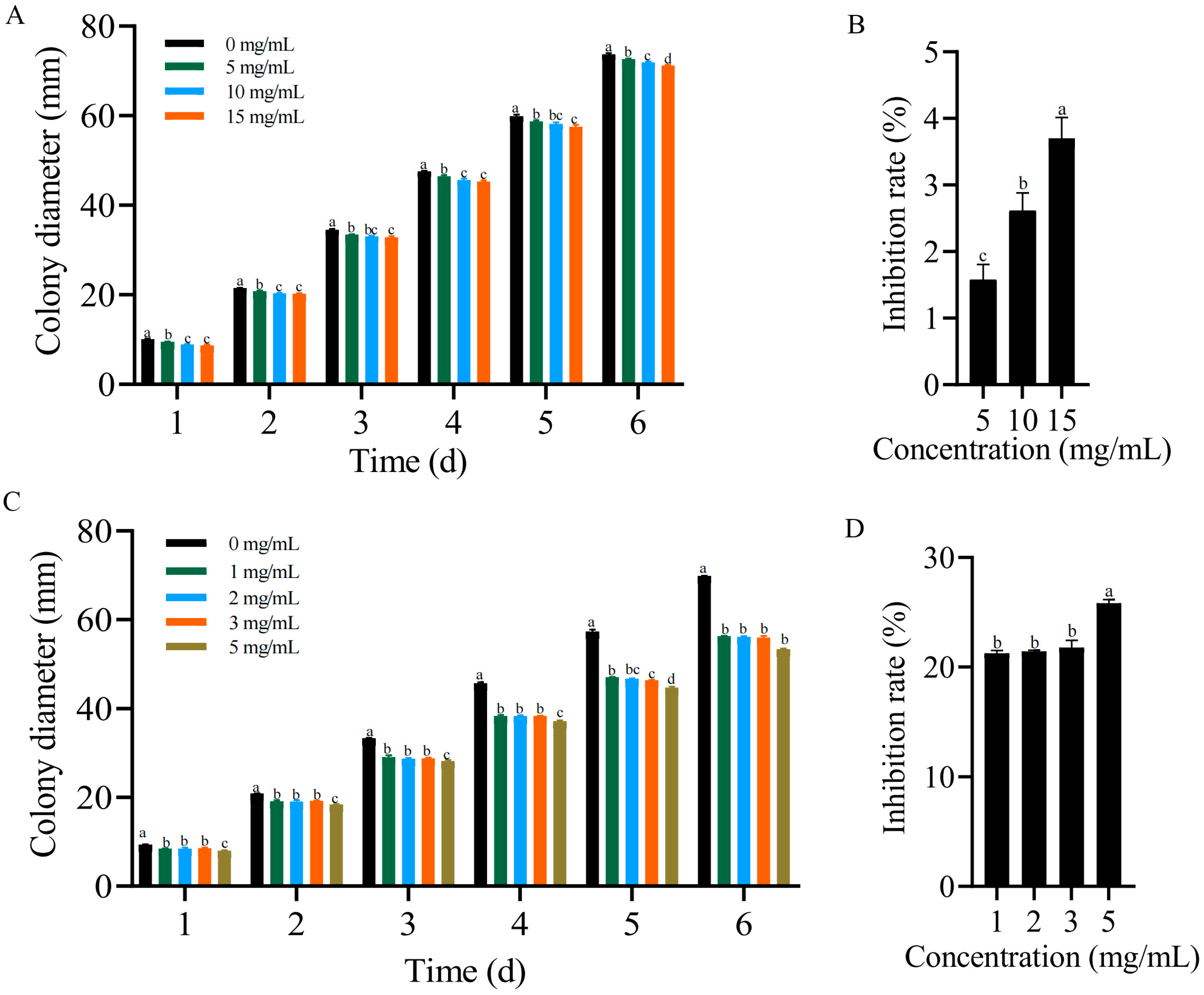
2.7. Antagonistic Effects of Formononetin and Biochanin A on F. solani In Vitro
2.8. Analysis of RPS25 Expression under Hormone Treatments
2.9. Statistical Analysis
3. Results
3.1. Reduction in the Plant Wilt Rate
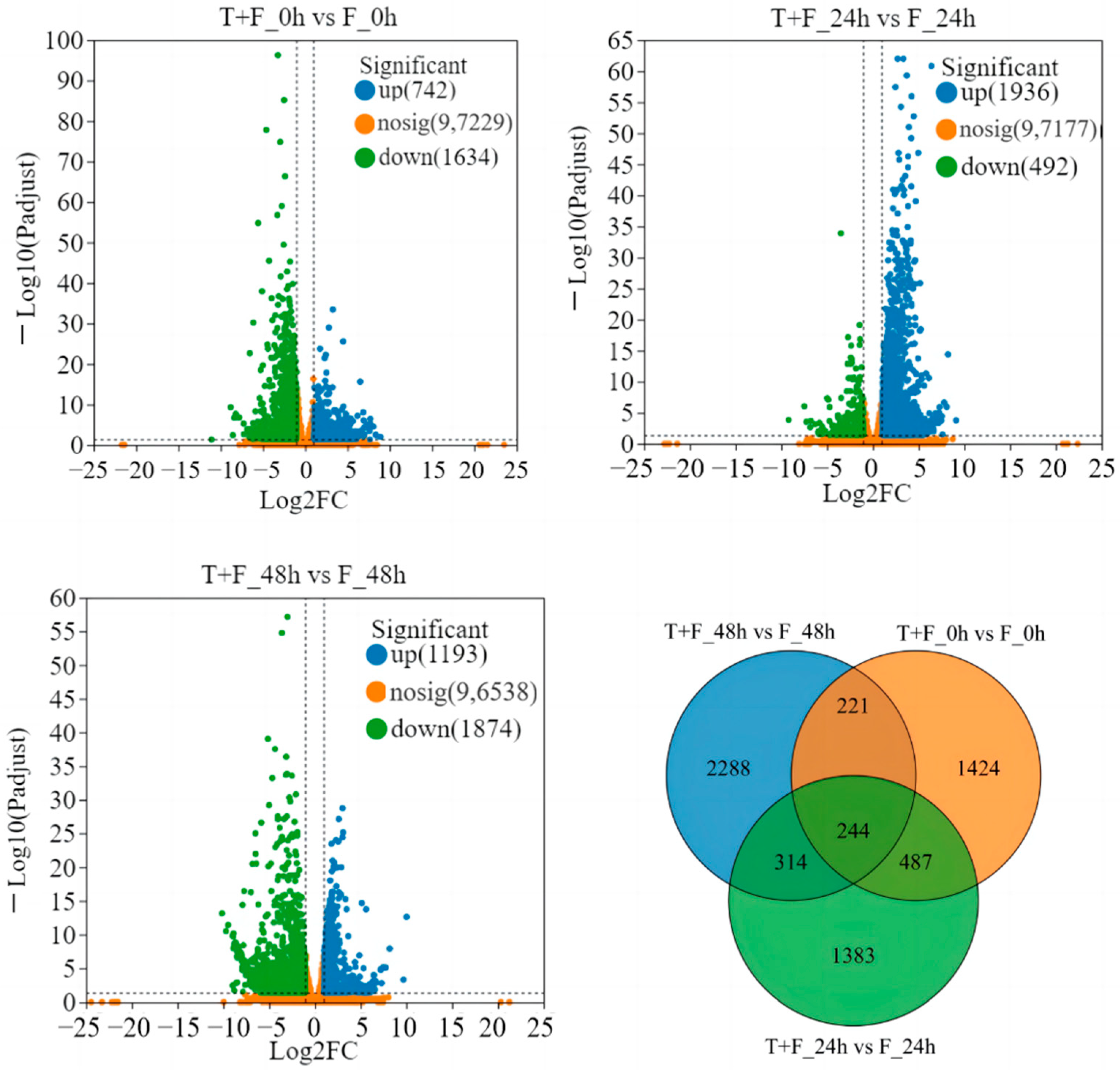
3.2. Transcriptomic Analysis and Identification of Differentially Expressed Genes
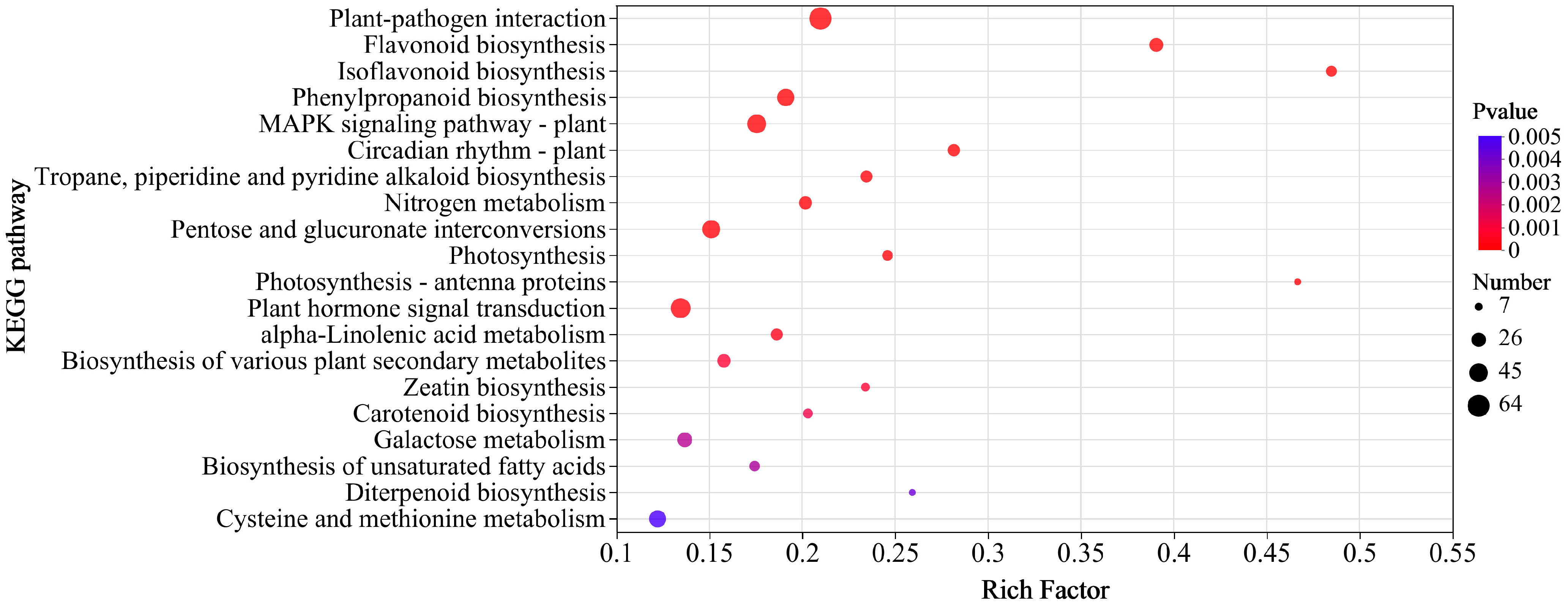
3.3. Functional Annotation and KEGG Pathway Enrichment Analysis of DEGs
3.4. Validation of RNA-Seq Data by RT–qPCR
3.5. Plant–Pathogen Interaction Pathway Analysis under T. harzianum Treatment
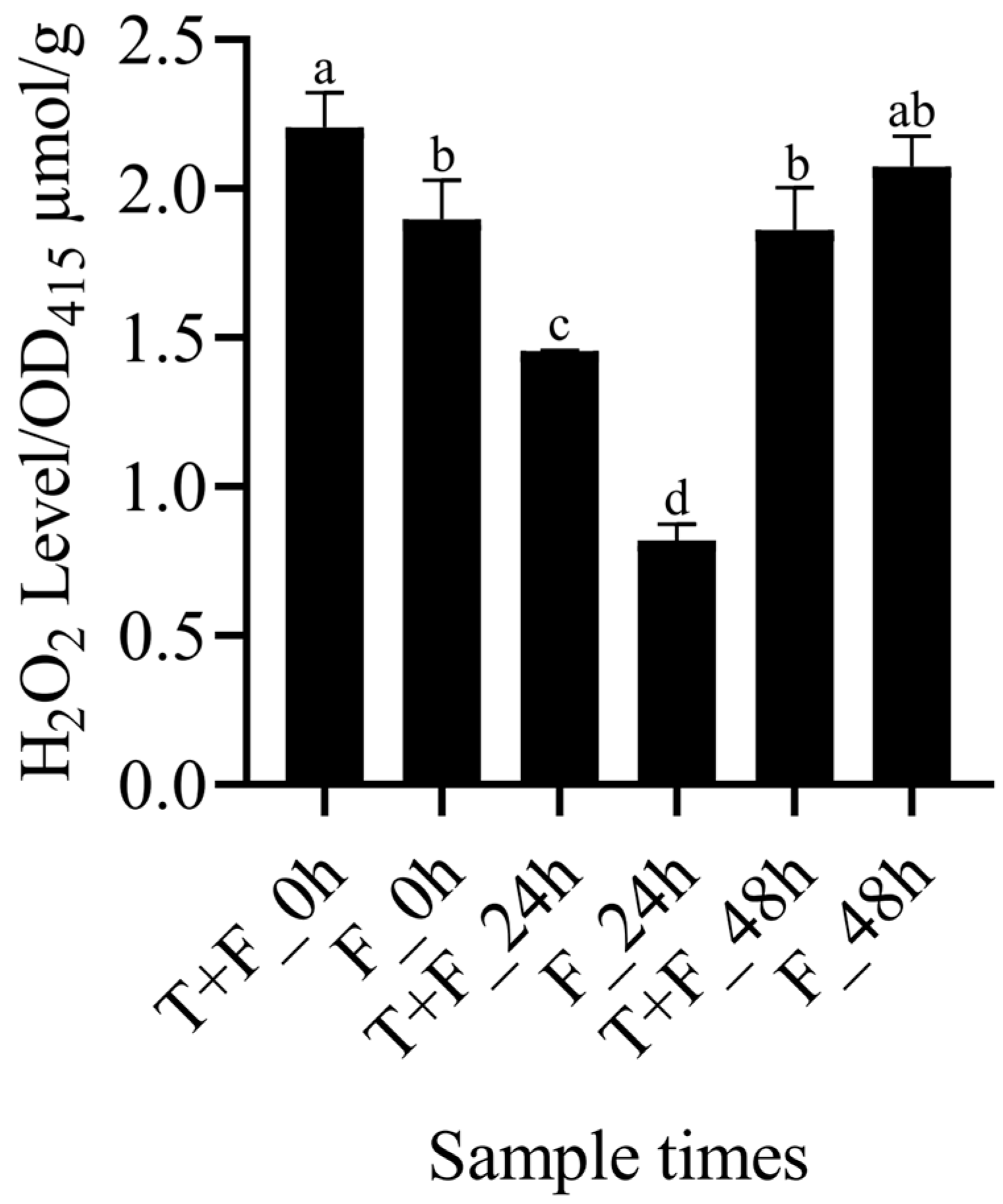
3.6. Assay of H2O2 Levels in the T + F and F Treatments
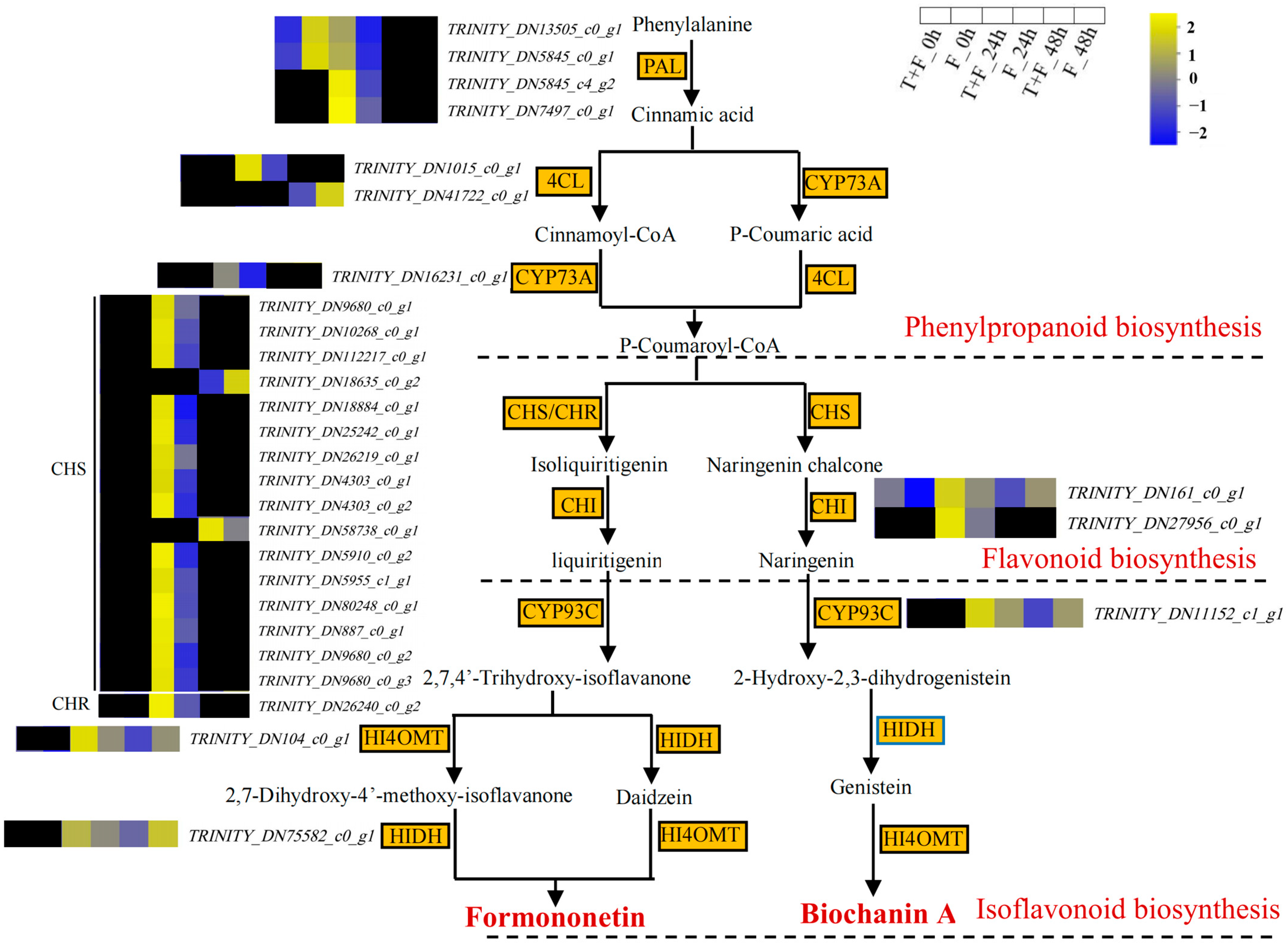
3.7. Formononetin and Biochanin A Biosynthesis Pathway Analysis under T. harzianum Treatment
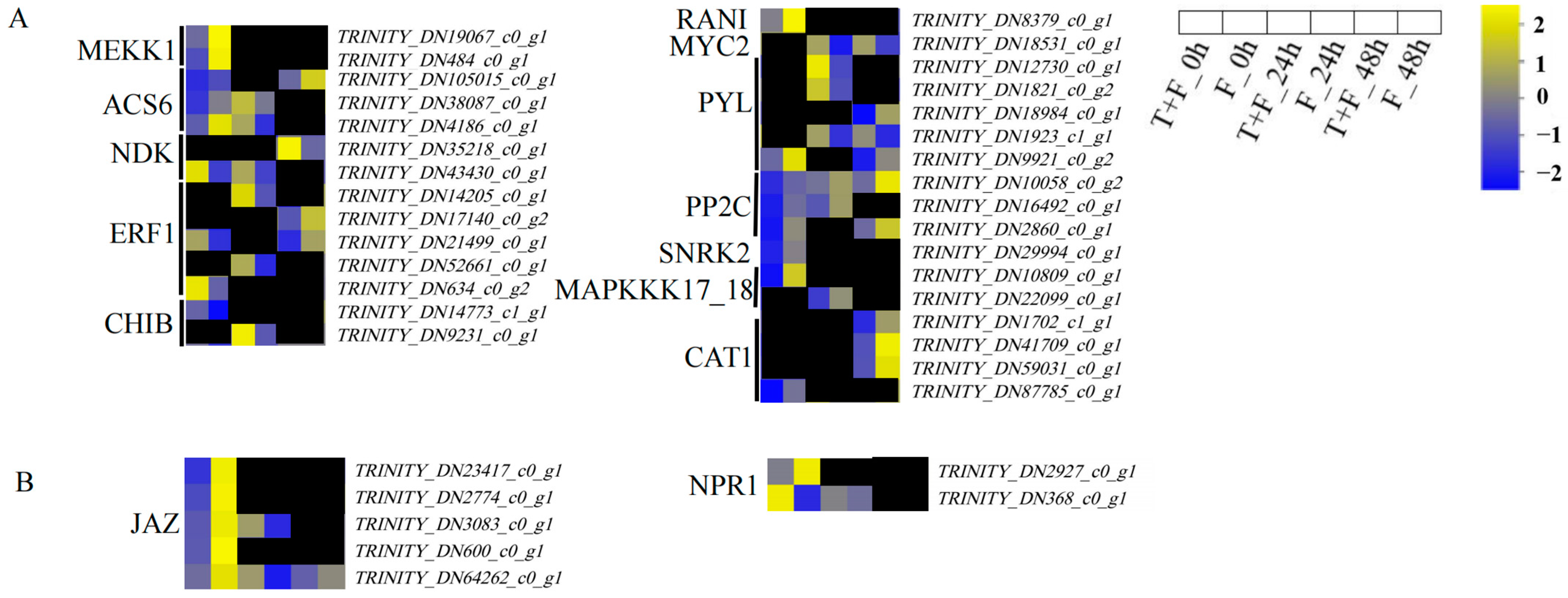
3.8. MAPK Signalling Pathway and Plant Hormone Signal Transduction Pathway Analysis under T. harzianum Treatment
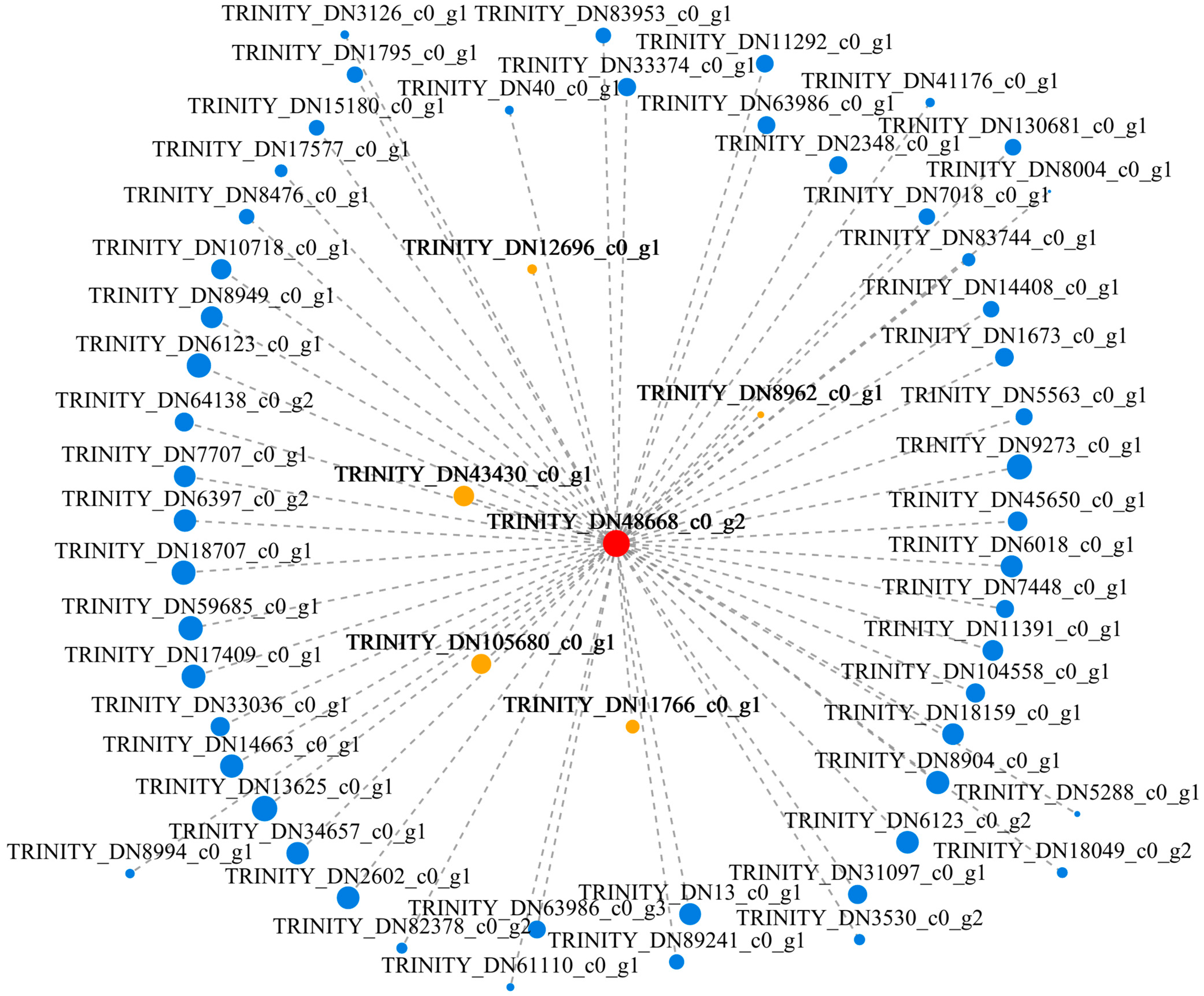
3.9. Gene Coexpression Network Analysis
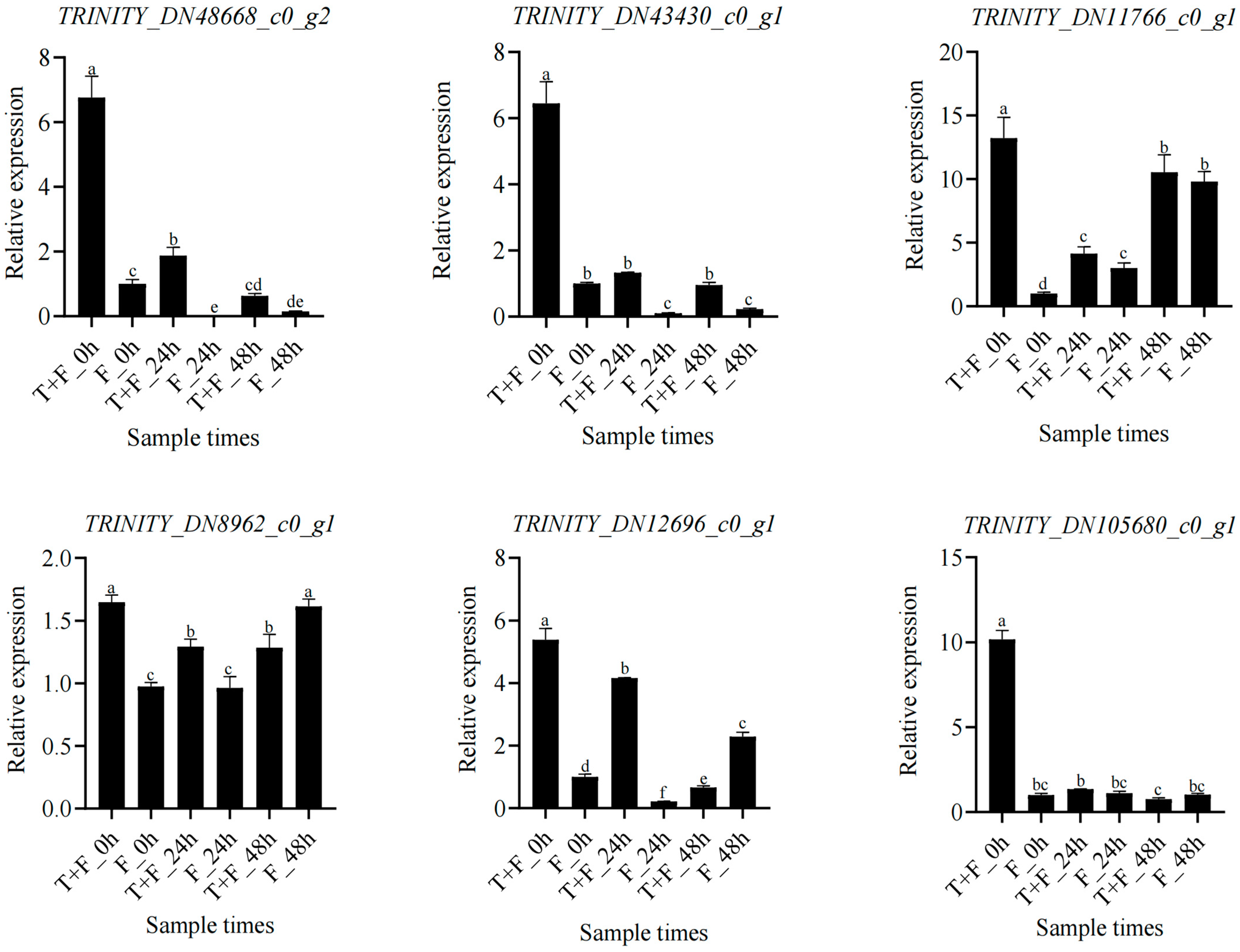
3.10. Expression Analysis of Hub Genes and Coexpressed Genes under T. harzianum Treatment
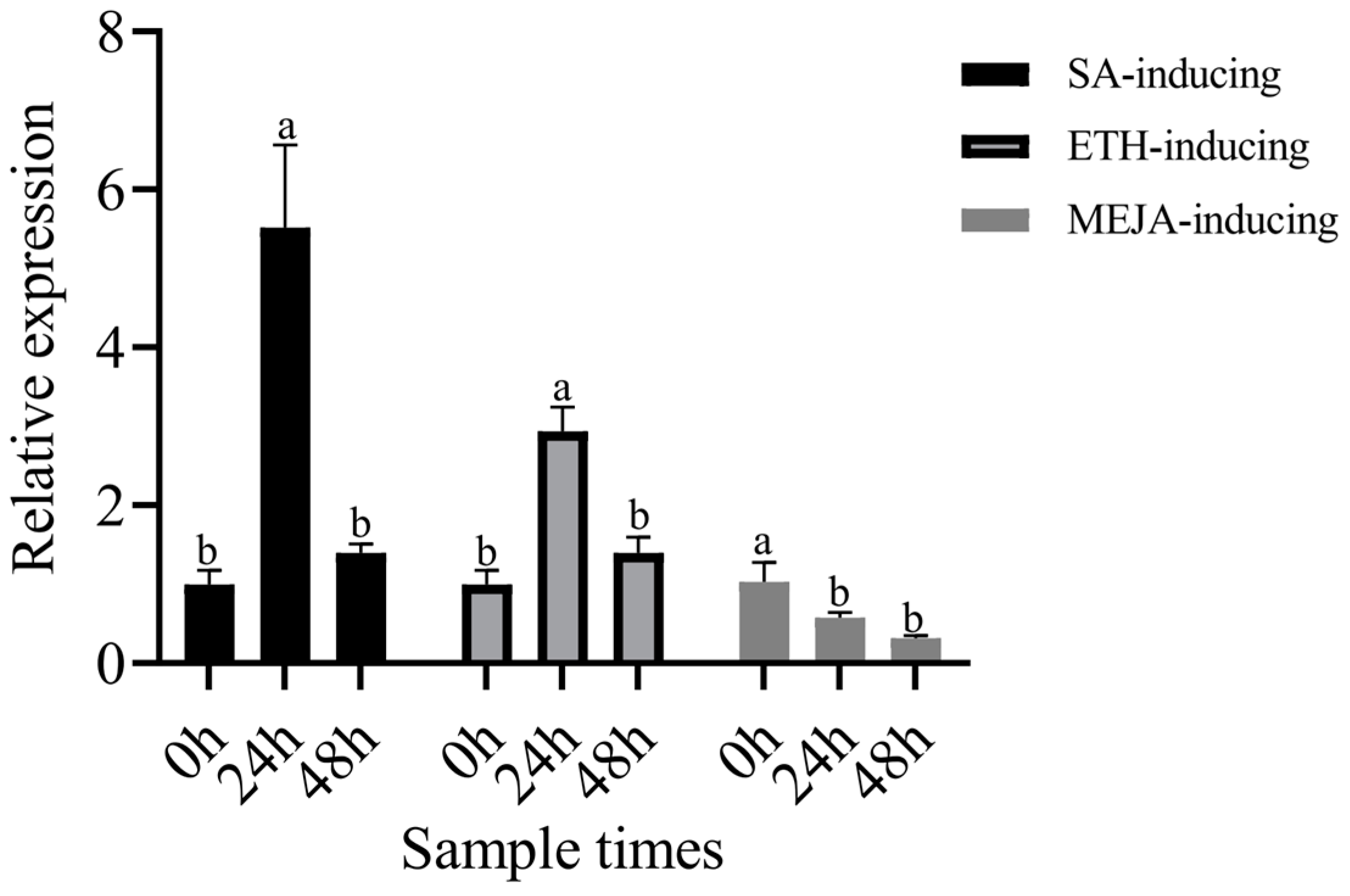
3.11. Expression Analysis of RPS25 Induced by the Exogenous Hormones SA, ETH, and MeJA
4. Discussion
4.1. Induction of PR1 by T. harzianum under F. solani Stress
4.2. H2O2 Induction by T. harzianum under F. solani Stress
4.3. Formononetin and Biochanin A Biosynthesis Pathways Induction by T. harzianum under F. solani Stress
4.4. CHIB Induction by T. harzianum under F. solani Stress
4.5. MYC2 and JAZ Induction by T. harzianum under F. solani Stress
4.6. Candidate Hub Gene Mining under T. harzianum Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, Z.Q.; Findlay, J.A. Constituents of Astragalus mongholicus. J. Nat. Prod. 1991, 54, 810–815. [Google Scholar] [CrossRef]
- Tang, W.C.; Eisenbrand, G. Chinese Drugs of Plant Origin; Springer: Berlin/Heidelberg, Germany, 1992; pp. 191–197. [Google Scholar]
- Fu, J.; Wang, Z.H.; Huang, L.F.; Zheng, S.H.; Wang, D.M.; Chen, S.L.; Zhang, H.T.; Yang, S.H. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus mongholicus (Huangqi). Phytother. Res. 2014, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Zhang, J.F.; Ma, J.X.; Liu, L.; Li, J.J.; Shen, T.; Tian, Y.Q. The major component of cinnamon oil as a natural substitute against Fusarium solani on Astragalus mongholicus. J. Appl. Microbiol. 2022, 132, 3125–3141. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Guan, Y.M.; Wang, Q.X.; Li, M.J.; Pan, X.X.; Zhang, Y.Y. Research progress on main diseases and control measures of Astragalus mongholicus. Spec. Wild Econ. Anim. Plant Res. 2019, 41, 101–107. [Google Scholar]
