From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus
Abstract
1. Introduction
1.1. Taxonomical Classification of the Genus Acidithiobacillus
1.2. Ecological Roles and Physiological Diversity
2. Industrial Application of Acidithiobacillus
| Trait | A. ferrooxidans | A. ferrivorans | A. ferriphilus | A. ferridurans | A. ferrianus | A. thiooxidans | A. caldus | A. sulfuriphilus | A. albertensis |
|---|---|---|---|---|---|---|---|---|---|
| % GC | 58.5 | 56.0 | 56.5 | 58.0 | 58.0 | 53.0 | 61.0 | 61.5 | 52.5 |
| Cell size (µm) | 1.0 × 0.5 | 2.4 × 0.5 | 1.0–2.0 | 1.0–2.0 | 1.2–2.5 | 1.0–2.0 × 0.5 | 1.2–1.9 × 0.7 | 1.5–2.5 × 0.5 | 1.0–2.0 × 0.4–0.6 |
| Motility | +/− | + | + | + | + | + | + | + | + |
| Growth pH (optimum) | 1.3–4.5 (2.0–2.5) | 1.9–3.4 (2.5) | 1.5 (2.0) | 1.4–3.0 (2.1) | 1.5–3.0 (2.2) | 0.5–5.5 (2.0–3.0) | 1.0–3.5 (2.0–2.5) | 1.8–7.0 (3.0) | 2.0–4.5 (3.5–4.0) |
| Growth T (°C) (optimum) | 10–37 (30–35) | 4–37 (28–33) | 5–33 (30) | 10–37 (29) | NR (28–30) | 10–37 (28–30) | 32–52 (40–45) | 15–30 (25–28) | 10–40 (25–30) |
| Growth on: | |||||||||
| Sulfur | + | + | + | + | + | + | + | + | + |
| Thiosulfate | + | + | + | + | + | + | + | + | + |
| Metal sulfides | + | + | + | + | +/- | + | - | - | - |
| Ferrous iron | + | + | + | + | + | - | - | - | - |
| Hydrogen | + | +/- | - | + | + | - | + | - | NR |
| References | [1,34,35] | [25,34,36] | [24,34] | [34,37,38] | [4] | [1,6,34] | [1,34,39] | [13] | [1,34,40] |
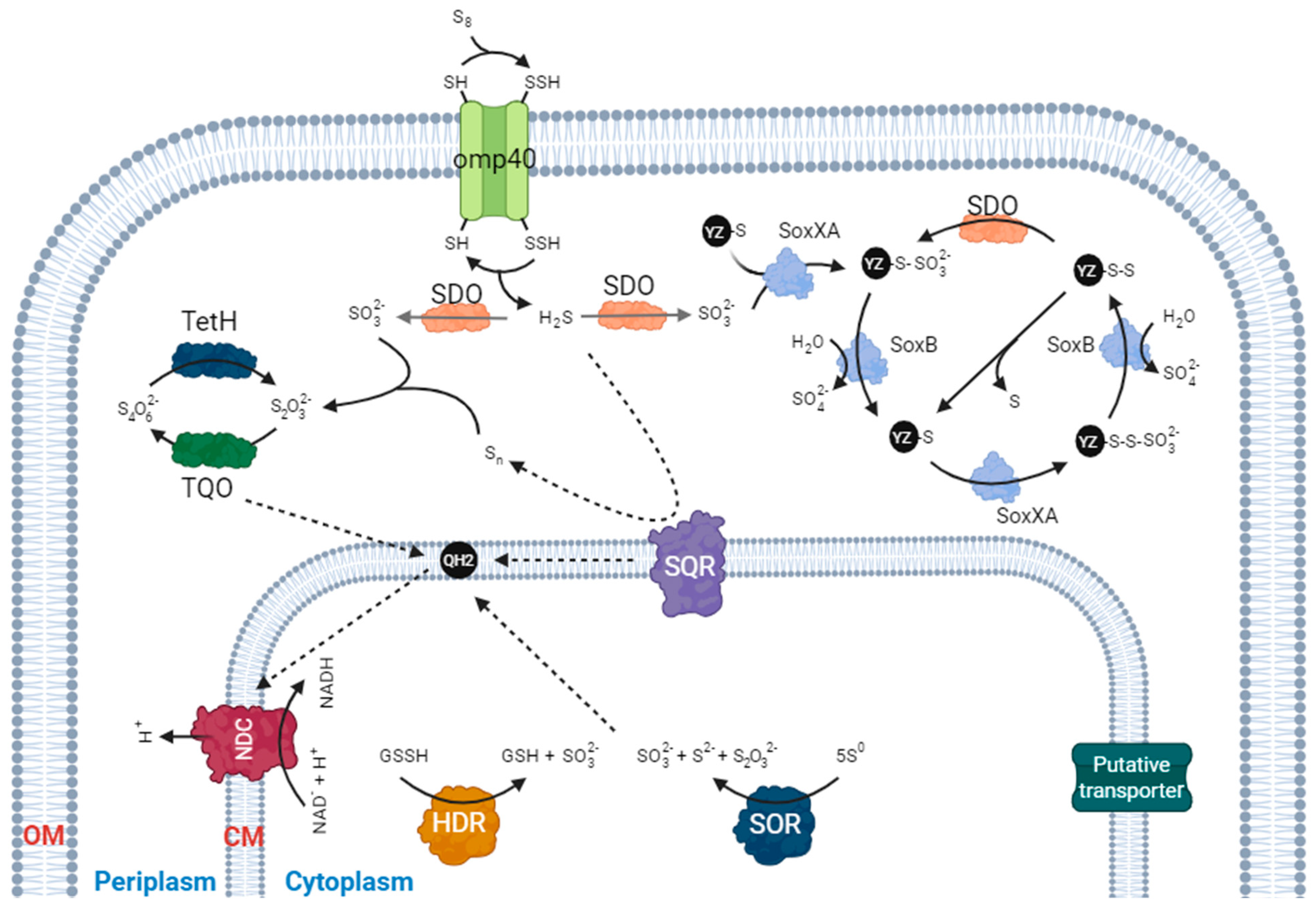
3. Elemental Sulfur Metabolism and Acidithiobacillus
3.1. Sulfur Dioxygenase (SDO)
3.2. Sulfur Oxygenase Reductase (SOR)
3.3. Heterodisulfide Reductase (HDR)-like System
4. Beyond Elemental Sulfur: Other Essential Pathways in Sulfur Metabolism
4.1. Thiosulfate
4.2. Sulfide Oxidation
4.3. Sulfite Oxidation
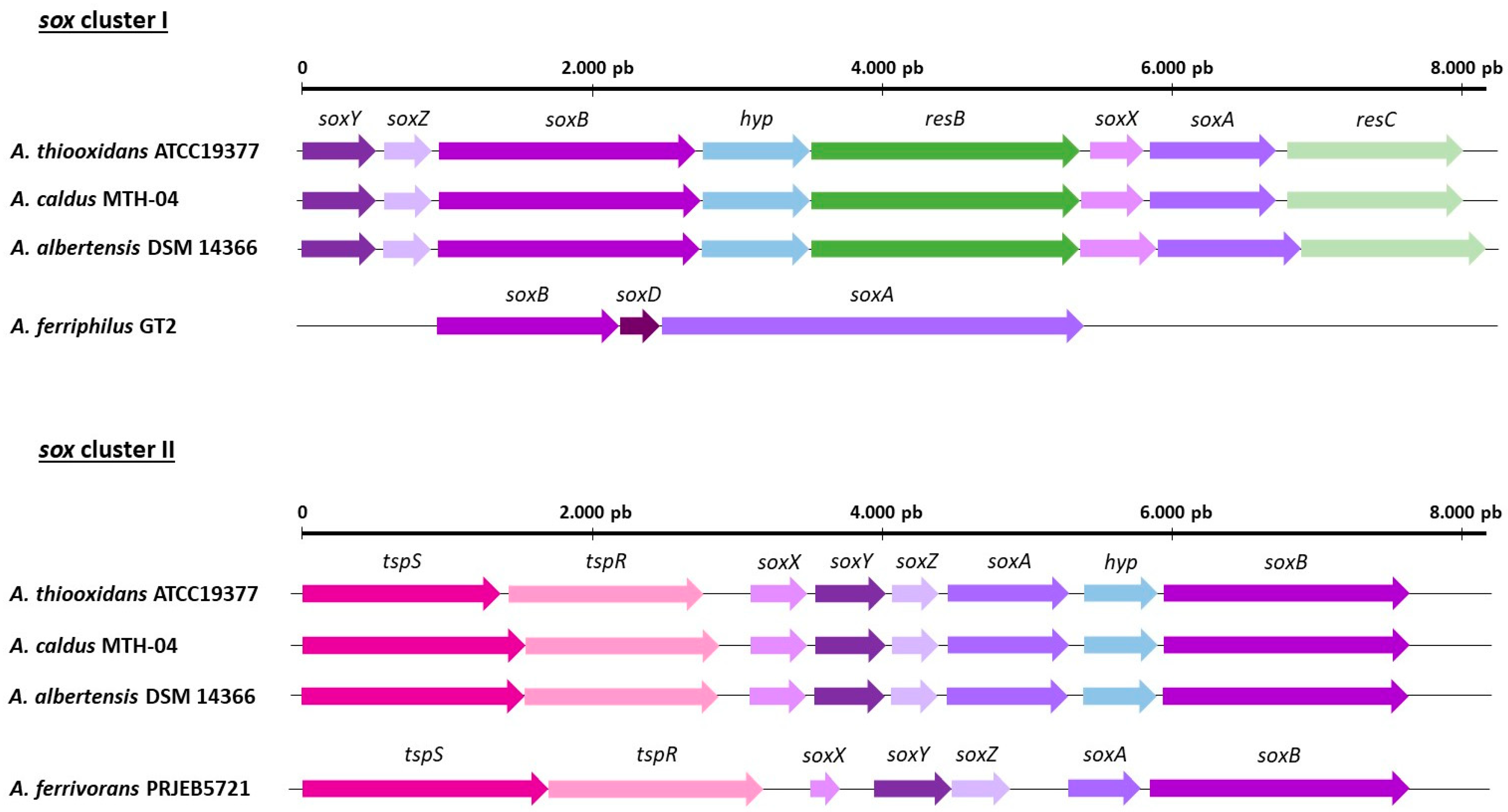
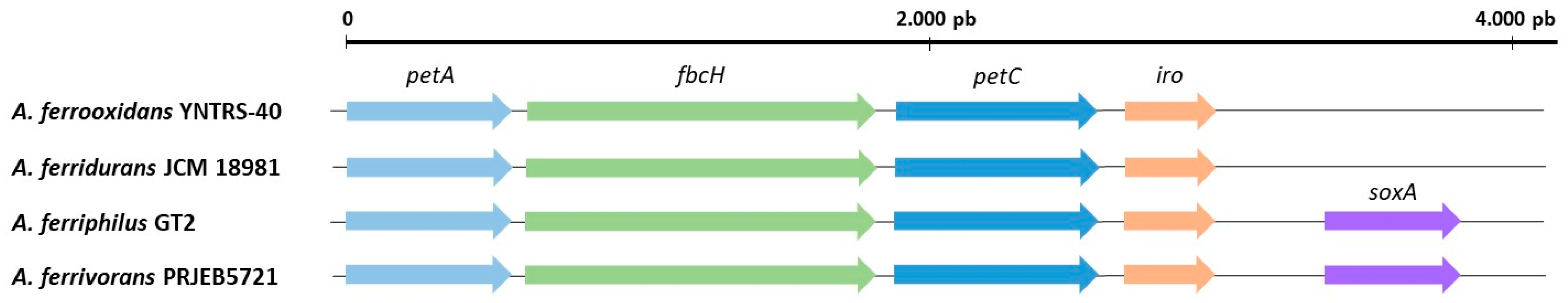
5. Genetic and Molecular Aspects of Acidithiobacillus
