Metabolic Transcriptional Activation in Ulcerative Colitis Identified Through scRNA-seq Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. PICO Statement
2.2. Public Datasets
2.2.1. Training Transcriptome Dataset of Ulcerative Colitis Tissues
2.2.2. Validation Transcriptome Dataset of Ulcerative Colitis Tissues
2.3. Mammalian Metabolic Transcriptional Program
2.4. Transcriptome Analyses
2.5. Elastic Net Machine Learning Tuning
2.6. Elastic Net (Enet) Expression Score Computing
2.7. KEGG Enrichment Network
3. Results
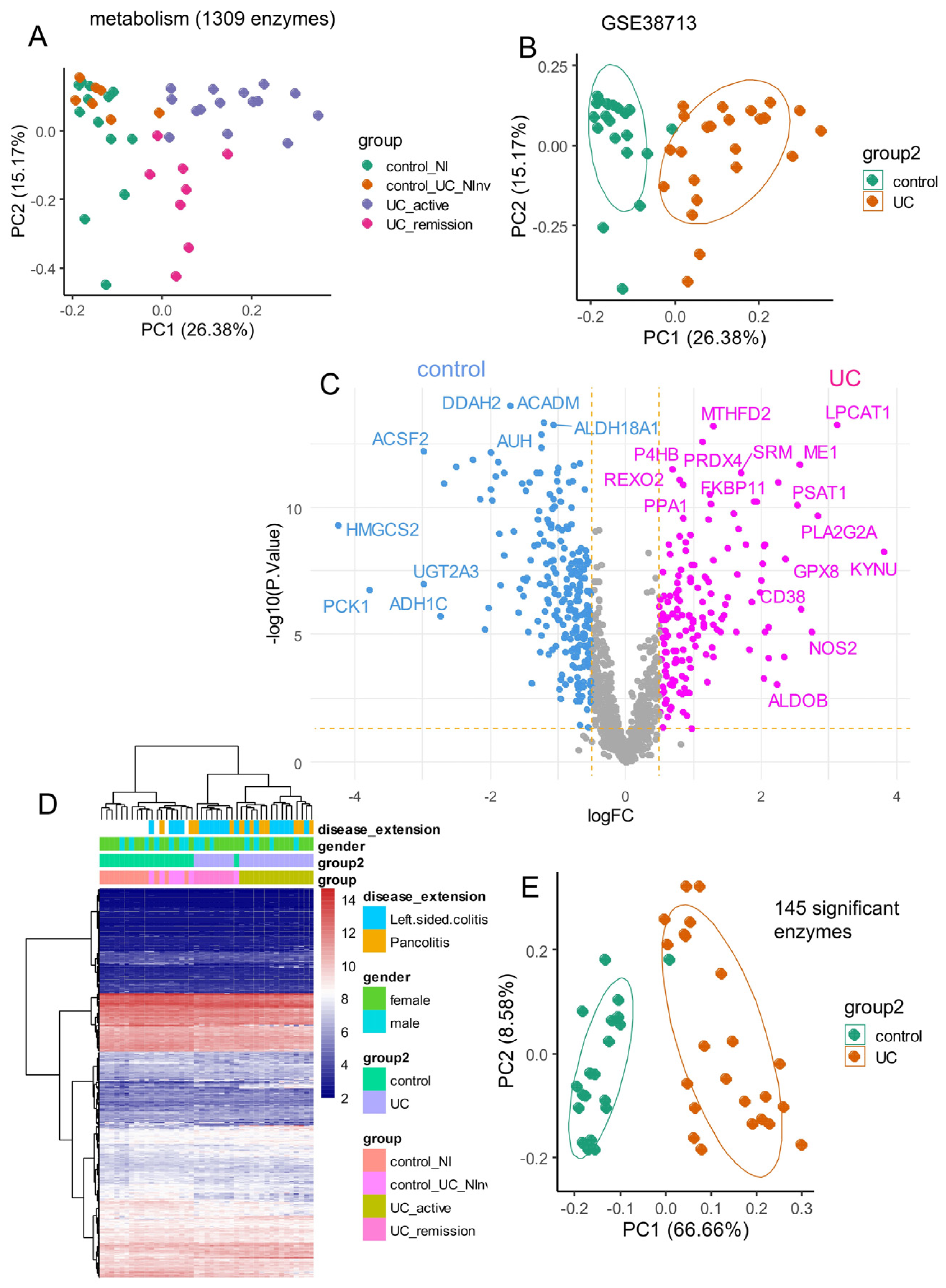
3.1. Metabolic Enzymes Regulated During Ulcerative Colitis
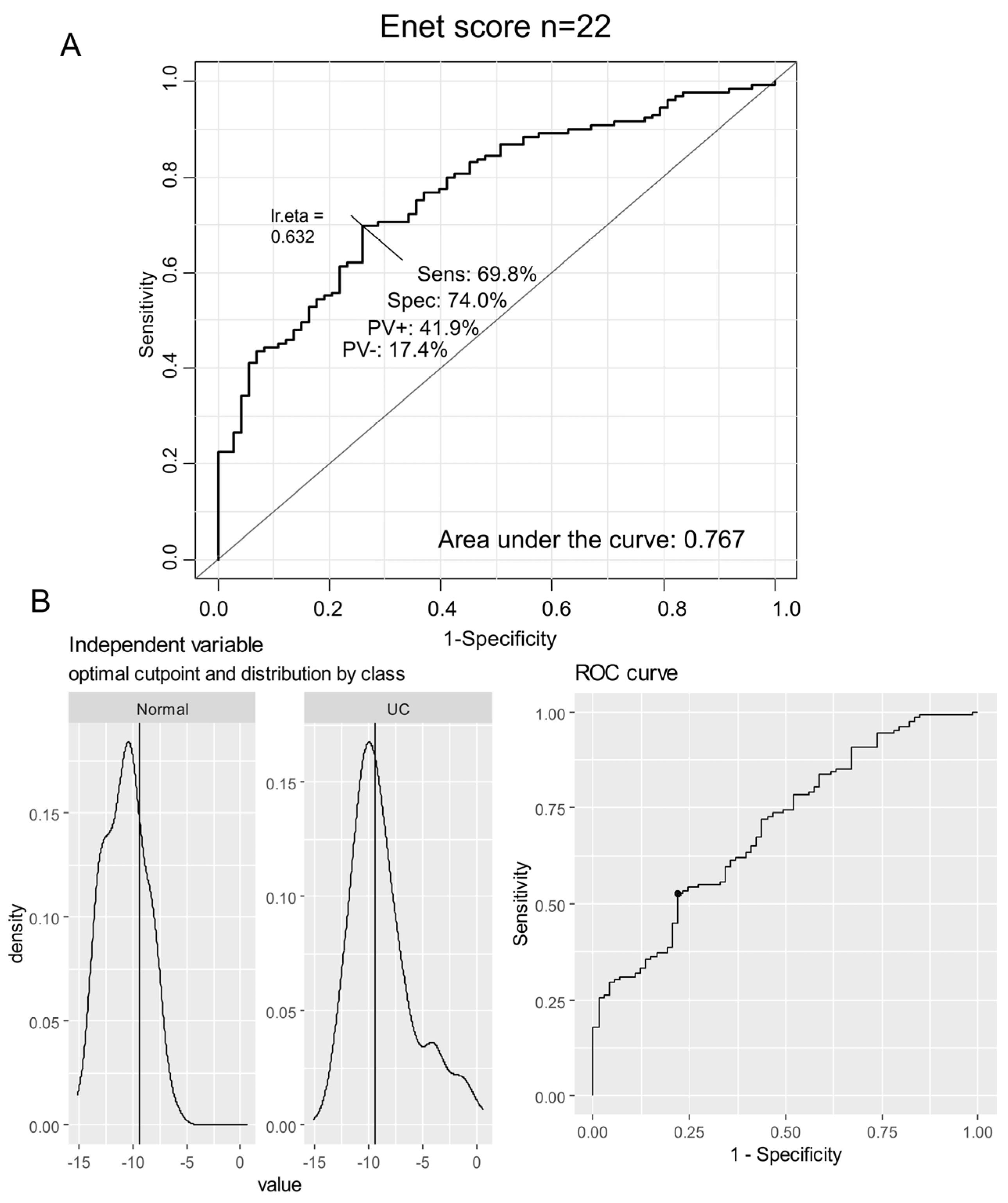
3.2. Elastic Net Validation of a 22-Enzyme Signature Activated in an Independent Cohort of Ulcerative Colitis
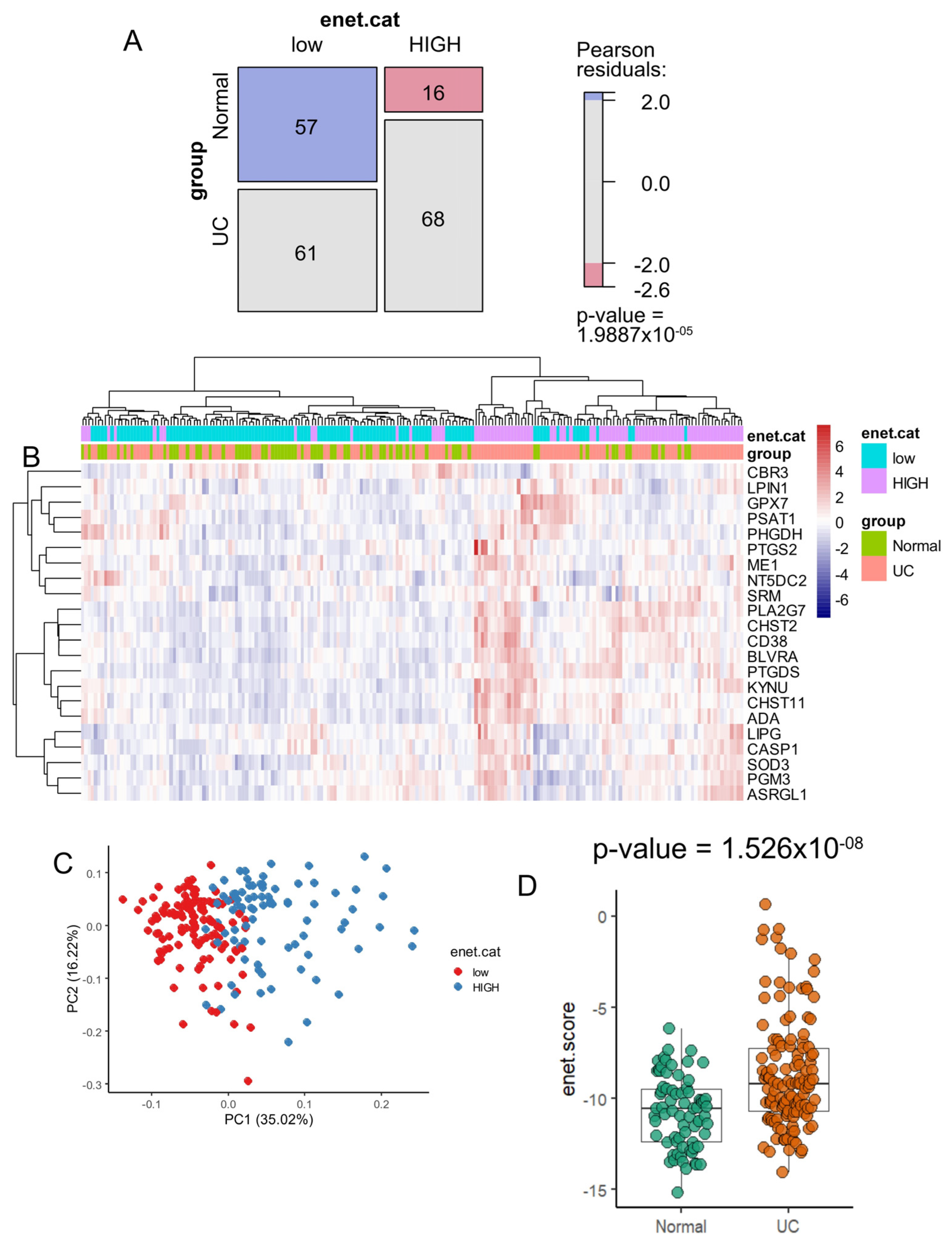
3.3. Enzyme Elastic Net Expression (Enet) Score Is a Significant Marker to Predict Ulcerative Colitis in Colon Tissue
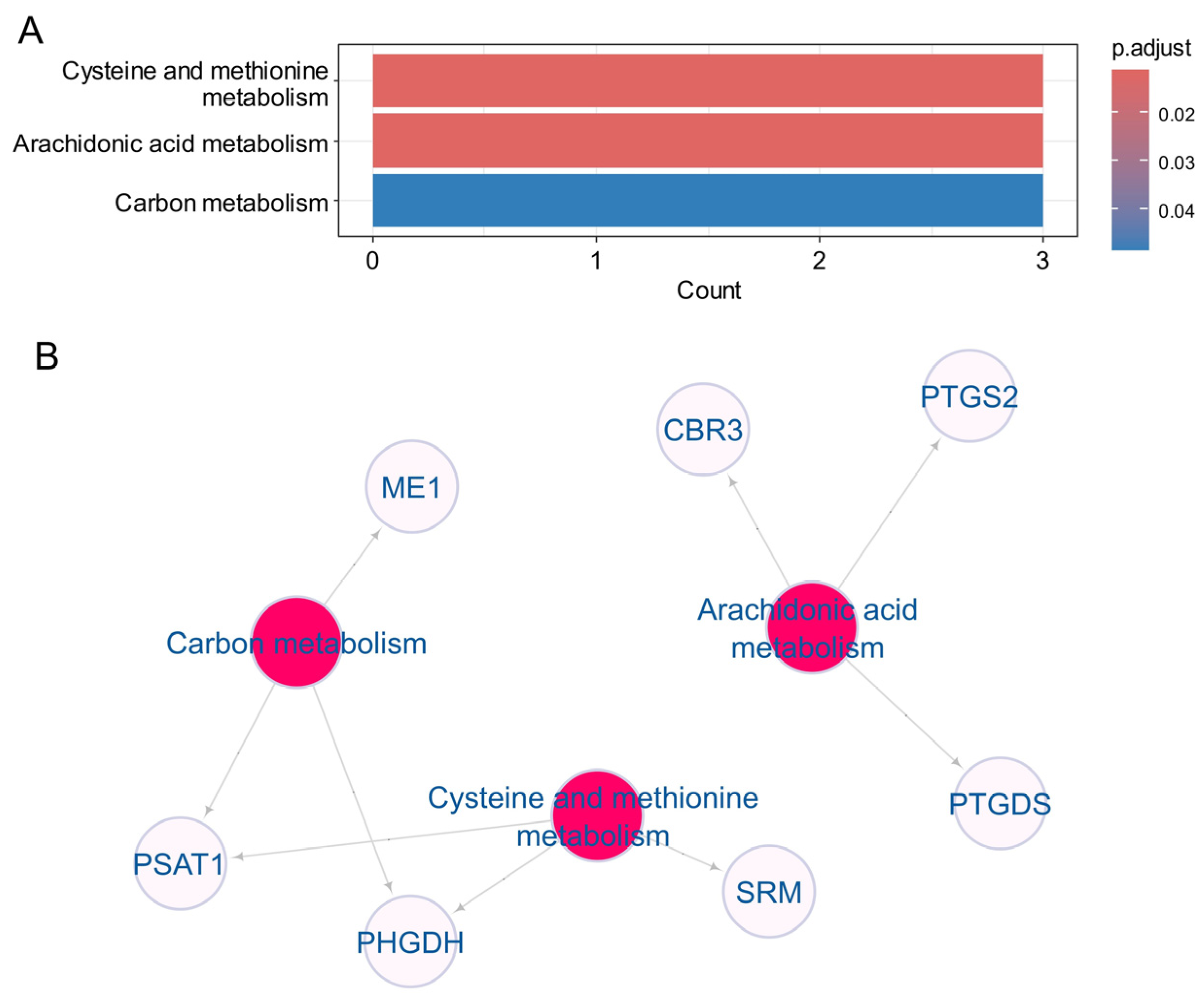
3.4. Heterogeneity Metabolic of the 22 Enet Upregulated Enzyme During Ulcerative Colitis
4. Discussion
Clinical Relevance and Utility of Findings
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, L.; Ha, C. Epidemiology and Pathogenesis of Ulcerative Colitis. Gastroenterol. Clin. N. Am. 2020, 49, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Chung, H.; Xu, Y.; Qiu, H. Trends in the Prevalence and Incidence of Ulcerative Colitis in Japan and the US. Int. J. Colorectal Dis. 2023, 38, 135. [Google Scholar] [CrossRef] [PubMed]
- Kaenkumchorn, T.; Wahbeh, G. Ulcerative Colitis: Making the Diagnosis. Gastroenterol. Clin. N. Am. 2020, 49, 655–669. [Google Scholar] [CrossRef]
- Voelker, R. What Is Ulcerative Colitis? JAMA 2024, 331, 716. [Google Scholar] [CrossRef] [PubMed]
- Ordás, I.; Eckmann, L.; Talamini, M.; Baumgart, D.C.; Sandborn, W.J. Ulcerative Colitis. Lancet 2012, 380, 1606–1619. [Google Scholar] [CrossRef]