- Wang, L.X.; Sun, X.R.; Bai, Q.J.; Qi, F.M. Pathogen identification of root rot in Astragallus mongholicus. Acta Agric. Boreali-Sin. 1994, 9, 107–109. [Google Scholar]
- Zhang, C.H.; Xu, J.P.; Wang, J.; Zhang, G.J.; Li, W.H.; Guo, L.P. Investigation and arrangement of ecological planting pattern of Astragalus radix in Inner Mongolia. Mod. Chin. Med. 2018, 20, 1212–1216. [Google Scholar]
- Shi, A.L.; Xing, Z.M.; Guo, Y.W.; Shang, Y.R.; Ma, C.Y. Field control effect of combination of fungicide and insecticide on root rot disease of Astragalusm embranaceus. J. Hebei Agric. Sci. 2016, 20, 55–59. [Google Scholar]
- Gao, Y.H.; Zhang, Y.; Cheng, X.Q.; Zheng, Z.H.; Wu, X.H.; Dong, X.H.; Hu, Y.G.; Wang, X.F. Agricultural Jiaosu: An eco-friendly and cost-effective control strategy for suppressing fusarium root rot disease in Astragalus mongholicus. Front. Microbiol. 2022, 13, 823704. [Google Scholar] [CrossRef]
- Behiry, S.; Soliman, S.A.; Massoud, M.A.; Abdelbary, M.; Kordy, A.M.; Abdelkhalek, A.; Heflish, A. Trichoderma pubescens elicit induced systemic resistance in tomato challenged by Rhizoctonia solani. J. Fungi 2023, 9, 167. [Google Scholar] [CrossRef]
- Erazo, J.G.; Palacios, S.A.; Pastor, N.; Giordano, F.D.; Rovera, M.; Reynoso, M.M.; Venisse, J.S.; Torres, A.M. Biocontrol mechanisms of Trichoderma harzianum ITEM 3636 against peanut brown root rot caused by Fusarium solani RC 386. Biol. Control 2021, 164, 104774. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Daranagama, D.A.; Phillips, A.J.; Kannangara, S.D.; Promputtha, I. Fungi vs. fungi in biocontrol: An overview of fungal antagonists applied against fungal plant pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 604923–604942. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Guzmán, P.; Kumar, A.; de Los Santos-Villalobos, S.; Parra-Cota, F.I.; Orozco-Mosqueda, M.D.C.; Fadiji, A.E.; Hyder, S.; Babalola, O.O.; Santoyo, G. Trichoderma species: Our best fungal allies in the biocontrol of plant diseases—A review. Plants 2023, 12, 432. [Google Scholar] [CrossRef]
