A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Cloning of IbMYB330 and Its Promoter
2.3. Bioinformatics Analysis
2.4. Expression Analysis of IbMYB330
2.5. Construction of Overexpression Vector
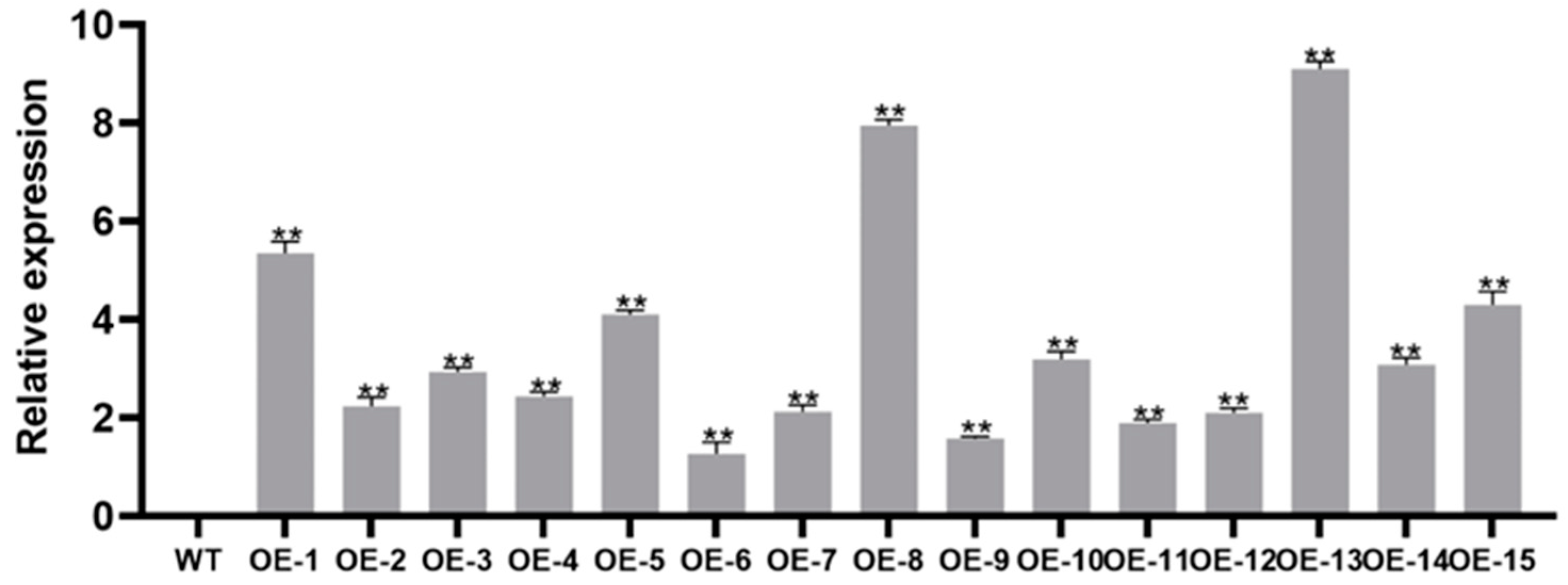
2.6. Production of Transgenic Plants
2.7. Statistical Analysis
3. Results
3.1. Isolation and Characterization of IbMYB330
3.2. Expression Analysis of the IbMYB330 in Sweet Potato
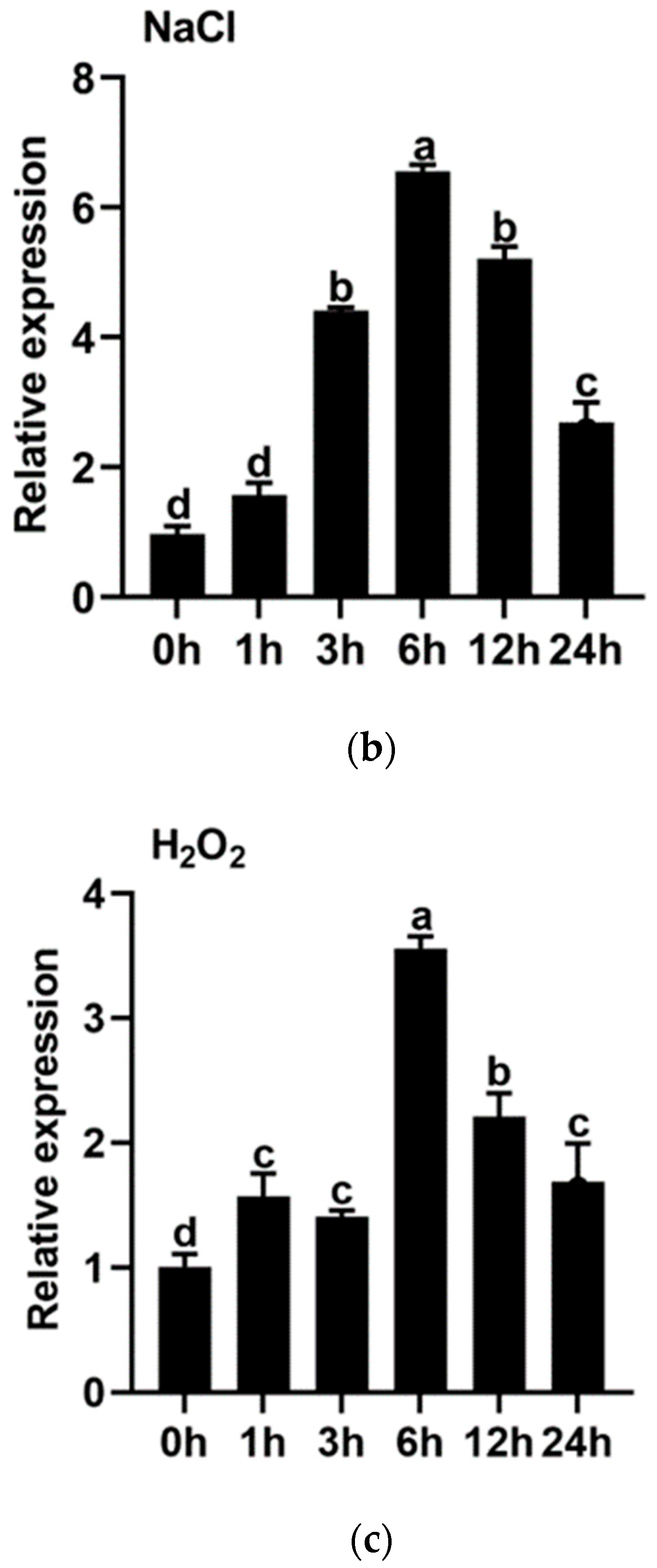
3.3. Expression Profiles of IbMYB330 under Abiotic Stresses
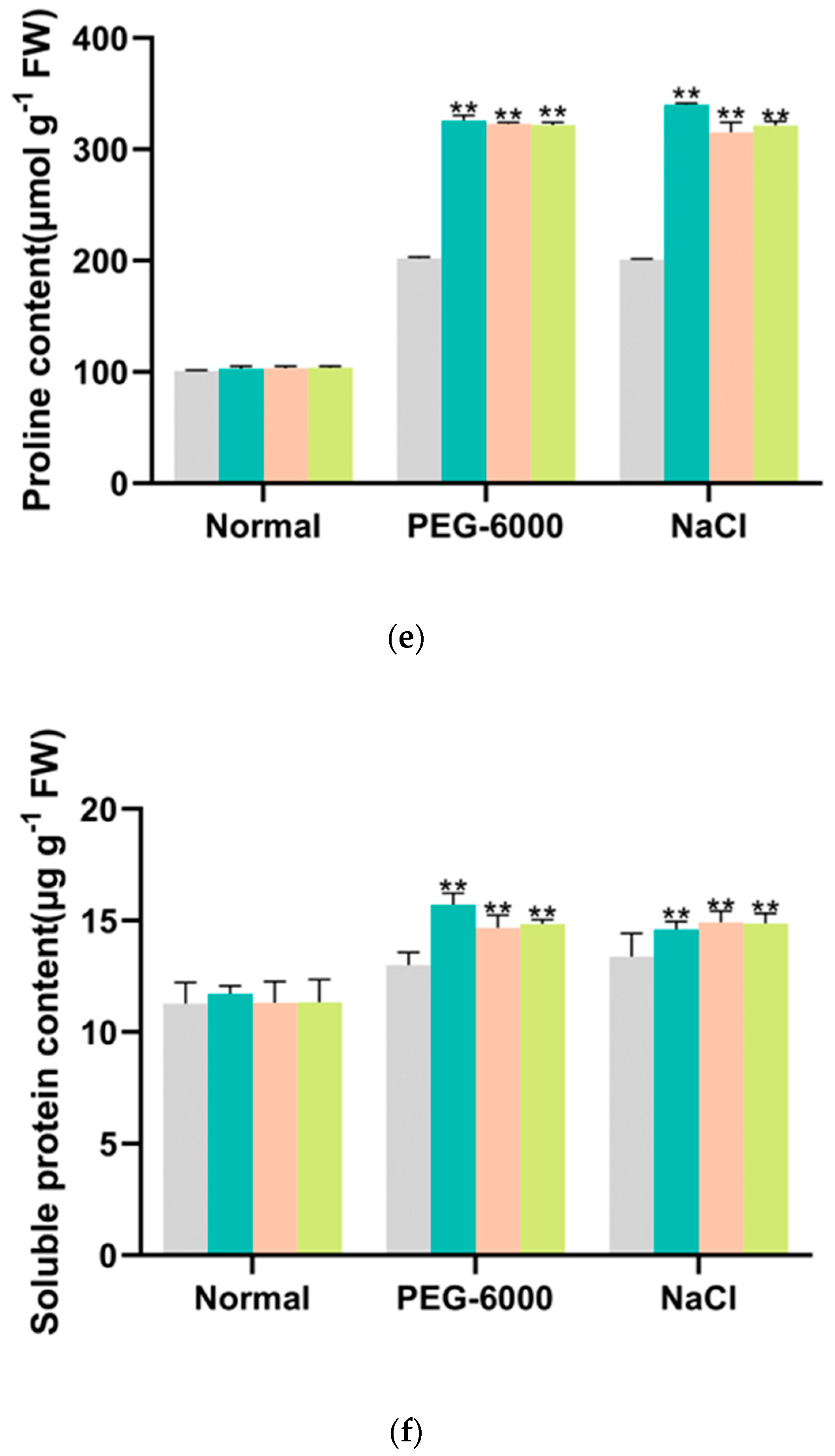
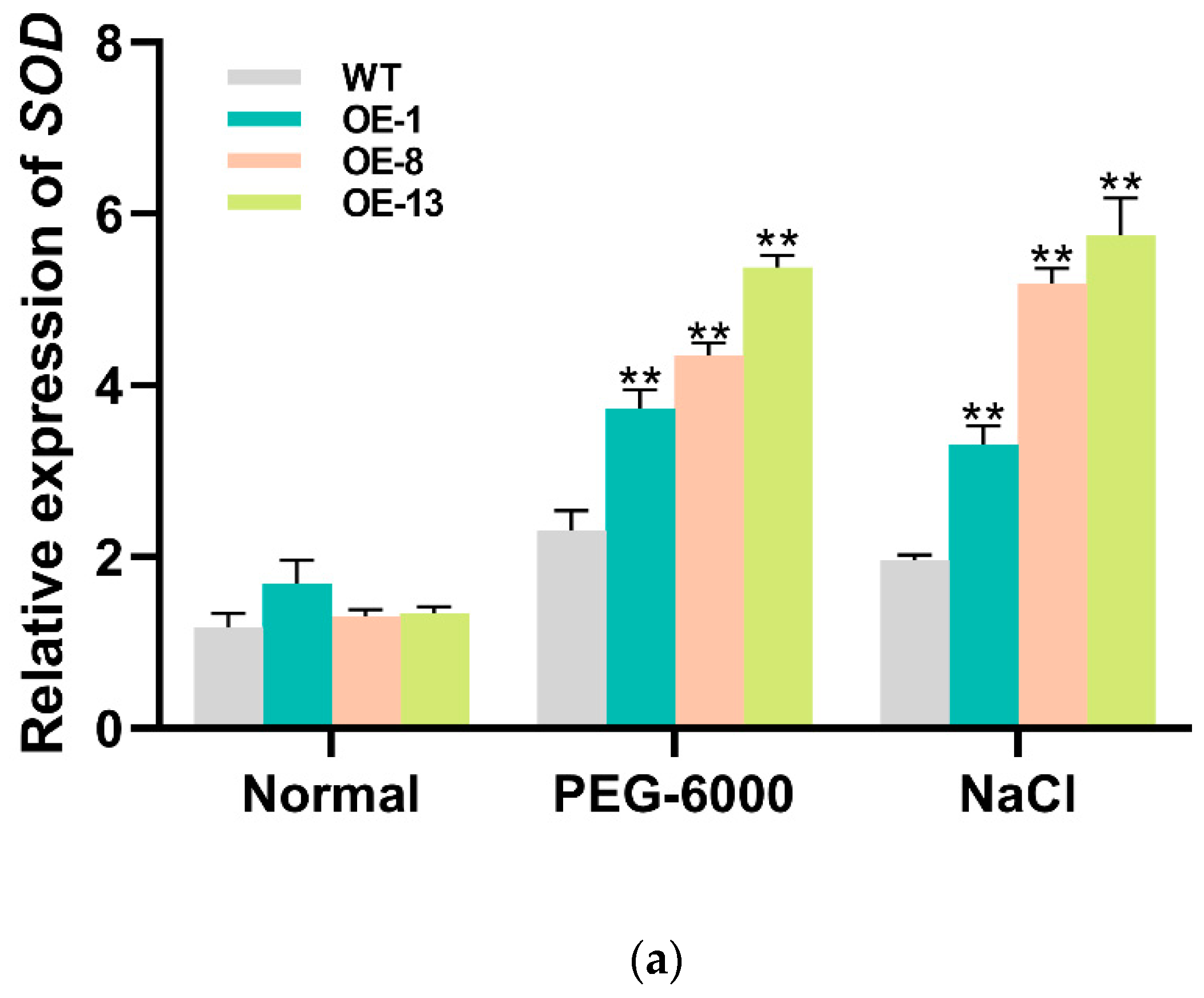
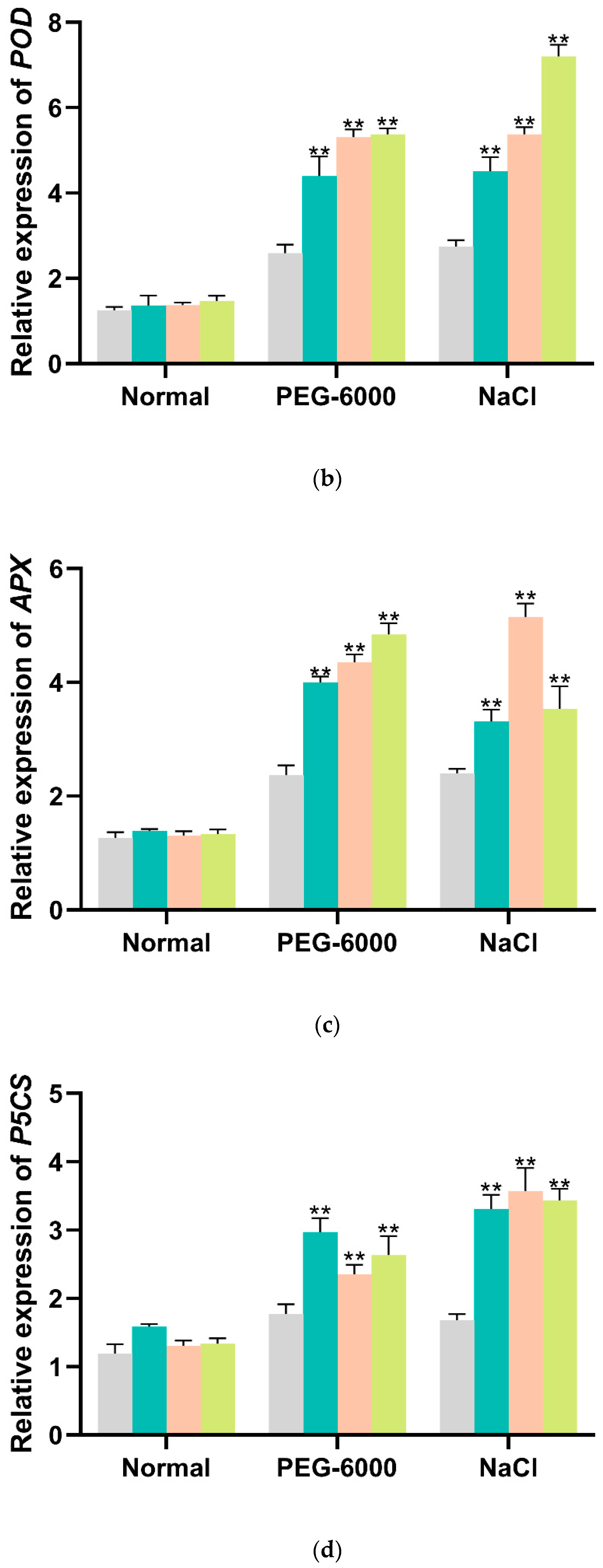
3.4. Overexpression of IbMYB330 Improves Tolerance to Drought and Salt Stress in Transgenic Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.X.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Xiong, L.M.; Li, W.B.; Zhu, J.K.; Zhu, J.H. The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 2011, 23, 1971–1984. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Niu, Y.L.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jiang, C.K.; Rao, G.Y. Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef]
- Zhu, Y.; Bao, Y. Genome-wide mining of MYB transcription factors in the anthocyanin biosynthesis pathway of Gossypium hirsutum. Biochem. Genet. 2021, 59, 678–696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Cui, Z.; Li, Y.X.; Kang, Y.Q.; Song, X.Q.; Wang, J.; Zhou, Y. Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 2021, 12, 714696. [Google Scholar] [CrossRef]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB transcription factors-functions outside the DNA-binding domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Peng, R.H.; Tian, Y.S.; Han, H.J.; Xu, J.; Yao, Q.H. Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.H.; Wang, Z.G.; Tang, X.F.; Yu, L.; Qi, G.; Wang, D.; Yan, X.F.; Kong, Y.Z.; Zhou, G.K. R2R3-MYB gene pairs in Populus: Evolution and contribution to secondary wall formation and flowering time. J. Exp. Bot. 2014, 65, 4255–4269. