Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response
Abstract
1. Introduction
2. Materials and Methods
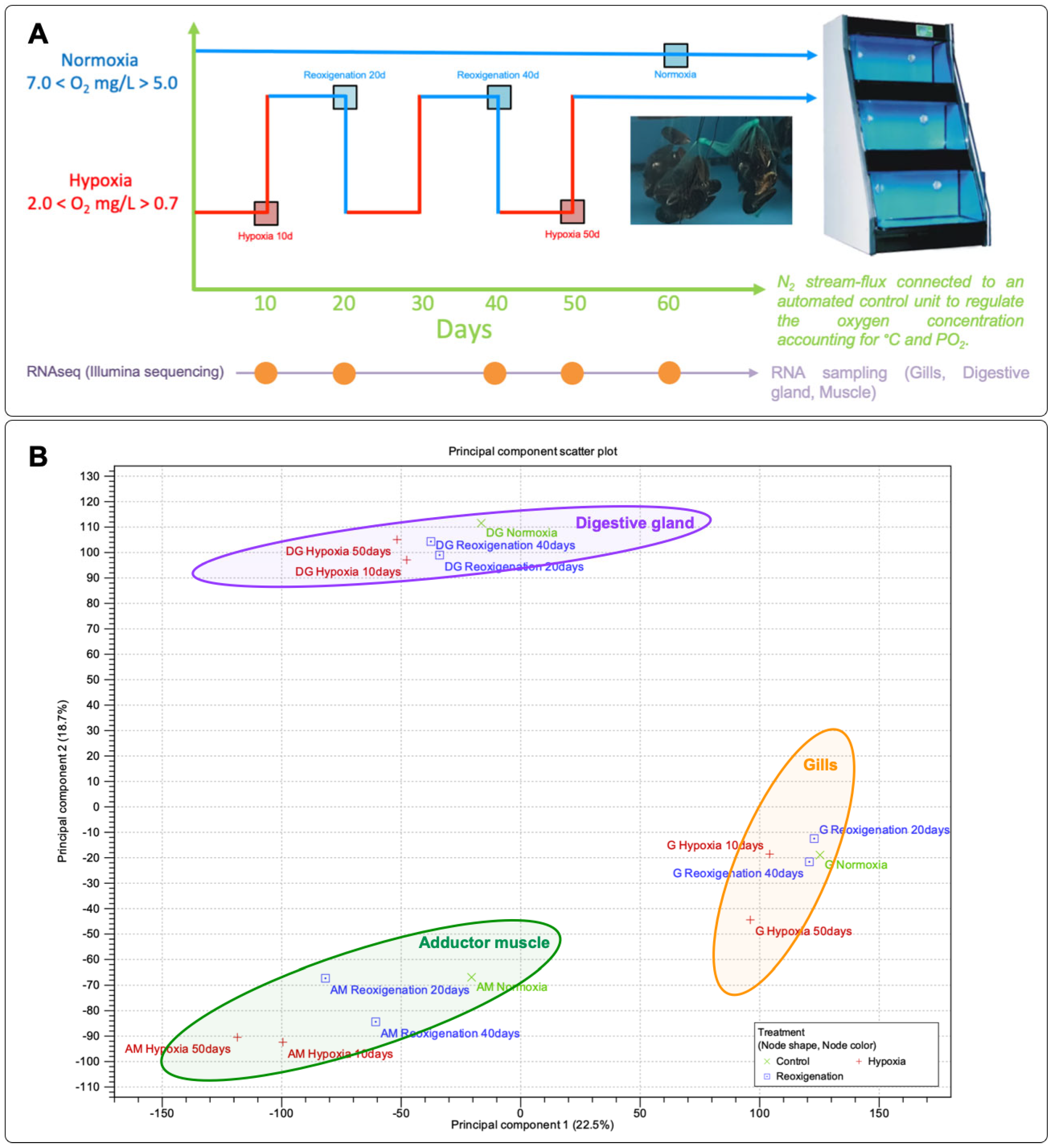
2.1. Mussel Acclimation, Hypoxia Challenge, and Sample Preparation
2.2. RNA Extraction and Library Preparation
2.3. Transcriptome Analysis and Gene Ontology Annotation
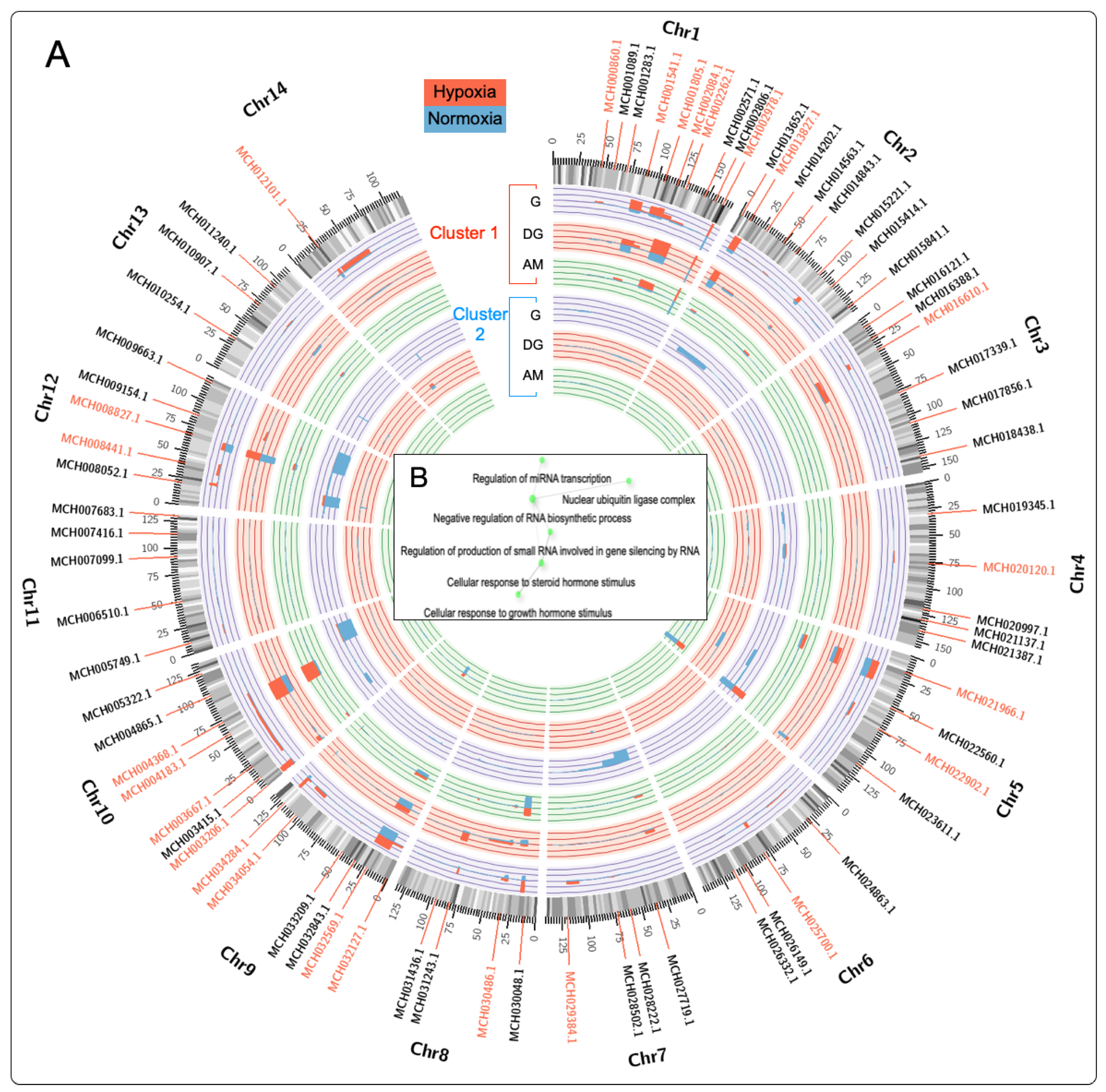
2.4. Chromosome Gene Expression (CGE) Analysis
2.5. Data Availability
3. Results
3.1. Principal Component Analysis (PCA) of Gene Expression Profiles in M. chilensis Tissues under Hypoxia and Reoxygenation
3.2. Differential Regulation of Transcripts under Normoxia and Hypoxia Conditions in Multiple Tissues of M. chilensis
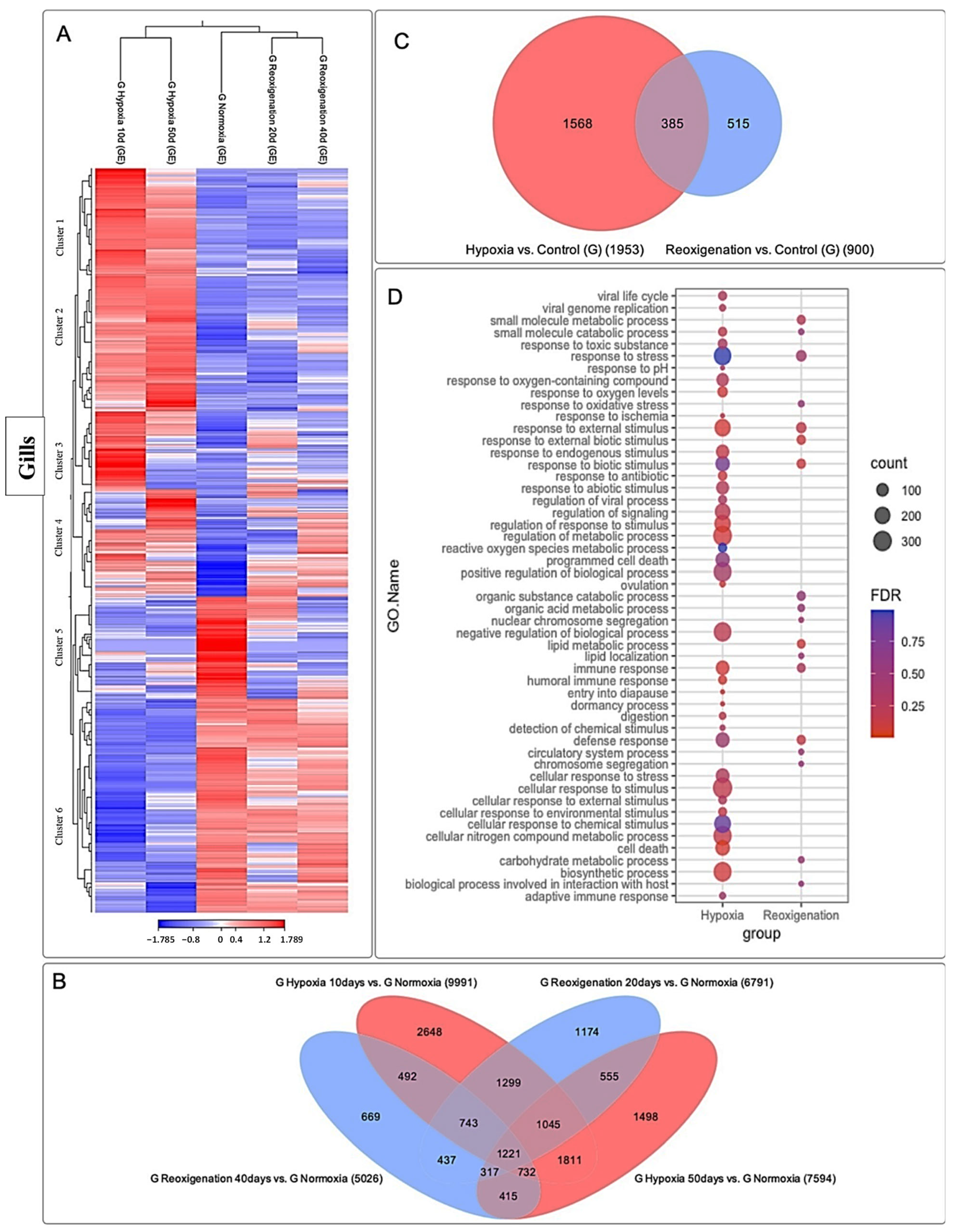
3.3. Differential Expression Analysis of Transcripts Expressed in M. chilensis Gills under Hypoxic and Reoxygenation Conditions
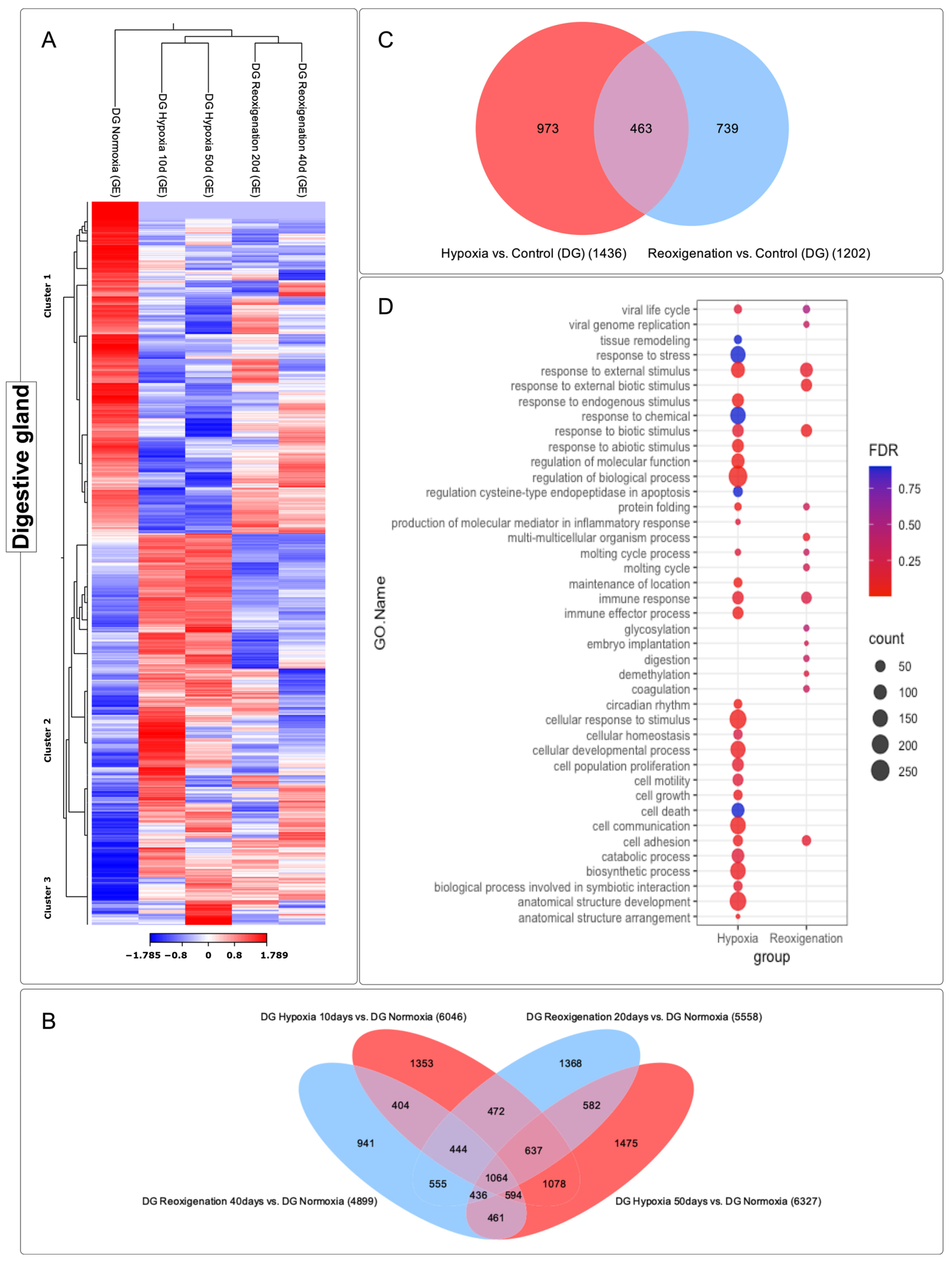
3.4. Differential Expression Analysis of Transcripts Observed in the Digestive Gland of M. chilensis under Hypoxic and Reoxygenation Conditions
3.5. GO Enrichment Analysis in the Digestive Gland of M. chilensis under Hypoxia and Reoxygenation Conditions
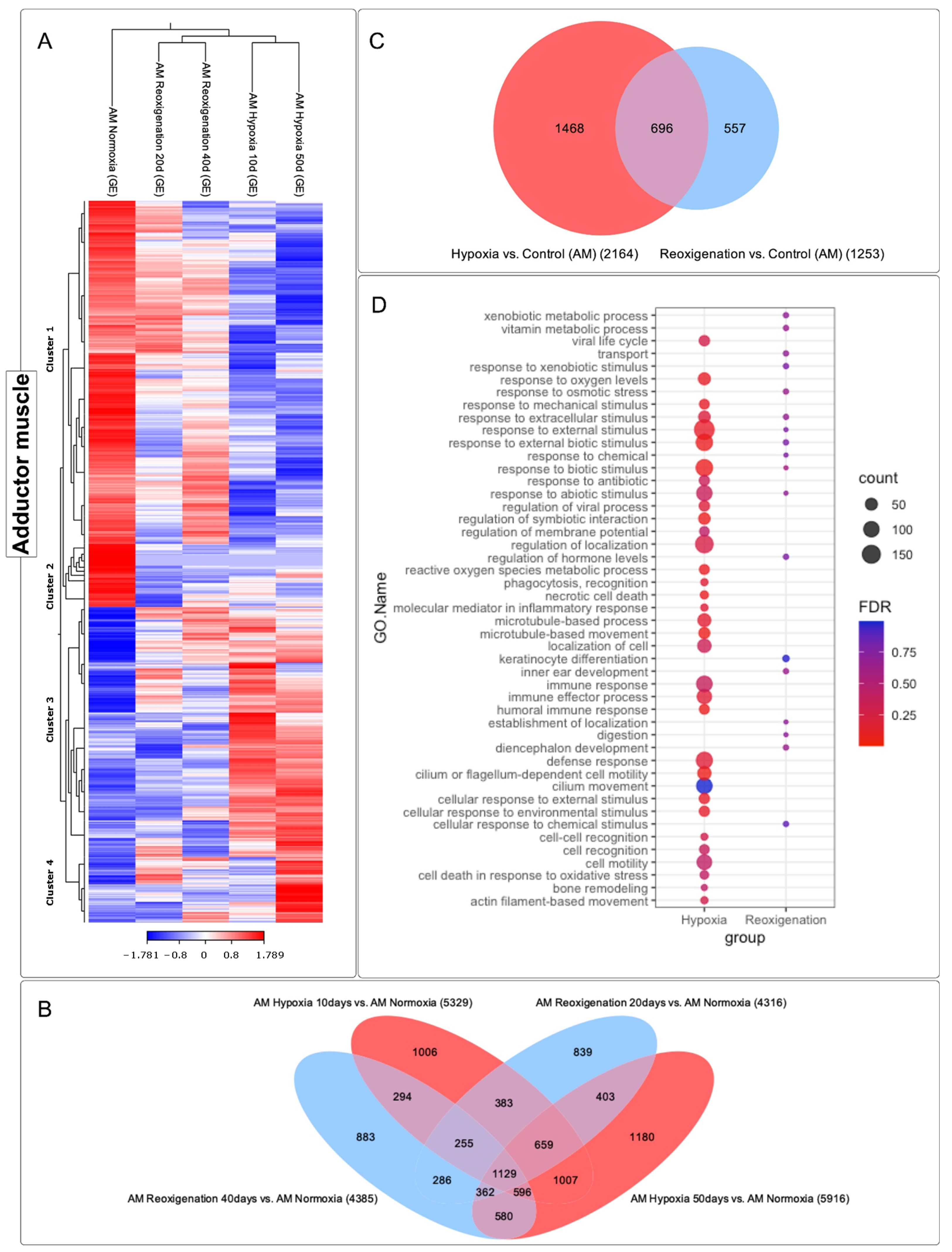
3.6. Differential Expression ANALYSIS of transcripts Observed in the Adductor Muscle of M. chilensis under Hypoxic and Reoxygenation Conditions
3.7. GO Enrichment Analysis in the Adductor Muscle of M. chilensis under Hypoxic and Reoxygenation Conditions
3.8. Identification and Expression of the mTOR Signaling Pathway in M. chilensis under Hypoxia
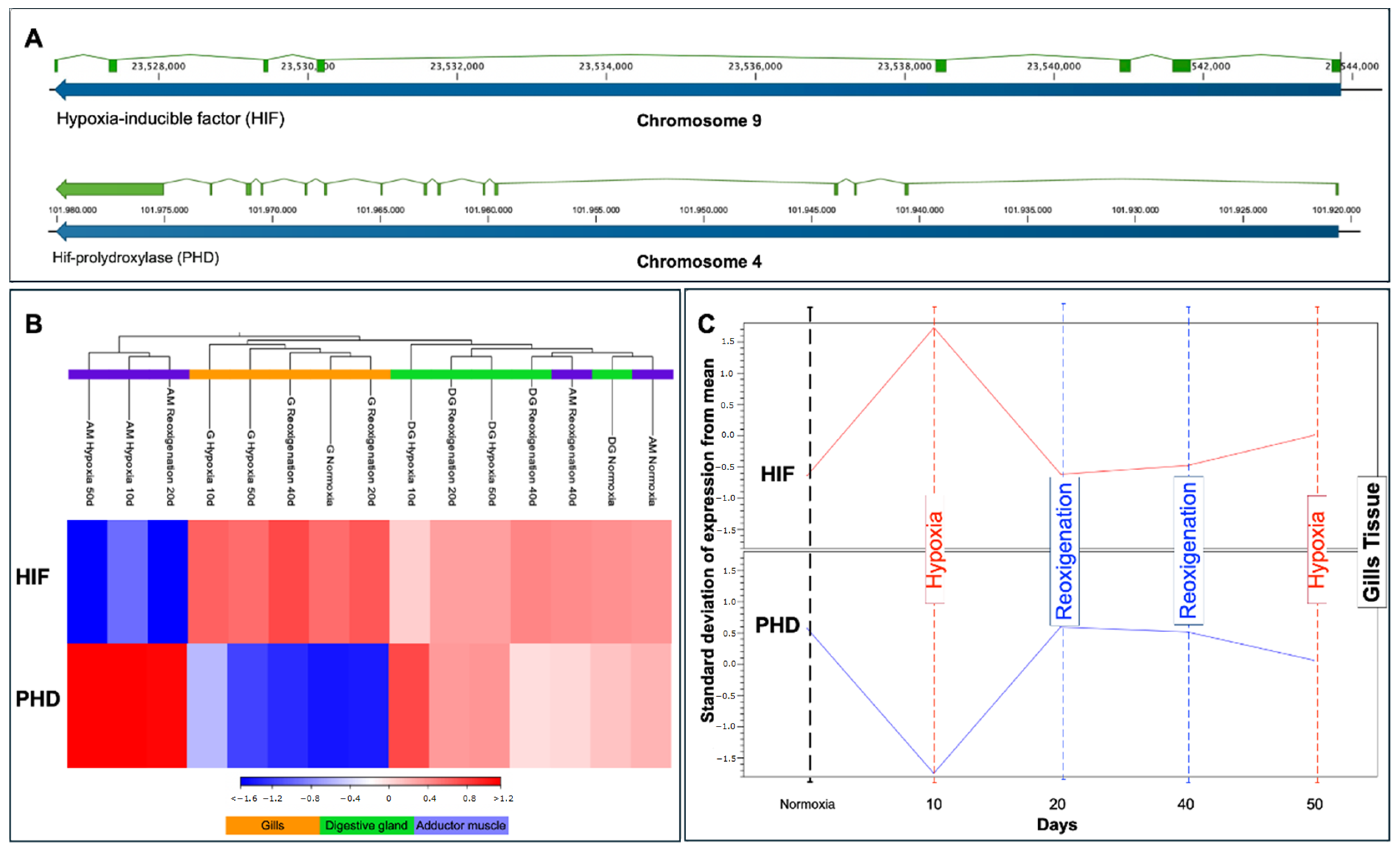
3.9. Transcriptional Response of HIF and PHD in Different Tissues of M. chilensis during Hypoxia and Reoxygenation Phases
3.10. Identifying and Expressing Transcripts in the Toll-like Receptor, Citrate Cycle (TCA), and Apoptosis Signaling Pathways in the Gills of M. chilensis under Hypoxia
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Calle, X.; Jiménez-Gallegos, D.; Muñoz-Córdova, F.; Sánchez, P.; Lavandero, S. Mecanismo sensor y de adaptación a los niveles de oxígeno y su implicancia en las enfermedades cardiovasculares: A propósito del Premio Nobel de Fisiología-Medicina 2019. Rev. Chil. Cardiol. 2019, 38, 225–235. [Google Scholar] [CrossRef]
- García, N.; Puentes, O.; Montalvo, J. Contaminación orgánica en el sector de la Bahía de Buena Vista cercano a la desembocadura de Río Guanó, Villa Clara, Cuba. Rev. Cuba. Química 2008, 20, 39–46. [Google Scholar]
- Haider, F.; Falfushynska, H.; Timm, S.; Sokolova, I. Effects of hypoxia and reoxygenation on intermediary metabolite homeostasis of marine bivalves Mytilus edulis and Crassostrea gigas. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020, 242, 110657. [Google Scholar] [CrossRef]
- Ali, J.; Yang, Y.; Pan, G. Oxygen micro-nanobubbles for mitigating eutrophication induced sediment pollution in freshwater bodies. J. Environ. Manag. 2023, 331, 117281. [Google Scholar] [CrossRef]