- Teng, Y.P.; Liang, Z.S.; Chen, R. Preliminary study of Trichoderma against the root rot disease of Astragallus. Acta Agric. Boreali-Occident. Sin. 2006, 15, 69–71. [Google Scholar]
- Singh, B.N.; Singh, A.; Singh, B.R.; Singh, H.B. Trichoderma harzianum elicits induced resistance in sunflower challenged by Rhizoctonia solani. J. Appl. Microbiol. 2014, 116, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma-plant-pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, G.E.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef]
- Vallad, G.E.; Goodman, R.M. Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 2004, 44, 1920–1934. [Google Scholar] [CrossRef]
- Saikia, R.; Srivastava, A.K.; Singh, K.; Arora, D.K.; Lee, M.W. Effect of iron availability on induction of systemic resistance to Fusarium wilt of Chickpea by Pseudomonas spp. Mycobiology 2005, 33, 35–40. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Abbas, A.; Mubeen, M.; Zheng, H.X.; Sohail, M.A.; Shakeel, Q.; Solanki, M.K.; Iftikhar, Y.; Sharma, S.; Kashyap, B.K.; Hussain, S.; et al. Trichoderma spp. genes involved in the biocontrol activity against Rhizoctonia solani. Front. Microbiol. 2022, 13, 884469. [Google Scholar] [CrossRef]
- Manganiello, G.; Sacco, A.; Ercolano, M.R.; Vinale, F.; Lanzuise, S.; Pascale, A.; Napolitano, M.; Lombardi, N.; Lorito, M.; Woo, S.L. Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic acid. Front. Microbiol. 2018, 9, 1966. [Google Scholar] [CrossRef] [PubMed]
- Amira, M.B.; Lopez, D.; Mohamed, A.T.; Khouaja, A.; Chaar, H.; Fumanal, B.; Gousset-Dupont, A.; Bonhomme, L.; Label, P.; Goupil, P.; et al. Beneficial effect of Trichoderma harzianum strain Ths97 in biocontrolling Fusarium solani causal agent of root rot disease in olive trees. Biol. Control 2017, 110, 70–78. [Google Scholar] [CrossRef]
- Howell, C.R.; Hanson, L.E.; Stipanovic, R.D.; Puckhaber, L.S. Induction of terpenoid synthesis in cotton roots and control of Rhizoctonia solani by seed treatment with Trichoderma virens. Phytopathology 2000, 90, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.F.; Bai, X.L.; Jiao, S.; Li, Y.M.; Li, P.R.; Yang, Y.; Zhang, H.; Wei, G.H. A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 2021, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Aamir, M.; Shanmugam, V.; Dubey, M.K.; Husain, F.M.; Adil, M.; Ansari, W.A.; Rai, A.; Sah, P. Transcriptomic characterization of Trichoderma harzianum T34 primed tomato plants: Assessment of biocontrol agent induced host specific gene expression and plant growth promotion. BMC Plant Biol. 2023, 23, 552. [Google Scholar] [CrossRef]