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Feng, B.R.; Yang, S.S.; Huang, Y.B.; Tang, Y.X. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.W.; Yi, D.X.; Shao, L.H.; Li, C. In silico genome-wide identification, phylogeny and expression analysis of the R2R3-MYB gene family in Medicago truncatula. J. Integr. Agric. 2017, 16, 1576–1591. [Google Scholar] [CrossRef]
- Abubakar, A.S.; Feng, X.; Gao, G.; Yu, C.; Chen, J.; Chen, K.; Wang, X.; Mou, P.; Shao, D.; Chen, P.; et al. Genome wide characterization of R2R3 MYB transcription factor from Apocynum venetum revealed potential stress tolerance and flavonoid biosynthesis genes. Genomics 2022, 114, 110275. [Google Scholar] [CrossRef]
- Dias, A.P.; Braun, E.L.; McMullen, M.D.; Grotewold, E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003, 131, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Yang, Y.; Li, Y.; Wang, G.; Peng, M. Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental-stress-induced abscission. Sci. Rep. 2016, 6, 32006. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Fan, S.L.; Yu, H.Y.; Lu, Y.X.; Wang, L.F. HbMYB44, a rubber tree MYB transcription factor with versatile functions in modulating multiple phytohormone signaling and abiotic stress responses. Front. Plant Sci. 2022, 13, 893896. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Jung, C.K.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Zhong, M.; Qu, L.; Yang, J.X.; Liu, X.Q.; Zhao, Q.; Liu, X.M.; Zhao, X.Y. AtMYB32 regulates the ABA response by targeting ABI3, ABI4 and ABI5 and the drought response by targeting CBF4 in Arabidopsis. Plant Sci. 2021, 310, 110983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, R.L.; Yang, X.P.; Ju, Q.; Li, W.Q.; Lu, S.Y.; Tran, L.P.; Xu, J. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Kang, J.Y.; Kim, S.Y. Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance. Mol. Cells 2011, 31, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.H.; Liu, N.; Wang, L.J.; Li, J.; Zheng, X.J.; Xiang, F.N.; Liu, Z.H. MYB94 and MYB96 additively inhibit callus formation via directly repressing LBD29 expression in Arabidopsis thaliana. Plant Sci. 2020, 293, 110323. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, G.; Biricolti, S.; Locatelli, F.; Baldoni, E.; Mattana, M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008, 27, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Jian, L.; Kang, K.Y.; Choi, Y.M.; Suh, M.C.; Paek, N.C. Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis. Plant J. 2022, 112, 339–351. [Google Scholar] [CrossRef] [PubMed]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef]
- Zhao, L.; Song, Z.B.; Wang, B.W.; Gao, Y.L.; Shi, J.L.; Sui, X.Y.; Chen, X.J.; Zhang, Y.H.; Li, Y.P. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco (Nicotiana tabacum L.). Front. Plant Sci. 2021, 12, 819247. [Google Scholar]
- Ahmad, I.; Soni, S.K.; Pandey, D. In-silico mining and characterization of MYB family genes in wilt-resistant hybrid guava (Psidium guajava× Psidium molle). J. Genet. Eng. Biot. 2023, 21, 74. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.L.; Zheng, J.L.; Sun, Z.; Fan, W.J.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Leite, C.E.C.; Souza, B.K.F.; Manfio, C.E.; Wamser, G.H.; Alves, D.P.; de Francisco, A. Sweet potato new varieties screening based on morphology, pulp color, proximal composition, and total dietary fiber content via factor analysis and principal component analysis. Front. Plant Sci. 2022, 13, 852709. [Google Scholar] [CrossRef] [PubMed]
- Mohanraj, R.; Sivasankar, S. Sweet potato (Ipomoea batatas [L.] Lam)—A valuable medicinal food: A rievew. J. Med. Food 2014, 17, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.X.; Nie, H.Z.; Wang, Y.Z.; Wang, X.Y.; Jarret, R.; Zhao, J.M.; Wang, H.X.; Yang, J. Exploring and exploiting genetics and genomics for sweetpotato improvement: Status and perspectives. Plant Commun. 2022, 3, 100332. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.H.; Del Rosario Herrera, M.; Crisovan, E.; Wu, S.; Fei, Z.; Khan, M.A.; Buell, C.R.; Gemenet, D.C. Transcriptomic analysis of sweet potato under dehydration stress identifies candidate genes for drought tolerance. Plant Direct. 