- Breitburg, D.; Levin, L.; Oschlies, A.; Grégoire, M.; Chavez, F.; Conley, D.; Garçon, V.; Gilbert, D.; Gutiérrez, D.; Isensee, K.; et al. Declining oxygen in the global ocean and coastal waters. Science 2018, 359, eaam7240. [Google Scholar] [CrossRef]
- Capet, A.; Beckers, J.; Grégoire, M. Drivers, mechanisms and long-term variability of seasonal hypoxia on the Black Sea northwestern shelf—Is there any recovery after eutrophication? Biogeosciences 2013, 10, 3943–3962. [Google Scholar] [CrossRef]
- Conley, D.; Carstensen, J.; Vaquer-Sunyer, R.; Duarte, C. Ecosystem thresholds with hypoxia. Hydrobiologia 2009, 629, 21–29. [Google Scholar] [CrossRef]
- Sagasti, A.; Schaffner, L.; Duffy, J. Effects of periodic hypoxia on mortality, feeding and predation in an estuarine epifaunal community. J. Exp. Mar. Biol. Ecol. 2001, 258, 257–283. [Google Scholar] [CrossRef]
- Silva, N.; Vargas, C. Hypoxia in Chilean Patagonian Fjords. Prog. Oceanogr. 2014, 129, 62–74. [Google Scholar] [CrossRef]
- Melzner, F.; Thomsen, J.; Koeve, W.; Oschlies, A.; Gutowska, M.; Bange, H.; Hansen, H.; Körtzinger, A. Future ocean acidification will be amplified by hypoxia in coastal habitats. Mar. Biol. 2013, 160, 1875–1888. [Google Scholar] [CrossRef]
- Scanes, E.; Scanes, P.; Ross, P. Climate change rapidly warms and acidifies Australian estuaries. Nat. Commun. 2020, 11, 1803. [Google Scholar] [CrossRef]
- Scanes, E.; Parker, L.; Seymour, J.; Siboni, N.; King, W.; Danckert, N.; Wegner, K.; Dove, C.; O’Connor, A.; Ross, P. Climate change alters the haemolymph microbiome of oysters. Mar. Pollut. Bull. 2021, 164, 111991. [Google Scholar] [CrossRef]
- Yáñez, E.; Lagos, N.; Norambuena, R.; Silva, C.; Letelier, J.; Muck, K.; Martin, G.; Benítez, S.; Broitman, B.; Contreras, H.; et al. Impacts of Climate Change on Marine Fisheries and Aquaculture in Chile. In Climate Change Impacts on Fisheries and Aquaculture: A Global Analysis; Phillips, B., Pérez-Ramírez, M., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; Volume I, pp. 239–332. [Google Scholar]
- Bianchi, T.; Arndt, S.; Austin, W.; Benn, D.; Bertrand, S.; Cui, X.; Faust, J.; Koziorowska-Makuch, K.; Moy, C.; Savage, C.; et al. Fjords as Aquatic Critical Zones (ACZs). Earth-Sci. Rev. 2020, 203, 103145. [Google Scholar] [CrossRef]
- Iriarte, J.; Pantoja, S.; Daneri, G. Oceanographic Processes in Chilean Fjords of Patagonia: From small to large-scale studies. Prog. Oceanogr. 2014, 129, 1–7. [Google Scholar] [CrossRef]
- Schmidtko, S.; Stramma, L.; Visbeck, M. Decline in global oceanic oxygen content during the past five decades. Nature 2017, 542, 335–339. [Google Scholar] [CrossRef]
- Nogueira, L.; Ferraz, D.; Trevisan, R.; Garcia, D.; Da Silva, D.; Luiz, A.; Alves, E. Hypoxia effects on oxidative stress and immunocompetence biomarkers in the mussel Perna perna (Mytilidae, Bivalvia). Mar. Environ. Res. 2017, 126, 109–115. [Google Scholar] [CrossRef]