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garrity, G.M.; Brenner, D.J.; Krieg, N.R.; Staley, J.T. Bergey’s Manual of Systematic Bacteriology: Volume Two, Part. B. The Gammaproteobacteria, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, 2005; ISBN 978-0-387-95041-9. [Google Scholar]
- Martinez, P.; Vera, M.; Bobadilla-Fazzini, R.A. Omics on Bioleaching: Current and Future Impacts. Appl. Microbiol. Biotechnol. 2015, 99, 8337–8350. [Google Scholar] [CrossRef]
- Moya-Beltrán, A.; Beard, S.; Rojas-Villalobos, C.; Issotta, F.; Gallardo, Y.; Ulloa, R.; Giaveno, A.; Degli Esposti, M.; Johnson, D.B.; Quatrini, R. Genomic Evolution of the Class Acidithiobacillia: Deep-Branching Proteobacteria Living in Extreme Acidic Conditions. ISME J. 2021, 15, 3221–3238. [Google Scholar] [CrossRef] [PubMed]
- Norris, P.R.; Falagán, C.; Moya-Beltrán, A.; Castro, M.; Quatrini, R.; Johnson, D.B. Acidithiobacillus ferrianus sp. nov.: An Ancestral Extremely Acidophilic and Facultatively Anaerobic Chemolithoautotroph. Extremophiles 2020, 24, 329–337. [Google Scholar] [CrossRef]
- Kucera, J.; Lochman, J.; Bouchal, P.; Pakostova, E.; Mikulasek, K.; Hedrich, S.; Janiczek, O.; Mandl, M.; Johnson, D.B. A Model of Aerobic and Anaerobic Metabolism of Hydrogen in the Extremophile Acidithiobacillus ferrooxidans. Front. Microbiol. 2020, 11, 610836. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A.; Joffe, J.S. Microorganisms Concerned in the Oxidation of Sulfur in the Soil: Thiobacillus thiooxidans, a New Sulfur-Oxidizing Organism Isolated from the Soil. J. Bacteriol. 1922, 7, 239–256. [Google Scholar] [CrossRef]
- Kelly, D.P.; Wood, A.P. Reclassification of Some Species of Thiobacillus to the Newly Designated Genera Acidithiobacillus gen. nov., Halothiobacillus Gen. Nov. and Thermithiobacillus Gen. Nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 511–516. [Google Scholar] [CrossRef]
- Inaba, Y.; Kernan, T.; West, A.; Banta, S.; West, A.C. Dispersion of Sulfur Enables Sulfur Oxidation before Iron Oxidation in Acidithiobacillus ferrooxidans: A Valuable Formulation for the Genetic Engineering Toolbox. Biotechnol. Bioeng. 2021, 118, 3225–3238. [Google Scholar] [CrossRef]