2018, 2, e00092. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.J.; Kim, M.D.; Deng, X.P.; Kwak, S.O.; Chen, W. Enhanced salt stress tolerance in transgenic potato plants expressing IbMYB1, a sweet potato transcription factor. J. Microbiol. Biotechnol. 2013, 23, 1737–1746. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Zhu, H.; He, S.Z.; Zhai, H.; Zhao, N.; Xing, S.H.; Wei, Z.H.; Liu, Q.C. A novel sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Fron. Plant Sci. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Zhao, H.Q.; Hu, Y.F.; Zhang, S.S.; He, S.Z.; Zhang, H.; Zhao, N.; Liu, Q.C.; Gao, S.P.; Zhai, H. Expression of the sweet potato MYB transcription factor IbMYB48 confers salt and drought tolerance in Arabidopsis. Genes 2022, 13, 1883. [Google Scholar] [CrossRef]
- Wang, C.L.; Hou, X.M.; Qi, N.N.; Li, C.X.; Luo, Y.; Hu, D.L.; Li, Y.H.; Liao, W.B. An optimized method to obtain high-quality RNA from different tissues in Lilium davidii var. unicolor. Sci. Rep. 2022, 12, 2825. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, X.R.; Zhi, Y.H.; Li, X.; Zhang, Q.; Niu, J.B.; Wang, J.; Zhai, H.; Zhao, N.; Li, J.G.; et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Bing, D.U.; Sun, S.; He, S.Z.; Zhao, N.; Liu, Q.C.; Zhai, H. A novel aldo-keto reductase gene, IbAKR, from sweet potato confers higher tolerance to cadmium stress in tobacco. Front. Agric. Sci. Eng. 2018, 5, 206–213. [Google Scholar]
- Zhang, L.; Donald, B.F. Connecting proline metabolism and signaling pathways in plant senescence. Front. Plant Sci. 2015, 6, 552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.C.; Deng, X.X.; Wang, J.G. The effects of mepiquat chloride (DPC) on the soluble protein content and the activities of protective enzymes in cotton in response to aphid feeding and on the activities of detoxifying enzymes in aphids. BMC Plant Biol. 2022, 22, 213. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Hrmova, M.; Hussain, S.S. Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.F.; Geng, F.; Cheng, Y.J.; Wang, J.Q. Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 166, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, Y.; Xu, L.; Wang, J.; Qi, H.; Guo, J.; Zhang, L.; Shen, J.; Wang, H.; Zhang, F.; et al. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice. Mol. Plant 2022, 15, 1227–1242. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.X.; Zou, J.J.; Wang, J.X.; Yang, K.Z.; Wang, X.Q.; Le, J. A rice R2R3-Type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC. Int. J. Mol. Sci. 2022, 23, 5873. [Google Scholar] [CrossRef]
- Chen, X.J.; Wang, P.J.; Gu, M.Y.; Lin, X.Y.; Hou, B.H.; Zheng, Y.C.; Sun, Y.; Jin, S.; Ye, N.X. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.J.; Lei, J.; Chai, S.S.; Jin, X.J.; Zou, Y.Y.; Sun, X.Q.; Mei, Y.Q.; Cheng, X.L.; Yang, X.S.; et al. IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco. Genes 2022, 13, 1476. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Yang, X.; Wang, X.; Li, Q.Y.; Guo, J.Y.; Ma, T.; Zhao, C.M.; Tang, Y.Y.; Qiao, L.X.; Wang, J.S.; et al. The sweetpotato β-amylase gene IbBAM1.1 enhances drought and salt stress resistance by regulating ROS homeostasis and osmotic balance. Plant Physiol. Biochem. 2021, 168, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell. Biol. 2022, 23, 633–679. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Ali Wani, O.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: From source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Krasylenko, Y.; Zeiner, A.; Samaj, J.; Takac, T. Signaling toward reactive oxygen species-scavenging enzymes in plants. Front. Plant Sci. 2020, 11, 618835. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Xue, M.F.; Zhang, X.F.; Chen, S.B. Genome-wide analysis of the metallothionein gene family in cassava reveals its role in response to physiological stress through the regulation of reactive oxygen species. BMC Plant Biol. 2023, 23, 227. [Google Scholar] [CrossRef]