- Kim, C.; Park, C.; Kim, E.; Nam, Y. Transcriptional modulation patterns of abalone Haliotis discus hannai hypoxia inducible factor-1α (HIF-1α) in interdependent crosstalk between hypoxia, infection, and environmental stresses. Aquac. Rep. 2021, 19, 100566. [Google Scholar] [CrossRef]
- Levin, L.; Ekau, W.; Gooday, A.; Jorissen, F.; Middelburg, J.; Naqvi, S.; Neira, C.; Rabalais, N.; Zhang, J. Effects of natural and human-induced hypoxia on coastal benthos. Biogeosciences 2009, 6, 2063–2098. [Google Scholar] [CrossRef]
- Grieshaber, M.; Hardewig, I.; Kreutzer, U.; Pörtner, H. Physiological and metabolic responses to hypoxia in invertebrates. Rev. Physiol. Biochem. Pharmacol. 1994, 125, 43–147. [Google Scholar] [CrossRef]
- Chu, J.; Curkan, C.; Tunnicliffe, V. Drivers of temporal beta diversity of a benthic community in a seasonally hypoxic fjord. R. Soc. Open Sci. 2018, 5, 172284. [Google Scholar] [CrossRef]
- Hernández-Miranda, E.; Veas, R.; Anabalón, V.; Quiñones, R. Short-term alteration of biotic and abiotic components of the pelagic system in a shallow bay produced by a strong natural hypoxia event. PLoS ONE 2017, 12, e0179023. [Google Scholar] [CrossRef]
- Tirpe, A.; Gulei, D.; Ciortea, S.; Crivii, C.; Berindan-Neagoe, I. Hypoxia: Overview on Hypoxia-Mediated Mechanisms with a Focus on the Role of HIF Genes. Int. J. Mol. Sci. 2019, 20, 6140. [Google Scholar] [CrossRef]
- Kodama, K.; Horiguchi, T. Effects of hypoxia on benthic organisms in Tokyo Bay, Japan: A review. Mar. Pollut. Bull. 2011, 63, 215–220. [Google Scholar] [CrossRef]
- Batie, M.; Del Peso, L.; Rocha, S. Hypoxia and Chromatin: A Focus on Transcriptional Repression Mechanisms. Biomedicines 2018, 6, 47. [Google Scholar] [CrossRef]
- Soldatov, A.; Gostyukhina, O.; Golovina, I. Antioxidant enzyme complex of tissues of the bivalve Mytilus galloprovincialis Lam. under normal and oxidative-stress conditions: A review. Appl. Biochem. Microbiol. 2007, 43, 556–562. [Google Scholar] [CrossRef]
- Vale, G.; Mehennaoui, K.; Cambier, S.; Libralato, G.; Jomini, S.; Domingos, R. Manufactured nanoparticles in the aquatic environment-biochemical responses on freshwater organisms: A critical overview. Aquat. Toxicol. 2016, 170, 162–174. [Google Scholar] [CrossRef]
- Pytharopoulou, S.; Sazakli, E.; Grintzalis, K.; Georgiou, C.; Leotsinidis, M.; Kalpaxis, D. Translational responses of Mytilus galloprovincialis to environmental pollution: Integrating the responses to oxidative stress and other biomarker responses into a general stress index. Aquat. Toxicol. 2008, 89, 18–27. [Google Scholar] [CrossRef]
- Falfushynska, H.; Piontkivska, H.; Sokolova, I. Effects of intermittent hypoxia on cell survival and inflammatory responses in the intertidal marine bivalves Mytilus edulis and Crassostrea gigas. J. Exp. Biol. 2020, 223, jeb217026. [Google Scholar] [CrossRef]
- Sokolova, I. Mitochondrial Adaptations to Variable Environments and Their Role in Animals’ Stress Tolerance. Integr. Comp. Biol. 2018, 58, 519–531. [Google Scholar] [CrossRef]