- Tatiya-Aphiradee, N.; Chatuphonprasert, W.; Jarukamjorn, K. Immune Response and Inflammatory Pathway of Ulcerative Colitis. J. Basic. Clin. Physiol. Pharmacol. 2018, 30, 1–10. [Google Scholar] [CrossRef]
- Mitsialis, V.; Wall, S.; Liu, P.; Ordovas-Montanes, J.; Parmet, T.; Vukovic, M.; Spencer, D.; Field, M.; McCourt, C.; Toothaker, J.; et al. Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology 2020, 159, 591–608.e10. [Google Scholar] [CrossRef]
- Lefevre, P.L.C.; Wang, Z.; Teft, W.; Zou, G.; Van Viegen, T.; Linggi, B.; Jairath, V.; Feagan, B.G.; Pai, R.K.; Vande Casteele, N. Identification of Immune Cell Markers Associated with Ulcerative Colitis Histological Disease Activity in Colonic Biopsies. J. Clin. Pathol. 2024, jcp-2023-209327. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, Y.; Hao, C.; Guo, X.; Yang, Q.; Du, J.; Hou, Y.; Cao, G.; Li, J.; Wang, H.; et al. The Significance of PD-1/PD-L1 Imbalance in Ulcerative Colitis. PeerJ 2023, 11, e15481. [Google Scholar] [CrossRef]
- Baxi, S.; Yang, A.; Gennarelli, R.L.; Khan, N.; Wang, Z.; Boyce, L.; Korenstein, D. Immune-Related Adverse Events for Anti-PD-1 and Anti-PD-L1 Drugs: Systematic Review and Meta-Analysis. BMJ 2018, 360, k793. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Yang, Y. Update of Gut Gas Metabolism in Ulcerative Colitis. Expert. Rev. Gastroenterol. Hepatol. 2024, 18, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Liu, X.J.; Hao, J.Y. Gut Microbiota in Ulcerative Colitis: Insights on Pathogenesis and Treatment. J. Dig. Dis. 2020, 21, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Niu, C.; Hu, X.-L.; Yuan, Z.-W.; Xiao, Y.; Ji, P.; Wei, Y.-M.; Hua, Y.-L. Pulsatilla Decoction Improves DSS-Induced Colitis via Modulation of Fecal-Bacteria-Related Short-Chain Fatty Acids and Intestinal Barrier Integrity. J. Ethnopharmacol. 2023, 300, 115741. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut Microbiota and IBD: Causation or Correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Tomasello, G.; Mazzola, M.; Leone, A.; Sinagra, E.; Zummo, G.; Farina, F.; Damiani, P.; Cappello, F.; Gerges Geagea, A.; Jurjus, A.; et al. Nutrition, Oxidative Stress and Intestinal Dysbiosis: Influence of Diet on Gut Microbiota in Inflammatory Bowel Diseases. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 2016, 160, 461–466. [Google Scholar] [CrossRef]
- Motley, R.J.; Rhodes, J.; Williams, G.; Tavares, I.A.; Bennett, A. Smoking, Eicosanoids and Ulcerative Colitis. J. Pharm. Pharmacol. 1990, 42, 288–289. [Google Scholar] [CrossRef]
- Casellas, F.; Borruel, N.; Papo, M.; Antolín, M.; Armengol, J.R.; Malagelada, J.R. Usefulness of Rectal Dialysis to Determine Intrarectal Eicosanoids Release in Ulcerative Colitis. Rev. Esp. Enferm. Dig. 1997, 89, 280–288. [Google Scholar]
- Hillingsø, J.G.; Kjeldsen, J.; Schmidt, P.T.; Rasmussen, T.N.; Fisher-Hansen, B.; Holst, J.J.; Lauristen, K.; Bukhave, K.; Rask-Madsen, J. Effects of Topical Ropivacaine on Eicosanoids and Neurotransmitters in the Rectum of Patients with Distal Ulcerative Colitis. Scand. J. Gastroenterol. 2002, 37, 325–329. [Google Scholar] [CrossRef]
- Yang, V.W. Eicosanoids and Inflammatory Bowel Disease. Gastroenterol. Clin. N. Am. 1996, 25, 317–332. [Google Scholar] [CrossRef]
- Hendel, S.K.; Kellermann, L.; Hausmann, A.; Bindslev, N.; Jensen, K.B.; Nielsen, O.H. Tuft Cells and Their Role in Intestinal Diseases. Front. Immunol. 2022, 13, 822867. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, A.T.; Neish, A.S.; Madara, J.L. Mechanisms of Active Intestinal Inflammation and Potential Down-Regulation via Lipoxins. Adv. Exp. Med. Biol. 2002, 507, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Schwanke, R.C.; Marcon, R.; Bento, A.F.; Calixto, J.B. EPA- and DHA-Derived Resolvins’ Actions in Inflammatory Bowel Disease. Eur. J. Pharmacol. 