- Zhou, J.H.; Qiao, J.Z.; Wang, J.; Quan, R.D.; Huang, R.F.; Qin, H. OsQHB improves salt tolerance by scavenging reactive oxygen species in rice. Front. Plant Sci. 2022, 13, 848891. [Google Scholar] [CrossRef]
- Chen, K.; Su, C.; Tang, W.; Zhou, Y.B.; Xu, Z.S.; Chen, J.; Li, H.Y.; Chen, M.; Ma, Y.Z. Nuclear transport factor GmNTF2B-1 enhances soybean drought tolerance by interacting with oxidoreductase GmOXR17 to reduce reactive oxygen species content. Plant J. 2021, 107, 740–759. [Google Scholar] [CrossRef]
- Abdel-Hameed, A.A.E.; Prasad, K.; Jiang, Q.; Reddy, A.S.N. Salt-Induced stability of SR1/CAMTA3 mRNA is mediated by reactive oxygen species and requires the 3′ end of its open reading frame. Plant Cell Physiol. 2020, 61, 748–760. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, A.I.; Rafudeen, M.S.; Gomaa, A.M.; Hasanuzzaman, M. Exogenous melatonin enhances the reactive oxygen species metabolism, antioxidant defense-related gene expression, and photosynthetic capacity of Phaseolus vulgaris L. to confer salt stress tolerance. Physiol. Plant 2021, 173, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Tan, P.P.; Chang, L.; Yue, Z.M.; Zeng, C.Z.; Li, M.; Liu, Z.X.; Dong, X.J.; Yan, M.L. Exogenous proline mitigates toxic effects of cadmium via the decrease of cadmium accumulation and reestablishment of redox homeostasis in Brassica juncea. BMC Plant Biol. 2022, 22, 182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Gong, X.Q.; Su, X.J.; Yu, H.X.; Cheng, S.Y.; Huang, J.W.; Li, D.Y.; Lei, Z.L.; Li, M.J.; Ma, F.W. The ubiquitin-binding protein MdRAD23D1 mediates drought response by regulating degradation of the proline-rich protein MdPRP6 in apple (Malus domestica). Plant Biotechnol. J. 2023, 21, 1560–1576. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.S.; Ahmad, A.; Hayat, S.; Tahir, I. Epibrassinolide and proline alleviate the photosynthetic and yield inhibition under salt stress by acting on antioxidant system in mustard. Plant Physiol. Biochem. 