- Storey, K.; Storey, J. Metabolic rate depression in animals: Transcriptional and translational controls. Biol. Rev. Camb. Philos. Soc. 2004, 79, 207–233. [Google Scholar] [CrossRef]
- Liao, C.; Liu, X.; Zhang, C.; Zhang, Q. Tumor hypoxia: From basic knowledge to therapeutic implications. Semin Cancer Biol 2023, 88, 172–186. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P. HIF-1 signaling: A key orchestrator of cancer radioresistance. Radiat. Med. Prot. 2020, 1, 7–14. [Google Scholar] [CrossRef]
- Jung-whan, K.; Tchernyshyov, I.; Semenza, L.; Dang, V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Bailey, P.; Nathan, J. Metabolic Regulation of Hypoxia-Inducible Transcription Factors: The Role of Small Molecule Metabolites and Iron. Biomedicines 2018, 6, 60. [Google Scholar] [CrossRef]
- Krzywinska, E.; Stockmann, C. Hypoxia, Metabolism and Immune Cell Function. Biomedicines 2018, 6, 56. [Google Scholar] [CrossRef]
- Sokolova, I.; Sokolov, E.; Haider, F. Mitochondrial Mechanisms Underlying Tolerance to Fluctuating Oxygen Conditions: Lessons from Hypoxia-Tolerant Organisms. Integr. Comp. Biol. 2019, 59, 938–952. [Google Scholar] [CrossRef]
- Fava, L.; Bock, F.; Geley, S.; Villunger, A. Caspase-2 at a glance. J. Cell Sci. 2012, 125, 5911–5915. [Google Scholar] [CrossRef]
- Movassagh, M.; Foo, R. Simplified apoptotic cascades. Heart Fail. Rev. 2008, 13, 111–119. [Google Scholar] [CrossRef]
- Adli, M.; Merkhofer, E.; Cogswell, P.; Baldwin, A. IKKalpha and IKKbeta each function to regulate NF-kappaB activation in the TNF-induced/canonical pathway. PLoS ONE 2010, 5, e9428. [Google Scholar] [CrossRef]
- Angelo, A.; Rovere-Querini, P.; Clementi, S.; Clementi, B. Cell Death: Tipping the Balance of Autoimmunity and Tissue Repair. Curr. Pharm. Des. 2008, 14, 269–277. [Google Scholar] [CrossRef]
- Kaltschmidt, B.; Kaltschmidt, C.; Hofmann, T.; Hehner, S.; Dröge, W.; Schmitz, M. The pro- or anti-apoptotic function of NF-κB is determined by the nature of the apoptotic stimulus. Eur. J. Biochem. 2000, 267, 3828–3835. [Google Scholar] [CrossRef]
- Moret, I.; Cerrillo, E.; Navarro-Puche, A.; Iborra, M.; Rausell, F.; Tortosa, L.; Beltrán, B. Estrés oxidativo en la enfermedad de Crohn. Gastroenterol. Hepatol. 2014, 37, 28–34. [Google Scholar] [CrossRef]
- Thornton, C.; Leaw, B.; Mallard, C.; Nair, S.; Jinnai, M.; Hagberg, H. Cell Death in the Developing Brain after Hypoxia-Ischemia. Front. Cell. Neurosci. 2017, 11, 248. [Google Scholar] [CrossRef]
- Carella, F.; Feist, S.; Bignell, J.; De Vico, G. Comparative pathology in bivalves: Aetiological agents and disease processes. J. Invertebr. Pathol. 2015, 131, 107–120. [Google Scholar] [CrossRef]