- Rana, K.; Rana, N.; Singh, B. Applications of Sulfur Oxidizing Bacteria. In Physiological and Biotechnological Aspects of Extremophiles; Elsevier: Amsterdam, The Netherlands, 2020; pp. 131–136. [Google Scholar]
- Nuñez, H.; Covarrubias, P.C.; Moya-Beltrán, A.; Issotta, F.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Detection, Identification and Typing of Acidithiobacillus Species and Strains: A Review. Res. Microbiol. 2016, 167, 555–567. [Google Scholar] [CrossRef]
- Hua, Z.-S.; Han, Y.-J.; Chen, L.-X.; Liu, J.; Hu, M.; Li, S.-J.; Kuang, J.-L.; Chain, P.S.; Huang, L.-N.; Shu, W.-S. Ecological Roles of Dominant and Rare Prokaryotes in Acid Mine Drainage Revealed by Metagenomics and Metatranscriptomics. ISME J. 2015, 9, 1280–1294. [Google Scholar] [CrossRef]
- Quatrini, R.; Escudero, L.V.; Moya-Beltrán, A.; Galleguillos, P.A.; Issotta, F.; Acosta, M.; Cárdenas, J.P.; Nuñez, H.; Salinas, K.; Holmes, D.S.; et al. Draft Genome Sequence of Acidithiobacillus thiooxidans CLST Isolated from the Acidic Hypersaline Gorbea Salt Flat in Northern Chile. Stand. Genom. Sci. 2017, 12, 84. [Google Scholar] [CrossRef]
- Falagán, C.; Moya-Beltrán, A.; Castro, M.; Quatrini, R.; Johnson, D.B. Acidithiobacillus sulfuriphilus sp. nov.: An Extremely Acidophilic Sulfur-Oxidizing Chemolithotroph Isolated from a Neutral PH Environment. Int. J. Syst. Evol. Microbiol. 2019, 69, 2907–2913. [Google Scholar] [CrossRef] [PubMed]
- González-Rosales, C.; Vergara, E.; Dopson, M.; Valdés, J.H.; Holmes, D.S. Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle. Front. Microbiol. 2022, 12, 822229. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Sharmin, S.; Yoshino, E.; Tokuhisa, M.; Kanao, T. Draft Genome Sequence of Acidithiobacillus sp. Strain SH, a Marine Acidophilic Sulfur-Oxidizing Bacterium. Genome Announc. 2018, 6, e01603-17. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, T.; Cairns, S.; Cowan, D.A.; Danson, M.J.; Hough, D.W.; Johnson, D.B.; Norris, P.R.; Raven, N.; Robinson, C.; Robson, R.; et al. A Microbiological Survey of Montserrat Island Hydrothermal Biotopes. Extremophiles 2000, 4, 305–313. [Google Scholar] [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Li, L.; Liu, Z.; Meng, D.; Liu, X.; Li, X.; Zhang, M.; Tao, J.; Gu, Y.; Zhong, S.; Yin, H. Comparative Genomic Analysis Reveals the Distribution, Organization, and Evolution of Metal Resistance Genes in the Genus Acidithiobacillus. Appl. Environ. Microbiol. 2019, 85, e02153-18. [Google Scholar] [CrossRef]
- Mirete, S.; Morgante, V.; González-Pastor, J.E. Acidophiles: Diversity and Mechanisms of Adaptation to Acidic Environments. In Adaption of Microbial Life to Environmental Extremes; Stan-Lotter, H., Fendrihan, S., Eds.; Springer International Publishing: Cham, Switzerland, 2017; p. 26. ISBN 978-3-319-48325-2. [Google Scholar]
- Nuñez, H.; Moya-Beltrán, A.; Covarrubias, P.C.; Issotta, F.; Cárdenas, J.P.; González, M.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Molecular Systematics of the Genus Acidithiobacillus: Insights into the Phylogenetic Structure and Diversification of the Taxon. Front. Microbiol. 2017, 8, 30. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Li, L.; Wei, G.; Zhang, D.; Liang, Y.; Miao, B. Phylogeny, Divergent Evolution, and Speciation of Sulfur-Oxidizing Acidithiobacillus Populations. BMC Genom. 2019, 20, 438. [Google Scholar] [CrossRef]
- Ibáñez, A.; Barreiro, C.; Diez-Galán, A.; Cobos, R.; Calvo-Peña, C.; Coque, J.J.R. Molecular Identification and Acid Stress Response of an Acidithiobacillus thiooxidans Strain Isolated from Rio Tinto (Spain). Int. J. Mol. Sci. 2023, 24, 13391. [Google Scholar] [CrossRef]
- Espejo, R.T.; Romero, P. Growth of Thiobacillus ferrooxidans on Elemental Sulfur. Appl. Environ. Microbiol. 1987, 53, 1907. [Google Scholar] [CrossRef]
- Falagán, C.; Johnson, D.B. Acidithiobacillus ferriphilus sp. nov., a Facultatively Anaerobic Iron- and Sulfur-Metabolizing Extreme Acidophile. Int. J. Syst. Evol. Microbiol. 2016, 66, 206–211. [Google Scholar] [CrossRef]