- Pradhan, D.A.; Makandar, R. Delineating host responses induced by Trichoderma in castor through comparative transcriptome analysis. Rhizosphere 2023, 27, 100745–100761. [Google Scholar] [CrossRef]
- Niu, J.P.; Zhao, J.; Guo, Q.; Zhang, H.Y.; Yue, A.Q.; Zhao, J.Z.; Yin, C.C.; Wang, M.; Du, W.J. WGCNA reveals hub genes and key gene regulatory pathways of the response of soybean to infection by Soybean mosaic virus. Genes 2024, 15, 566. [Google Scholar] [CrossRef]
- Niu, J.P.; Yan, X.; Shi, Z.Y.; Song, S.J.; Liu, J.J.; Du, J.; Yuan, Y.X.; Shi, J.; Liang, J.P. Isolation and identification of pathogens causing Astragalus mongholicus root rot and analysis of its inhibition effects. J. Shanxi Agric. Sci. 2023, 51, 690–695. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.P.; Liu, L.M.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.X.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. IMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.B.; Li, Y.Q.; Liao, H.; Yang, Y.J. De novo transcriptome assembly of the bamboo snout beetle Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock. Biotechnol. Biofuels 2018, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Liang, J.P.; Li, W.Q.; Jia, X.Y.; Zhang, Y.; Zhao, J.P. Transcriptome sequencing and characterization of Astragalus mongholicus var. mongholicus root reveals key genes involved in flavonoids biosynthesis. Genes Genom. 2020, 42, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of flavonoids in plant resistance: A review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Wang, L.J.; Li, C.F.; Luo, K.M. Biosynthesis and metabolic engineering of isoflavonoids in model plants and crops: A review. Front. Plant Sci. 2024, 15, 1384091. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.Y.; Wei, C.H.; Amo, A.; Ahmed, B.; Yang, X.Z.; Zhang, X. Phenylpropanoid pathway engineering: An emerging approach towards plant defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef]
- Cristina, M.S.; Petersen, M.; Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [CrossRef]
- Ding, L.N.; Li, Y.T.; Wu, Y.Z.; Li, T.; Geng, R.; Cao, J.; Zhang, W.; Tan, X.L. Plant disease resistance-related signaling pathways: Recent progress and future prospects. Int. J. Mol. Sci. 2022, 23, 16200. [Google Scholar] [CrossRef]