2016, 785, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Planell, N.; Lozano, J.J.; Mora-Buch, R.; Masamunt, M.C.; Jimeno, M.; Ordás, I.; Esteller, M.; Ricart, E.; Piqué, J.M.; Panés, J.; et al. Transcriptional Analysis of the Intestinal Mucosa of Patients with Ulcerative Colitis in Remission Reveals Lasting Epithelial Cell Alterations. Gut 2013, 62, 967–976. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Noble, C.L.; Abbas, A.R.; Cornelius, J.; Lees, C.W.; Ho, G.-T.; Toy, K.; Modrusan, Z.; Pal, N.; Zhong, F.; Chalasani, S.; et al. Regional Variation in Gene Expression in the Healthy Colon Is Dysregulated in Ulcerative Colitis. Gut 2008, 57, 1398–1405. [Google Scholar] [CrossRef]
- Corcoran, C.C.; Grady, C.R.; Pisitkun, T.; Parulekar, J.; Knepper, M.A. From 20th Century Metabolic Wall Charts to 21st Century Systems Biology: Database of Mammalian Metabolic Enzymes. Am. J. Physiol.-Ren. Physiol. 2017, 312, F533–F542. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Desterke, C.; Xiang, Y.; Elhage, R.; Duruel, C.; Chang, Y.; Hamaï, A. Ferroptosis Inducers Upregulate PD-L1 in Recurrent Triple-Negative Breast Cancer. Cancers 2023, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R. Using the Caret Package. J. Stat. Soft. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Soft. 2023, 28, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Ogutu, J.O.; Schulz-Streeck, T.; Piepho, H.-P. Genomic Selection Using Regularized Linear Regression Models: Ridge Regression, Lasso, Elastic Net and Their Extensions. BMC Proc. 2012, 6 (Suppl. S2), S10. [Google Scholar] [CrossRef] [PubMed]