- Connon, R.; D’Abronzo, L.; Hostetter, N.; Javidmehr, A.; Roby, D.D.; Evans, A.; Loge, F.; Werner, I. Transcription Profiling in Environmental Diagnostics: Health Assessments in Columbia River Basin Steelhead (Oncorhynchus mykiss). Environ. Sci. Technol. 2012, 46, 6081–6087. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Wang, Y.; Day, R.; Yang, H.; Zhang, Z. Immunity-related genes and signaling pathways under hypoxic stresses in Haliotis diversicolor: A transcriptome analysis. Sci. Rep. 2019, 9, 19741. [Google Scholar] [CrossRef]
- Gallardo-Escarate, C.; Valenzuela-Munoz, V.; Gustavo, N.; Valenzuela-Miranda, D.; Tapia, F.; Yevenes, M.; Gajardo, G.; Toro, J.; Oyarzun, P.; Arriagada, G.; et al. Chromosome-Level Genome Assembly of the Blue Mussel Mytilus chilensis Reveals Molecular Signatures Facing the Marine Environment. Genes 2023, 14, 876. [Google Scholar] [CrossRef]
- Yévenes, M.; Núñez-Acuña, G.; Gallardo-Escárate, C.; Gajardo, G. Adaptive mitochondrial genome functioning in ecologically different farm-impacted natural seedbeds of the endemic blue mussel Mytilus chilensis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 42, 100955. [Google Scholar] [CrossRef]
- Núñez-Acuña, G.; Gallardo-Escárate, C. Identification of immune-related SNPs in the transcriptome of Mytilus chilensis through high-throughput sequencing. Fish Shellfish Immunol. 2013, 35, 1899–1905. [Google Scholar] [CrossRef]
- Detree, C.; Nunez-Acuna, G.; Roberts, S.; Gallardo-Escarate, C. Uncovering the Complex Transcriptome Response of Mytilus chilensis against Saxitoxin: Implications of Harmful Algal Blooms on Mussel Populations. PLoS ONE 2016, 11, e0165231. [Google Scholar] [CrossRef] [PubMed]
- Malachowicz, M.; Wenne, R. Mantle transcriptome sequencing of Mytilus spp. and identification of putative biomineralization genes. PeerJ 2019, 6, e6245. [Google Scholar] [CrossRef]
- Lohrmann, K.; Bustos, E.; Rojas, R.; Navarrete, F.; Robotham, H.; Bignell, J. Histopathological assessment of the health status of Mytilus chilensis (Hupé 1854) in southern Chile. Aquaculture 2019, 503, 40–50. [Google Scholar] [CrossRef]
- Osores, S.; Lagos, N.; San Martín, V.; Manríquez, P.; Vargas, C.; Torres, R.; Navarro, J.; Poupin, J.; Saldías, G.; Lardies, M. Plasticity and inter-population variability in physiological and life-history traits of the mussel Mytilus chilensis: A reciprocal transplant experiment. J. Exp. Mar. Biol. Ecol. 2017, 490, 1–12. [Google Scholar] [CrossRef]
- Ríos, V.; Ocampo, N.; Astorga, M. Proximal chemical composition and morphometry of the ribbed (Aulacomya ater, Molina 1782) and the blue (Mytilus chilensis, Hupé, 1854) mussels commercialized in the Region of Magallanes. An. Inst. Patagon. 2018, 46, 49–58. [Google Scholar] [CrossRef]
- Navarro, J.; Duarte, C.; Manríquez, P.; Lardies, M.; Torres, R.; Acuña, K.; Vargas, C.; Lagos, N. Ocean warming and elevated carbon dioxide: Multiple stressor impacts on juvenile mussels from southern Chile. ICES J. Mar. Sci. 2016, 73, 764–771. [Google Scholar] [CrossRef]
- Subpesca. Mejillón. Available online: https://www.subpesca.cl/portal/616/w3-article-843.html#presentacion (accessed on 1 May 2023).