- Chen, S.C.; Zhao, H.J.; Wang, Z.H.; Zheng, C.X.; Zhao, P.Y.; Guan, Z.H.; Qin, H.Y.; Liu, A.R.; Lin, X.M.; Ahammed, G.J. Trichoderma harzianum-induced resistance against Fusarium oxysporum involves regulation of nuclear DNA content, cell viability and cell cycle-related genes expression in cucumber roots. Eur. J. Plant Pathol. 2017, 147, 43–53. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, X.B.; Sun, Q.X.; Mou, Y.F.; Wang, J.; Yan, C.X.; Yuan, C.L.; Li, C.J.; Shan, S.H. Genome-wide identification of the LRR-RLK gene family in peanut and functional characterization of AhLRR-RLK265 in salt and drought stresses. Int. J. Biol. Macromol. 2024, 254, 127829. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Cui, X.X.; Lin, S.; Gan, S.P.; Xing, H.; Dou, D.L. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Navazio, L.; Szabo, I. The contribution of organelles to plant intracellular calcium signalling. J. Exp. Bot. 2018, 69, 4175–4193. [Google Scholar] [CrossRef] [PubMed]
- Navazio, L.; Baldan, B.; Moscatiello, R.; Zuppini, A.; Woo, S.L.; Mariani, P.; Lorito, M. Calcium-mediated perception and defense responses activated in plant cells by metabolite mixtures secreted by the biocontrol fungus Trichoderma atroviride. BMC Plant Biol. 2007, 7, 41. [Google Scholar] [CrossRef]
- Moscatiello, R.; Sello, S.; Ruocco, M.; Barbulova, A.; Cortese, E.; Nigris, S.; Baldan, B.; Chiurazzi, M.; Mariani, P.; Lorito, M.; et al. The hydrophobin HYTLO1 secreted by the biocontrol fungus Trichoderma longibrachiatum triggers a NAADP-mediated calcium signalling pathway in Lotus japonicus. Int. J. Mol. Sci. 2018, 19, 2596. [Google Scholar] [CrossRef]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzym. Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef]
- Gallou, A.; Cranenbrouck, S.; Declerck, S. Trichoderma harzianum elicits defence response genes in roots of potato plantlets challenged by Rhizoctonia solani. Eur. J. Plant Pathol. 2009, 124, 219–230. [Google Scholar] [CrossRef]
- Morán-Diez, E.; Rubio, B.; Domínguez, S.; Hermosa, R.; Monte, E.; Nicolás, C. Transcriptomic response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 2012, 169, 614–620. [Google Scholar] [CrossRef]