- Thiele, C.; Hirschfeld, G. Cutpointr: Improved Estimation and Validation of Optimal Cutpoints in R. J. Stat. Soft. 2021, 98, 1–27. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Zeileis, A.; Meyer, D.; Hornik, K. Residual-Based Shadings for Visualizing (Conditional) Independence. J. Comput. Graph. Stat. 2007, 16, 507–525. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of Biological Networks and Gene Expression Data Using Cytoscape. Nat. Protoc. 2007, 2, 2366–2382. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Deng, R.; Wang, P.; Lu, X.; Zhao, X.; Wang, X.; Cai, R.; Lin, J. LIPG: An Inflammation and Cancer Modulator. Cancer Gene Ther. 2021, 28, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Shan, Y.; Wang, G.; Wu, Z.; Wang, H.; Wu, S.; Yin, Z.; Wei, J.; Bao, W. Integrated of Multi-Omics and Molecular Docking Reveal PHGDH, PSAT1 and PSPH in the Serine Synthetic Pathway as Potential Targets of T-2 Toxin Exposure in Pig Intestinal Tract. Int. J. Biol. Macromol. 2023, 253, 126647. [Google Scholar] [CrossRef]
- Wu, B.; Qiang, L.; Zhang, Y.; Fu, Y.; Zhao, M.; Lei, Z.; Lu, Z.; Wei, Y.-G.; Dai, H.; Ge, Y.; et al. The Deubiquitinase OTUD1 Inhibits Colonic Inflammation by Suppressing RIPK1-Mediated NF-κB Signaling. Cell Mol. Immunol. 2022, 19, 276–289. [Google Scholar] [CrossRef]
- Piedra-Quintero, Z.L.; Wilson, Z.; Nava, P.; Guerau-de-Arellano, M. CD38: An Immunomodulatory Molecule in Inflammation and Autoimmunity. Front. Immunol. 2020, 11, 597959. [Google Scholar] [CrossRef]
- Cai, B.; Zhao, J.; Zhang, Y.; Liu, Y.; Ma, C.; Yi, F.; Zheng, Y.; Zhang, L.; Chen, T.; Liu, H.; et al. USP5 Attenuates NLRP3 Inflammasome Activation by Promoting Autophagic Degradation of NLRP3. Autophagy 2022, 18, 990–1004. [Google Scholar] [CrossRef]
- Martín-Vázquez, E.; Cobo-Vuilleumier, N.; López-Noriega, L.; Lorenzo, P.I.; Gauthier, B.R. The PTGS2/COX2-PGE2 Signaling Cascade in Inflammation: Pro or Anti? A Case Study with Type 1 Diabetes Mellitus. Int. J. Biol. Sci. 2023, 19, 4157–4165. [Google Scholar] [CrossRef]
- Chen, Y.-I.; Wei, P.-C.; Hsu, J.-L.; Su, F.-Y.; Lee, W.-H. NPGPx (GPx7): A Novel Oxidative Stress Sensor/Transmitter with Multiple Roles in Redox Homeostasis. Am. J. Transl. Res. 2016, 8, 1626–1640. [Google Scholar]