- Oyarzún, P.; Toro, E.; Jaramillo, R.; Guiñez, R.; Briones, C.; Astorga, M. Gonadal cycle of the mussel Mytilus chilensis (Bivalvia: Mytilidae) at two localities in southern of Chile. Lat. Am. J. Aquat. Res. 2011, 39, 512–525. [Google Scholar] [CrossRef]
- Salas-Yanquin, L.; Navarro, J.; Pechenik, J.; Montory, J.; Chaparro, O. Volcanic ash in the water column: Physiological impact on the suspension-feeding bivalve Mytilus chilensis. Mar. Pollut. Bull. 2018, 127, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Molinet, C.; Diaz, M.; Marin, S.; Astorga, M.; Ojeda, M.; Cares, L.; Asencio, E. Relation of mussel spatfall on natural and artificial substrates: Analysis of ecological implications ensuring long-term success and sustainability for mussel farming. Aquaculture 2017, 467, 211–218. [Google Scholar] [CrossRef]
- IFOP. Innovaciones en la Tecnología de Cultivo de Chorito (Mytilus chilensis), Tendientes a Mejorar la Calidad y Rentabilidad de la Actividad Mitícola en la X Región. Available online: https://www.ifop.cl/wp-content/contenidos/uploads/biblioteca/libros_digitales/Curso_Cultivo_de_choritos.pdf (accessed on 9 January 2023).
- Subpesca. Informe Sectorial de Pesca y Acuicultura Consolidado (2020–2021). Available online: https://www.subpesca.cl/portal/618/articles-114306_documento.pdf (accessed on 1 October 2023).
- ProChile. Chile se Convierte en el Mayor Proveedor Mundial de 28 Productos Liderados por Cobre, Cerezas y Salmón. Available online: https://www.prochile.gob.cl/noticias/detalle-noticia/2021/08/12/chile-se-convierte-en-el-mayor-proveedor-mundial-de-28-productos-liderados-por-cobre-cerezas-y-salm%C3%B3n (accessed on 3 October 2023).
- Blanc, J.; Molinet, C.; Subiabre, R.; Díaz, P. Cadmium determination in Chilean blue mussels Mytilus chilensis: Implications for environmental and agronomic interest. Mar. Pollut. Bull. 2018, 129, 913–917. [Google Scholar] [CrossRef]
- Yevenes, M.; Lagos, N.; Farías, L.; Vargas, C. Greenhouse gases, nutrients and the carbonate system in the Reloncaví Fjord (Northern Chilean Patagonia): Implications on aquaculture of the mussel, Mytilus chilensis, during an episodic volcanic eruption. Sci. Total Environ. 2019, 669, 49–61. [Google Scholar] [CrossRef]
- Walker, C.; Beneden, R.; Muttray, A.; Böttger, A.; Kelley, M.; Tucker, A.; Kelley, T. Chapter One—p53 Superfamily Proteins in Marine Bivalve Cancer and Stress Biology; Michael, L., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 59, pp. 1–36. [Google Scholar]
- Venier, P.; De Pittà, C.; Pallavicini, A.; Marsano, F.; Varotto, L.; Romualdi, C.; Dondero, F.; Viarengo, A.; Lanfranchi, G. Development of mussel mRNA profiling: Can gene expression trends reveal coastal water pollution? Mutat. Res./Fundam. Mol. Mech. Mutagen. 2006, 602, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Baqueiro–Cárdenas, E.; Borabe, L.; Goldaracena–Islas, C.; Rodríguez–Navarro, J. Mollusks and pollution. A review. Rev. Mex. Biodivers. 2007, 78, 1–7. [Google Scholar]
- Hernández-Miranda, E.; Quiñones, R.; Aedo, G.; Valenzuela, A.; Mermoud, N.; Román, C.; Yañez, F. A major fish stranding caused by a natural hypoxic event in a shallow bay of the eastern South Pacific Ocean. J. Fish Biol. 2010, 76, 1543–1564. [Google Scholar] [CrossRef]
- Hernández-Miranda, E.; Veas, R.; Labra, F.; Salamanca, M.; Quiñones, R. Response of the epibenthic macrofaunal community to a strong upwelling-driven hypoxic event in a shallow bay of the southern Humboldt Current System. Mar. Environ. Res. 2012, 79, 16–28. [Google Scholar] [CrossRef]
- Labra, F.; Hernández-Miranda, E.; Quiñones, R. Dynamic relationships between body size, species richness, abundance, and energy use in a shallow marine epibenthic faunal community. Ecol. Evol. 2015, 5, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.; Husmann, G.; Thorne, M.; Burns, G.; Truebano, M.; Peck, L.; Abele, D.; Philipp, E. Hypoxia impacts large adults first: Consequences in a warming world. Glob. Change Biol. 2013, 19, 2251–2263. [Google Scholar] [CrossRef]
- Gu, H.; Shang, Y.; Clements, J.; Dupont, S.; Wang, T.; Wei, S.; Wang, X.; Chen, J.; Huang, W.; Hu, M.; et al. Hypoxia aggravates the effects of ocean acidification on the physiological energetics of the blue mussel Mytilus edulis. Mar. Pollut. Bull. 2019, 149, 110538. [Google Scholar] [CrossRef] [PubMed]
- Piontkivska, H.; Chung, S.; Ivanina, A.; Sokolov, E.; Techa, S.; Sokolova, I. Molecular characterization and mRNA expression of two key enzymes of hypoxia-sensing pathways in eastern oysters Crassostrea virginica (Gmelin): Hypoxia-inducible factor α (HIF-α) and HIF-prolyl hydroxylase (PHD). Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 103–114. [Google Scholar] [CrossRef]
- Coxe, N.; Casas, S.; Marshall, D.; La Peyre, M.; Kelly, M.; La Peyre, J. Differential hypoxia tolerance of eastern oysters from the northern Gulf of Mexico at elevated temperature. J. Exp. Mar. Biol. Ecol. 2023, 559, 151840. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, T.; Zhang, Q.; Xue, R.; Qu, Y.; Wang, Q.; Dong, Z.; Zhao, J. Different patterns of hypoxia aggravate the toxicity of polystyrene nanoplastics in the mussels Mytilus galloprovincialis: Environmental risk assessment of plastics under global climate change. Sci. Total Environ. 2022, 818, 151818. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, F.; Sun, S. The survival and responses of blue mussel Mytilus edulis to 16-day sustained hypoxia stress. Mar. Environ. Res. 2022, 176, 105601. [Google Scholar] [CrossRef] [PubMed]
- Gostyukhina, O.; Yu, A.; Chelebieva, E.; Vodiasova, E.; Lantushenko, A.; Kladchenko, E. Adaptive potential of the Mediterranean mussel Mytilus galloprovincialis to short-term environmental hypoxia. Fish Shellfish Immunol. 2022, 131, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Kotsyuba, E.; Dyachuk, V. Effect of Air Exposure-Induced Hypoxia on Neurotransmitters and Neurotransmission Enzymes in Ganglia of the Scallop Azumapecten farreri. Int. J. Mol. Sci. 2022, 23, 2027. [Google Scholar] [CrossRef] [PubMed]
- Salmond, N.; Wing, S. Sub-lethal and lethal effects of chronic ammonia exposure and hypoxia on a New Zealand bivalve. J. Exp. Mar. Biol. Ecol. 2022, 549, 151696. [Google Scholar] [CrossRef]