- Yang, D.; Chen, T.; Wu, Y.S.; Tang, H.Q.; Yu, J.Y.; Dai, X.Q.; Zheng, Y.X.; Wan, X.R.; Yang, Y.; Tan, X.D. Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum. BMC Genom. 2024, 25, 200–236. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Y.; Shao, J.; Luo, M.Y.; Chen, D.S.; Tang, D.Z.; Shi, H. Brassionsteroid-signaling kinase1 associates with and is required for cysteine protease response to dehydration 19-mediated disease resistance in Arabidopsis. Plant Sci. 2024, 342, 112033. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; Luca, V.D.; Despres, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Kinkema, M.; Fan, W.H.; Dong, X.N. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 20, 2339–2350. [Google Scholar] [CrossRef]
- Li, Y.Q.; Liu, Y.A.; Jin, L.B.; Peng, R.Y. Crosstalk between Ca2+ and other regulators assists plants in responding to abiotic stress. Plants 2022, 11, 1351. [Google Scholar] [CrossRef]
- Marcec, M.J.; Tanaka, K. Crosstalk between calcium and ROS signaling during flg22-triggered immune response in Arabidopsis leaves. Plants 2022, 11, 14. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Bujarski, J.J.; Frankowska-Łukawska, J.; Torres, M.A. Respiratory burst oxidase homologs RBOHD and RBOHF as key modulating components of response in turnip mosaic virus—Arabidopsis thaliana (L.) Heyhn System. Int. J. Mol. Sci. 2020, 21, 8510. [Google Scholar] [CrossRef]
- Sehrish, A.; Wei, Y.; Yuan, Y.; Khan, M.T.; Qin, L.F.; Powell, C.A.; Chen, B.S.; Zhang, M.Q. Gene expression profiling of reactive oxygen species (ROS) and antioxidant defense system following Sugarcane mosaic virus (SCMV) infection. BMC Plant Biol. 2020, 20, 532. [Google Scholar]
- Garg, N.; Manchanda, G. ROS generation in plants: Boon or bane? Plant Biosyst. 2009, 143, 81–96. [Google Scholar] [CrossRef]
- Huang, J.L.; Gu, M.; Lai, Z.B.; Fan, B.F.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z.X. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Dutra, J.M.; Espitia, P.J.P.; Batista, R.A. Formononetin: Biological effects and uses—A review. Food Chem. 2021, 359, 129975. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.X.; Shi, X.C.; Xu, D.; Laborda, P.; Wu, G.C.; Liu, F.Q.; Laborda, P.; Wang, S.Y. Antibacterial mechanism of biochanin A and its efficacy for the control of Xanthomonas axonopodis pv. glycines in soybean. Pest Manag. Sci. 2021, 77, 1668–1673. [Google Scholar] [CrossRef]
- Weidenbörner, M.; Hindorf, H.; Jha, H.C.; Tsotsonos, P.; Egge, H. Antifungal activity of isoflavonoids in different reduced stages on Rhizoctonia solani and Sclerotium rolfsii. Phytochemistry 1990, 29, 801–803. [Google Scholar] [CrossRef]