- Kwon, M.-J.; Lee, K.-Y.; Lee, H.-W.; Kim, J.-H.; Kim, T.-Y. SOD3 Variant, R213G, Altered SOD3 Function, Leading to ROS-Mediated Inflammation and Damage in Multiple Organs of Premature Aging Mice. Antioxid. Redox Signal 2015, 23, 985–999. [Google Scholar] [CrossRef]
- Wu, Y.; Ran, L.; Yang, Y.; Gao, X.; Peng, M.; Liu, S.; Sun, L.; Wan, J.; Wang, Y.; Yang, K.; et al. Deferasirox Alleviates DSS-Induced Ulcerative Colitis in Mice by Inhibiting Ferroptosis and Improving Intestinal Microbiota. Life Sci. 2023, 314, 121312. [Google Scholar] [CrossRef]
- Narentuya; Takeda-Uchimura, Y.; Foyez, T.; Zhang, Z.; Akama, T.O.; Yagi, H.; Kato, K.; Komatsu, Y.; Kadomatsu, K.; Uchimura, K. GlcNAc6ST3 Is a Keratan Sulfate Sulfotransferase for the Protein-Tyrosine Phosphatase PTPRZ in the Adult Brain. Sci. Rep. 2019, 9, 4387. [Google Scholar] [CrossRef]
- Chang, W.-M.; Li, L.-J.; Chiu, I.-A.; Lai, T.-C.; Chang, Y.-C.; Tsai, H.-F.; Yang, C.-J.; Huang, M.-S.; Su, C.-Y.; Lai, T.-L.; et al. The Aberrant Cancer Metabolic Gene Carbohydrate Sulfotransferase 11 Promotes Non-Small Cell Lung Cancer Cell Metastasis via Dysregulation of Ceruloplasmin and Intracellular Iron Balance. Transl. Oncol. 2022, 25, 101508. [Google Scholar] [CrossRef] [PubMed]

| GSE38713 | Level | Control (n = 20) | UC (n = 23) | Total (n = 43) | p-Value |
|---|---|---|---|---|---|
| gender | male | 7 (35.0) | 7 (30.4) | 14 (32.6) | |
| female | 13 (65.0) | 16 (69.6) | 29 (67.4) | 1.0000 | |
| group | control_NI (non-inflammatory) | 13 (65.0) | 0 (0.0) | 13 (30.2) | |
| UC_remission | 0 (0.0) | 8 (34.8) | 8 (18.6) | ||
| UC_active | 0 (0.0) | 15 (65.2) | 15 (34.9) | ||
| control_UC_Ninv (non-involved) | 7 (35.0) | 0 (0.0) | 7 (16.3) | <1 × 10−4 | |
| age_years | mean (sd) | 41 (10.3) | 44.8 (10.4) | 43 (10.4) | 0.2201 |