- Andreyeva, A.; Gostyukhina, O.; Kladchenko, E.; Afonnikov, D.; Rasskazov, D.; Lantushenko, A.; Vodiasova, E. Hypoxia exerts oxidative stress and changes in expression of antioxidant enzyme genes in gills of Mytilus galloprovincialis (Lamarck, 1819). Mar. Biol. Res. 2021, 17, 369–379. [Google Scholar] [CrossRef]
- Khan, F.; Chen, H.; Gu, H.; Wang, T.; Dupont, S.; Kong, H.; Shang, Y.; Wang, X.; Lu, W.; Hu, M.; et al. Antioxidant responses of the mussel Mytilus coruscus co-exposed to ocean acidification, hypoxia and warming. Mar. Pollut. Bull. 2021, 162, 111869. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Kong, H.; Chang, X.; Dupont, S.; Chen, H.; Deng, Y.; Hu, M.; Wang, Y. Gonadal antioxidant responses to seawater acidification and hypoxia in the marine mussel Mytilus coruscus. Environ. Sci. Pollut. Res. 2021, 28, 53847–53856. [Google Scholar] [CrossRef]
- Sokolov, E.; Adzigbli, L.; Markert, S.; Bundgaard, A.; Fago, A.; Becher, D.; Hirschfeld, C.; Sokolova, I. Intrinsic Mechanisms Underlying Hypoxia-Tolerant Mitochondrial Phenotype During Hypoxia-Reoxygenation Stress in a Marine Facultative Anaerobe, the Blue Mussel Mytilus edulis. Front. Mar. Sci. 2021, 8, 773734. [Google Scholar] [CrossRef]
- Altschul, S.; Madden, T.; Schaffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1998, 12, 3389–3402. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.; Britto, R.; Cukura, A.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef]
- Aleksander, S.; Balhoff, J.; Carbon, S.; Cherry, J.; Drabkin, H.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.; Hill, D.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Tatusov, R.; Natale, D.; Garkavtsev, I.; Tatusova, T.; Shankavaram, U.; Rao, B.; Kiryutin, B.; Galperin, M.; Fedorova, N.; Koonin, E. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ge, S.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Valenzuela-Muñoz, V.; Gallardo-Escárate, C.; Benavente, B.; Valenzuela-Miranda, D.; Núñez-Acuña, G.; Escobar-Sepulveda, H.; Váldes, J. Whole-Genome Transcript Expression Profiling Reveals Novel Insights into Transposon Genes and Non-Coding RNAs during Atlantic Salmon Seawater Adaptation. Biology 2022, 11, 1. [Google Scholar] [CrossRef]
- Cui, Z.; Cui, Y.; Zang, T.; Wang, Y. Genome analysis interacCircos: An R package based on JavaScript libraries for the generation of interactive circos plots. Bioinformatics 2021, 37, 3642–3644. [Google Scholar] [CrossRef]
- Lee, S.; Choi, E.; Kim, T.; Hwang, J.; Lee, J. AtHAD1, A haloacid dehalogenase-like phosphatase, is involved in repressing the ABA response. Biochem. Biophys. Res. Commun. 2022, 587, 119–125. [Google Scholar] [CrossRef]
- Du, Z.; Deng, S.; Wu, Z.; Wang, C. Genome-wide analysis of haloacid dehalogenase genes reveals their function in phosphate starvation responses in rice. PLoS ONE 2021, 16, e0245600. [Google Scholar] [CrossRef]
- Pandey, B.; Mehra, P.; Verma, L.; Bhadouria, J.; Giri, J. OsHAD1, a Haloacid Dehalogenase-Like APase, Enhances Phosphate Accumulation. Plant Physiol. 2017, 174, 2316–2332. [Google Scholar] [CrossRef]
- Levin, L. Oxygen Minimum Zone Benthos: Adaptation and Community Response to Hypoxia. In Oceanography and Marine Biology, An Annual Review, 1st ed.; Gibson, R., Atkinson, A., Eds.; Taylor & Francis: London, UK, 2003; Volume 41, pp. 1–45. [Google Scholar]
- Rabalais, N.; Cai, W.; Carstensen, J.; Conley, D.; Fry, B.; Hu, X.; Quiñones-Rivera, Z.; Rosenberg, R.; Slomp, C.; Turner, R.; et al. Eutrophication-driven deoxygenation in the coastal ocean. Oceanography 2014, 27, 172–183. [Google Scholar] [CrossRef]
- Nie, H.; Wang, H.; Jiang, K.; Yan, X. Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: Potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genom. 2020, 21, 318. [Google Scholar] [CrossRef]
- Yan, X.; Nie, H.; Huo, Z.; Ding, J.; Li, Z.; Yan, L.; Jiang, L.; Mu, Z.; Wang, H.; Meng, X.; et al. Clam Genome Sequence Clarifies the Molecular Basis of Its Benthic Adaptation and Extraordinary Shell Color Diversity. iScience 2019, 19, 1225–1237. [Google Scholar] [CrossRef]
- Philipp, E.; Wessels, W.; Gruber, H.; Strahl, J.; Wagner, A.; Ernst, I.; Rimbach, G.; Kraemer, L.; Schreiber, S.; Abele, D.; et al. Gene Expression and Physiological Changes of Different Populations of the Long-Lived Bivalve Arctica islandica under Low Oxygen Conditions. PLoS ONE 2012, 7, e44621. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, S.; Liu, T.; Chen, M.; Li, W.; Zhang, X. The transcriptomic responses of the ark shell, Anadara broughtonii, to sulfide and hypoxia exposure. Mol. Biol. Rep. 2019, 46, 4245–4257. [Google Scholar] [CrossRef]
- Lee, J.; Bae, S.; Jeong, J.; Kim, S.; Kim, K. Hypoxia-inducible factor (HIF-1)α: Its protein stability and biological functions. Exp. Mol. Med. 2004, 36, 1–12. [Google Scholar] [CrossRef]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.; Torrealba, N.; Paredes, F.; Wang, Z.; Zorzano, A.; Hill, J.; Jaimovich, E.; et al. Endoplasmic Reticulum and the Unfolded Protein Response: Dynamics and Metabolic Integration. In International Review of Cell and Molecular Biology; Jeon, K., Ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; Volume 301, pp. 215–290. [Google Scholar]
- Steffen, J.; Falfushynska, H.; Piontkivska, H.; Sokolova, I. Molecular Biomarkers of the Mitochondrial Quality Control Are Differently Affected by Hypoxia-Reoxygenation Stress in Marine Bivalves Crassostrea gigas and Mytilus edulis. Front. Mar. Sci. 2020, 7, 604411. [Google Scholar] [CrossRef]
- Sun, S.; Zhao, G.; Jia, M.; Jiang, Q.; Li, S.; Wang, H.; Li, W.; Wang, Y.; Bian, X.; Zhao, Y.; et al. Stay in touch with the endoplasmic reticulum. Sci. China Life Sci. 2024, 67, 230–257. [Google Scholar] [CrossRef]
- Di Conza, G.; Ho, P.; Cubillos-Ruiz, J.; Huang, S. Control of immune cell function by the unfolded protein response. Nat. Rev. Immunol. 2023, 23, 546–562. [Google Scholar] [CrossRef]
- Adams, C.; Kopp, M.; Larburu, N.; Nowak, P.; Ali, M. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front. Mol. Biosci. 2019, 6, 11. [Google Scholar] [CrossRef]
- Noulsri, E.; Lerdwana, S. Reducing erythroblast apoptosis in β-thalassemia via unfolded protein response (UPR) signaling. Med. Hypotheses 2023, 177, 111117. [Google Scholar] [CrossRef]
- Wang, L.; Alzayady, K.; Yule, D. Proteolytic fragmentation of inositol 1,4,5-trisphosphate receptors: A novel mechanism regulating channel activity? J. Physiol. 2016, 594, 2867–2876. [Google Scholar] [CrossRef]