- Cho, S.M.; Shin, S.H.; Kim, K.S.; Kim, Y.C.; Eun, M.Y.; Cho, B.H. Enhanced expression of a gene encoding a nucleoside diphosphate kinase 1 (OsNDPK1) in rice plants upon infection with bacterial pathogens. Mol. Cells 2004, 18, 390–395. [Google Scholar] [CrossRef]
- Gan, P.F.; Tang, C.L.; Lu, Y.; Ren, C.R.; Nasab, H.R.; Kun, X.F.; Wang, X.D.; Li, L.Z.; Kang, Z.S.; Wang, X.J.; et al. Quantitative phosphoproteomics reveals molecular pathway network in wheat resistance to stripe rust. Stress Biol. 2024, 4, 32–47. [Google Scholar] [CrossRef]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.; Wang, R.; Liu, Y.; Zhang, S. Mitogen-activated protein kinase 3 and 6 regulate botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Diretto, G.; Digilio, M.C.; Woo, S.L.; Giuliano, G.; Molisso, D.; Pennacchio, F.; Lorito, M.; Rao, R. Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 2019, 10, 745. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.X.; Zhang, L.C.; Ji, Y.L.; Tan, D.M.; Yuan, H.; Wang, A.D. The jasmonate-activated transcription factor MdMYC2 regulates ethylene response factor and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Oñate-Sánchez, L.; Anderson, J.P.; Young, J.; Singh, K.B. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007, 143, 400–409. [Google Scholar] [CrossRef]
- Song, C.; Cao, Y.P.; Dai, J.; Li, G.H.; Manzoor, M.A.; Chen, C.W.; Deng, H. The multifaceted roles of MYC2 in plants: Toward transcriptional reprogramming and stress tolerance by jasmonate signaling. Front. Plant Sci. 2022, 13, 868874. [Google Scholar] [CrossRef]
- Wu, F.M.; Deng, L.; Zhai, Q.Z.; Zhao, J.H.; Chen, Q.; Li, C.Y. Mediator subunit MED25 couples alternative splicing of JAZ genes with fine-tuning of jasmonate signaling. Plant Cell 2020, 32, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssieres, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.J.; Mijiti, G.; Huang, Y.; Fan, H.J.; Wang, Y.C.; Liu, Z.H. Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536. Sci. Rep. 2018, 8, 7974. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Van, D.E.S.; Van, L.L.C.; Pieterse, C.M. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef]
- Ayash, M.; Abukhalaf, M.; Thieme, D.; Proksch, C.; Heilmann, M.; Schattat, M.H.; Hoehenwarter, W. LC-MS based draft map of the Arabidopsis thaliana nuclear proteome and protein import in pattern triggered immunity. Front. Plant Sci. 2021, 12, 744103. [Google Scholar] [CrossRef]