2019, 135, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Meng, R.; Liu, Y.; Chen, S.; Jiang, J.; Wang, L.; Zhao, S.; Wang, Z.; Fang, W.; Chen, F.; et al. Heterografted chrysanthemums enhance salt stress tolerance by integrating reactive oxygen species, soluble sugar, and proline. Hortic. Res. 2022, 9, uhac073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, R.H.; Song, Z.; Fu, W.D.; Yun, L.L.; Gao, J.H.; Hu, G.; Wang, Z.H.; Wu, H.W.; Zhang, G.L.; et al. Iris lactea var. chinensis plant drought tolerance depends on the response of proline metabolism, transcription factors, transporters and the ROS-scavenging system. BMC Plant Biol. 2023, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Adamipour, N.; Khosh-Khui, M.; Salehi, H.; Razi, H.; Karami, A.; Moghadam, A. Metabolic and genes expression analyses involved in proline metabolism of two rose species under drought stress. Plant Physiol. Biochem. 2020, 155, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.J.; Xie, Y.H.; Jiang, H.; Wang, X.; Hao, Y.J.; Zhang, Z.; You, C.X. The ankyrin repeat-containing protein MdANK2B regulates salt tolerance and ABA sensitivity in Malus domestica. Plant Cell Rep. 2021, 40, 405–419. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Liu, Y.J.; Zhang, J.C.; Wang, X.F.; You, C.X.; Hao, Y.J. MdABI5 works with its interaction partners to regulate abscisic acid-mediated leaf senescence in apple. Plant J. 2021, 105, 1566–1581. [Google Scholar] [CrossRef]
- Cheng, L.Q.; Li, X.X.; Huang, X.; Ma, T.; Liang, Y.; Ma, X.Y.; Peng, X.J.; Jia, J.T.; Chen, S.Y.; Chen, Y.; et al. Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 70, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.D.; Jiang, Y.L.; Liang, Y.N.; Chen, L.; Chen, W.J.; Cheng, B.J. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef] [PubMed]
 ), O. sativa (
), O. sativa ( ), Z. mays (
), Z. mays ( ), I. triloba (
), I. triloba ( ), I. nil (
), I. nil ( ), G. hirsutum (
), G. hirsutum ( ), G. raimondii (
), G. raimondii ( ), S. tuberosum (
), S. tuberosum ( ), and N. tabacum (
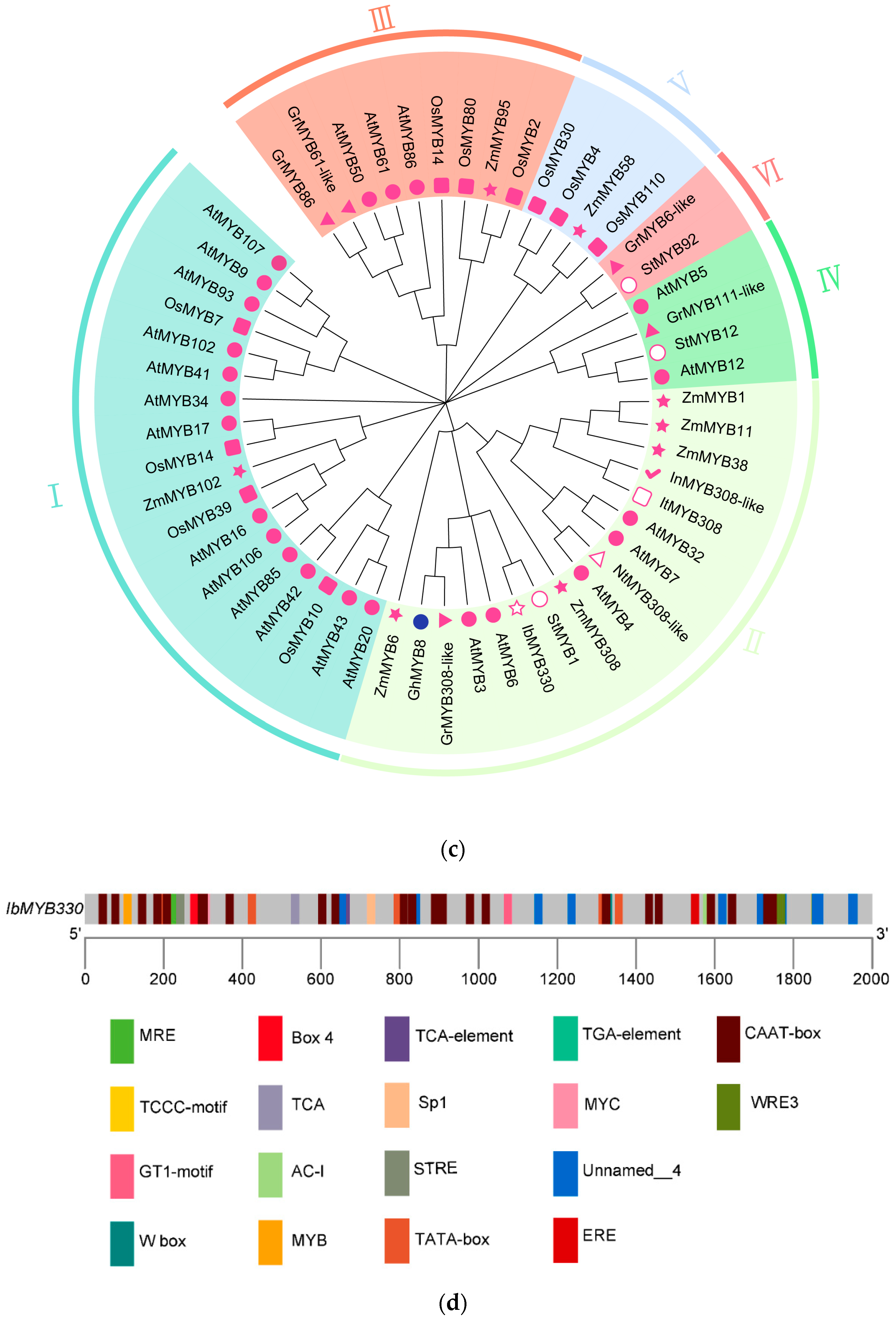
), and N. tabacum ( ). (d) Cis-acting elements in the promoter of IbMYB330.
). (d) Cis-acting elements in the promoter of IbMYB330.