| disease_extension | Left.sided.colitis | 5 (71.4) | 14 (60.9) | 19 (63.3) | |
| Pancolitis | 2 (28.6) | 9 (39.1) | 11 (36.7) | 0.9524 | |
| missing | 13 | 0 | 13 | ||
| treatment | Azathioprine | 4 (57.1) | 11 (47.8) | 15 (50.0) | |
| 5.ASA | 3 (42.9) | 9 (39.1) | 12 (40.0) | ||
| No.treatment | 0 (0.0) | 2 (8.7) | 2 (6.7) | ||
| Systemic.steroids | 0 (0.0) | 1 (4.3) | 1 (3.3) | 0.7952 | |
| missing | 13 | 0 | 13 | ||
| evolution_time_years | mean (sd) | 7.3 (4.5) | 8.6 (7.1) | 8.3 (6.5) | 0.6439 |
| missing | 13 | 0 | 13 |
| GSE11223 | Level | Low (n = 61) | High (n = 68) | Total (n = 129) | p-Value |
|---|---|---|---|---|---|
| tissue | UC Inflamed terminal ileum | 0 (0.0) | 1 (1.5) | 1 (0.8) | |
| UC Inflamed sigmoid colon | 9 (14.8) | 23 (33.8) | 32 (24.8) | ||
| UC Inflamed descending colon | 7 (11.5) | 12 (17.6) | 19 (14.7) | ||
| UC Uninflamed ascending colon | 13 (21.3) | 8 (11.8) | 21 (16.3) | ||
| UC Uninflamed sigmoid colon | 13 (21.3) | 12 (17.6) | 25 (19.4) | ||
| UC Uninflamed terminal ileum | 1 (1.6) | 4 (5.9) | 5 (3.9) | ||
| UC Uninflamed descending colon | 10 (16.4) | 5 (7.4) | 15 (11.6) | ||
| UC Inflamed ascending colon | 8 (13.1) | 3 (4.4) | 11 (8.5) | 0.0350474 | |
| age_diagnosis | mean (sd) | 36.5 (15) | 37.2 (14.6) | 36.9 (14.8) | 0.8027403 |
| joint_problems | FALSE | 61 (100.0) | 66 (97.1) | 127 (98.4) | |
| TRUE | 0 (0.0) | 2 (2.9) | 2 (1.6) | 0.5246148 | |
| uc_fare_up | TRUE | 2 (3.3) | 10 (14.7) | 12 (9.3) | |
| FALSE | 59 (96.7) | 58 (85.3) | 117 (90.7) | 0.0539442 | |
| family_history | FALSE | 51 (83.6) | 65 (95.6) | 116 (89.9) | |
| TRUE | 10 (16.4) | 3 (4.4) | 13 (10.1) | 0.0495199 | |
| Ucss:colonoscopic index of severity | mean (sd) | 2 (1.9) | 3.7 (3.3) | 2.9 (2.9) | 0.0001998 |
| ibd_relative | mean (sd) | 0.2 (0.5) | 0.1 (0.3) | 0.1 (0.4) | 0.1188165 |
| progression | NEW | 2 (3.3) | 9 (13.2) | 11 (8.5) | |
| FAILURE OF THERAPY | 15 (24.6) | 22 (32.4) | 37 (28.7) | ||