- Fakih, Z.; Plourde, M.B.; Germain, H. Differential participation of plant ribosomal proteins from the small ribosomal subunit in protein translation under stress. Biomolecules 2023, 13, 1160. [Google Scholar] [CrossRef]
- Carroll, A.J. The Arabidopsis cytosolic ribosomal proteome: From form to function. Front. Plant Sci. 2013, 4, 32. [Google Scholar] [CrossRef]
- Rajamäki, M.L.; Xi, D.H.; Sikorskaite-Gudziuniene, S.; Valkonen, J.P.T.; Whitham, S.A. Differential requirement of the ribosomal protein S6 and ribosomal protein S6 kinase for plant-virus accumulation and interaction of S6 kinase with potyviral VPg. Mol. Plant-Microbe Interact. 2017, 30, 374–384. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, C.; Dittman, J.D.; Whitham, S.A. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology 2009, 390, 163–173. [Google Scholar] [CrossRef]
- Solano, C.M.T.; Adame-García, J.; Gregorio-Jorge, J.; Jiménez-Jacinto, V.; Vega-Alvarado, L.; Iglesias-Andreu, L.; Escobar-Hernández, E.E.; Luna-Rodríguez, M. Increase in ribosomal proteins activity: Translational reprogramming in Vanilla planifolia Jacks., against Fusarium infection. BioRxiv 2019. [Google Scholar] [CrossRef]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Roatti, B.; Michele, P.; Cesare, G.; Ilaria, P. Abiotic stresses affect Trichoderma harzianum T39-induced resistance to downy mildew in grapevine. Phytopathology 2013, 103, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Pani, S.; Kumar, A.; Sharma, A. Trichoderma harzianum: An overview. Bull. Environ. Pharmacol. Life Sci. 2021, 10, 32–39. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, J.; Yan, X.; Bai, Y.; Li, W.; Lu, G.; Wang, Y.; Liu, H.; Shi, Z.; Liang, J. Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani. Genes 2024, 15, 1180. https://doi.org/10.3390/genes15091180
Niu J, Yan X, Bai Y, Li W, Lu G, Wang Y, Liu H, Shi Z, Liang J. Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani. Genes. 2024; 15(9):1180. https://doi.org/10.3390/genes15091180
Chicago/Turabian StyleNiu, Jingping, Xiang Yan, Yuguo Bai, Wandi Li, Genglong Lu, Yuanyuan Wang, Hongjun Liu, Zhiyong Shi, and Jianping Liang. 2024. "Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani" Genes 15, no. 9: 1180. https://doi.org/10.3390/genes15091180
APA StyleNiu, J., Yan, X., Bai, Y., Li, W., Lu, G., Wang, Y., Liu, H., Shi, Z., & Liang, J. (2024). Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani. Genes, 15(9), 1180. https://doi.org/10.3390/genes15091180