- Kokott-Vuong, A.; Jung, J.; Fehr, A.; Kirschfink, N.; Noristani, R.; Voigt, A.; Reich, A.; Schulz, J.; Huber, M.; Habib, P. Increased Post-Hypoxic Oxidative Stress and Activation of the PERK Branch of the UPR in Trap1-Deficient Drosophila melanogaster Is Abrogated by Metformin. Int. J. Mol. Sci. 2021, 22, 11586. [Google Scholar] [CrossRef]
- Colgan, S.; Hashimi, A.; Austin, R. Endoplasmic reticulum stress and lipid dysregulation. Expert Rev. Mol. Med. 2011, 13, e4. [Google Scholar] [CrossRef]
- Stevenson, J.; Huang, E.; Olzmann, J. Endoplasmic Reticulum-Associated Degradation and Lipid Homeostasis. In Annual Review of Nutrition; Stover, P., Ed.; Annual Reviews: Palo Alto, CA, USA, 2016; Volume 36, pp. 511–542. [Google Scholar]
- Gerhardtova, I.; Jankech, T.; Majerova, P.; Piestansky, J.; Olesova, D.; Kovac, A.; Jampilek, J. Recent Analytical Methodologies in Lipid Analysis. Int. J. Mol. Sci. 2024, 25, 2249. [Google Scholar] [CrossRef]
- Storrie, B. Maintenance of Golgi apparatus structure in the face of continuous protein recycling to the endoplasmic reticulum: Making ends meet. In International Review of Cytology—A Survey of Cell Biology; Jeon, K., Ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2005; Volume 244, pp. 69–94. [Google Scholar]
- Hetz, C.; Zhang, K.; Kaufman, R. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, Y.; Sun, L. Emerging mechanisms of the unfolded protein response in therapeutic resistance: From chemotherapy to Immunotherapy. Cell Commun. Signal. 2024, 22, 89. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.; Méthe, D.; Stewart-Clark, S.; Clark, F. Size and site specific transcriptomic responses of blue mussel (Mytilus edulis) to acute hypoxia. Mar. Genom. 2023, 71, 101060. [Google Scholar] [CrossRef] [PubMed]
- Soldatov, A.A.; Andreenko, T.I.; Sysoeva, I.V.; Sysoev, A.A. Tissue specificity of metabolism in bivalve mollusc Anadara inaequivalvis Br. under conditions of experimental anoxia. Comp. Ontog. Biochem. 2009, 45, 284–289. [Google Scholar] [CrossRef]
- Dezwaan, A.; Cortesi, P.; Vandenthillart, G.; Roos, J.; Storey, K. Differential sensitivities to hypoxia by two anoxia-tolerant marine molluscs: A biochemical analysis. Mar. Biol. 1991, 111, 343–351. [Google Scholar] [CrossRef]
- Cook, K.; Shen, H.; McKelvey, K.; Gee, H.; Hau, E. Targeting Glucose Metabolism of Cancer Cells with Dichloroacetate to Radiosensitize High-Grade Gliomas. Int. J. Mol. Sci. 2021, 22, 7265. [Google Scholar] [CrossRef] [PubMed]
- Gillen, S.; Waldron, J.; Bushell, M. Codon optimality in cancer. Oncogene 2021, 40, 6309–6320. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cubillos-Ruiz, J. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 2021, 21, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Gebert, M.; Slawski, J.; Kalinowski, L.; Collawn, J.; Bartoszewski, R. The Unfolded Protein Response: A Double-Edged Sword for Brain Health. Antioxidants 2023, 12, 1648. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller-Herzog, C.; Hall, M. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012, 22, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Marcondes-de-Castro, I.; Reis-Barbosa, P.; Marinho, T.; Aguila, M.; Mandarim-de-Lacerda, C. AMPK/mTOR pathway significance in healthy liver and non-alcoholic fatty liver disease and its progression. J. Gastroenterol. Hepatol. 2023, 38, 1868–1876. [Google Scholar] [CrossRef]
- Giannetto, A.; Maisano, M.; Cappello, T.; Oliva, S.; Parrino, V.; Natalotto, A.; De Marco, G.; Barberi, C.; Romeo, O.; Mauceri, A.; et al. Hypoxia-Inducible Factor α and Hif-prolyl Hydroxylase Characterization and Gene Expression in Short-Time Air-Exposed Mytilus galloprovincialis. Mar. Biotechnol. 2015, 17, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Thomas, Y.; Flye-Sainte-Marie, J.; Chabot, D.; Aguirre-Velarde, A.; Marques, G.; Pecquerie, L. Effects of of hypoxia on metabolic functions in marine organisms: Observed patterns and modelling assumptions within the context of Dynamic Energy Budget (DEB) theory. J. Sea Res. 2019, 143, 231–242. [Google Scholar] [CrossRef]
- Wu, R. Hypoxia: From molecular responses to ecosystem responses. Mar. Pollut. Bull. 2002, 45, 35–45. [Google Scholar] [CrossRef]
- Woo, S.; Denis, V.; Won, H.; Shin, K.; Lee, G.; Lee, T.; Yum, S. Expressions of oxidative stress-related genes and antioxidant enzyme activities in Mytilus galloprovincialis (Bivalvia, Mollusca) exposed to hypoxia. Zool. Stud. 2013, 52, 15. [Google Scholar] [CrossRef]
- Aguirre-Velarde, A.; Thouzeau, G.; Jean, F.; Mendo, J.; Cueto-Vega, R.; Kawazo-Delgado, M.; Vásquez-Spencer, J.; Herrera-Sanchez, D.; Vega-Espinoza, A.; Flye-Sainte-Marie, J. Chronic and severe hypoxic conditions in Paracas Bay, Pisco, Peru: Consequences on scallop growth, reproduction, and survival. Aquaculture 2019, 512, 734259. [Google Scholar] [CrossRef]
- Sui, Y.; Hu, M.; Shang, Y.; Wu, F.; Huang, X.; Dupont, S.; Storch, D.; Pörtner, H.; Li, J.; Lu, W.; et al. Antioxidant response of the hard shelled mussel Mytilus coruscus exposed to reduced pH and oxygen concentration. Ecotoxicol. Environ. Saf. 2017, 137, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Parisi, M.; Mauro, M.; Sarà, G.; Cammarata, M. Temperature increases, hypoxia, and changes in food availability affect immunological biomarkers in the marine mussel Mytilus galloprovincialis. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2017, 187, 1117–1126. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montúfar-Romero, M.; Valenzuela-Muñoz, V.; Valenzuela-Miranda, D.; Gallardo-Escárate, C. Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response. Genes 2024, 15, 658. https://doi.org/10.3390/genes15060658
Montúfar-Romero M, Valenzuela-Muñoz V, Valenzuela-Miranda D, Gallardo-Escárate C. Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response. Genes. 2024; 15(6):658. https://doi.org/10.3390/genes15060658
Chicago/Turabian StyleMontúfar-Romero, Milton, Valentina Valenzuela-Muñoz, Diego Valenzuela-Miranda, and Cristian Gallardo-Escárate. 2024. "Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response" Genes 15, no. 6: 658. https://doi.org/10.3390/genes15060658
APA StyleMontúfar-Romero, M., Valenzuela-Muñoz, V., Valenzuela-Miranda, D., & Gallardo-Escárate, C. (2024). Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response. Genes, 15(6), 658. https://doi.org/10.3390/genes15060658