| DISEASE IN REMISSION | 44 (72.1) | 37 (54.4) | 81 (62.8) | 0.0492499 | |
| smoking_status | ex | 27 (44.3) | 24 (35.3) | 51 (39.5) | |
| unknown | 0 (0.0) | 3 (4.4) | 3 (2.3) | ||
| never | 30 (49.2) | 32 (47.1) | 62 (48.1) | ||
| current | 4 (6.6) | 9 (13.2) | 13 (10.1) | 0.1871728 | |
| smoking_amount | 15–24 | 8 (13.1) | 6 (8.8) | 14 (10.9) | |
| unknown | 32 (52.5) | 35 (51.5) | 67 (51.9) | ||
| 5–14 | 14 (23.0) | 15 (22.1) | 29 (22.5) | ||
| 25-over | 4 (6.6) | 4 (5.9) | 8 (6.2) | ||
| 0–4 | 3 (4.9) | 8 (11.8) | 11 (8.5) | 0.6708940 | |
| anatomic_location | terminal ileum | 1 (1.6) | 5 (7.4) | 6 (4.7) | |
| sigmoid colon | 22 (36.1) | 35 (51.5) | 57 (44.2) | ||
| descending colon | 17 (27.9) | 17 (25.0) | 34 (26.4) | ||
| ascending colon | 21 (34.4) | 11 (16.2) | 32 (24.8) | 0.0384038 | |
| inflammation_status | Inflamed | 24 (39.3) | 39 (57.4) | 63 (48.8) | |
| Uninflamed | 37 (60.7) | 29 (42.6) | 66 (51.2) | 0.0619667 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desterke, C.; Fu, Y.; Francés, R.; Mata-Garrido, J. Metabolic Transcriptional Activation in Ulcerative Colitis Identified Through scRNA-seq Analysis. Genes 2024, 15, 1412. https://doi.org/10.3390/genes15111412
Desterke C, Fu Y, Francés R, Mata-Garrido J. Metabolic Transcriptional Activation in Ulcerative Colitis Identified Through scRNA-seq Analysis. Genes. 2024; 15(11):1412. https://doi.org/10.3390/genes15111412
Chicago/Turabian StyleDesterke, Christophe, Yuanji Fu, Raquel Francés, and Jorge Mata-Garrido. 2024. "Metabolic Transcriptional Activation in Ulcerative Colitis Identified Through scRNA-seq Analysis" Genes 15, no. 11: 1412. https://doi.org/10.3390/genes15111412
APA StyleDesterke, C., Fu, Y., Francés, R., & Mata-Garrido, J. (2024). Metabolic Transcriptional Activation in Ulcerative Colitis Identified Through scRNA-seq Analysis. Genes, 15(11), 1412. https://doi.org/10.3390/genes15111412