 ), O. sativa (
), O. sativa ( ), Z. mays (
), Z. mays ( ), I. triloba (
), I. triloba ( ), I. nil (
), I. nil ( ), G. hirsutum (
), G. hirsutum ( ), G. raimondii (
), G. raimondii ( ), S. tuberosum (
), S. tuberosum ( ), and N. tabacum (
), and N. tabacum ( ). (d) Cis-acting elements in the promoter of IbMYB330.
). (d) Cis-acting elements in the promoter of IbMYB330.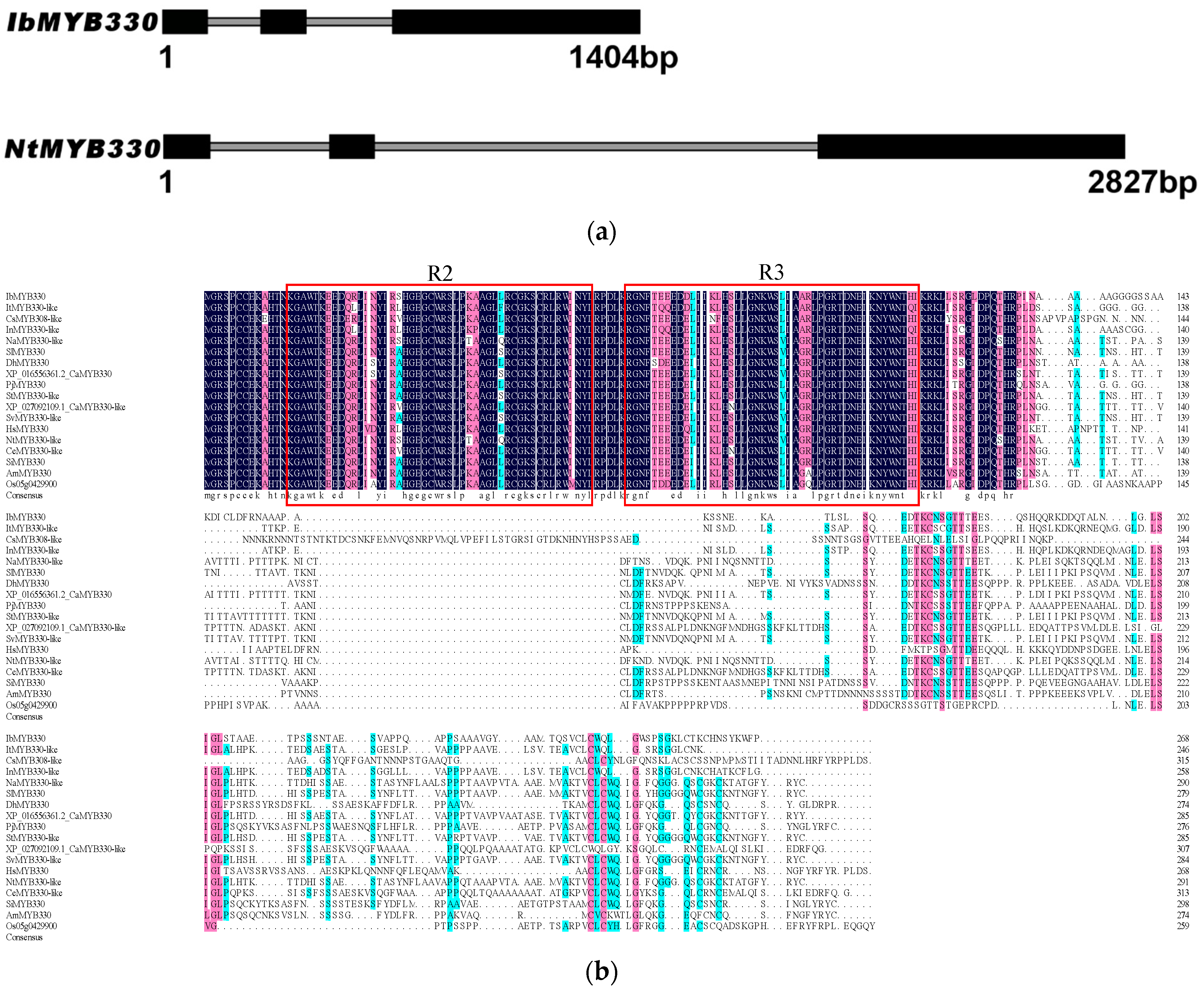







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Lei, J.; Jin, X.; Chai, S.; Jiao, C.; Yang, X.; Wang, L. A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco. Genes 2024, 15, 693. https://doi.org/10.3390/genes15060693
Wang C, Lei J, Jin X, Chai S, Jiao C, Yang X, Wang L. A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco. Genes. 2024; 15(6):693. https://doi.org/10.3390/genes15060693
Chicago/Turabian StyleWang, Chong, Jian Lei, Xiaojie Jin, Shasha Chai, Chunhai Jiao, Xinsun Yang, and Lianjun Wang. 2024. "A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco" Genes 15, no. 6: 693. https://doi.org/10.3390/genes15060693
APA StyleWang, C., Lei, J., Jin, X., Chai, S., Jiao, C., Yang, X., & Wang, L. (2024). A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco. Genes, 15(6), 693. https://doi.org/10.3390/genes15060693