- Hallberg, K.B.; González-Toril, E.; Johnson, D.B. Acidithiobacillus ferrivorans sp. nov.; Facultatively Anaerobic, Psychrotolerant Iron-, and Sulfur-Oxidizing Acidophiles Isolated from Metal Mine-Impacted Environments. Extremophiles 2010, 14, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, X.; Tao, J.; Ma, L.; Xiao, Y.; Liang, Y.; Liu, X.; Yin, H. Comparative Genomics of the Extreme Acidophile Acidithiobacillus thiooxidans Reveals Intraspecific Divergence and Niche Adaptation. Int. J. Mol. Sci. 2016, 17, 1355. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Wan, D.; He, K. 16S RDNA and 16S–23S Internal Transcribed Spacer Sequence Analyses Reveal Inter- and Intraspecific Acidithiobacillus Phylogeny. Microbiology 2008, 154, 2397–2407. [Google Scholar] [CrossRef][Green Version]
- Sriaporn, C.; Campbell, K.A.; Van Kranendonk, M.J.; Handley, K.M. Genomic Adaptations Enabling Acidithiobacillus Distribution across Wide-Ranging Hot Spring Temperatures and pHs. Microbiome 2021, 9, 135. [Google Scholar] [CrossRef]
- Feng, G.; Chen, Z.; Zhu, P.; Yan, L.; Hao, X.; Xiao, Y. The Potential Roles of Free and Attached Microbial Community in Decreasing Cadmium Level from Cadmium-Contaminated Soils by Mixotrophic Acidophiles of Different Scale-Up Cultivation Stages. Minerals 2023, 13, 546. [Google Scholar] [CrossRef]
- Jerez, C.A. Biomining of Metals: How to Access and Exploit Natural Resource Sustainably. Microb. Biotechnol. 2017, 10, 1191–1193. [Google Scholar] [CrossRef]
- Johnson, B.D. Biomining—Biotechnologies for Extracting and Recovering Metals from Ores and Waste Materials. Curr. Opin. Biotechnol. 2014, 30, 24–31. [Google Scholar] [CrossRef]
- Rawlings, D.E. Biomining (Mineral Bioleaching, Mineral Biooxidation). In Encyclopedia of Geobiology. Encyclopedia of Earth Sciences Series; Reitner, J., Thiel, V., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 182–185. [Google Scholar]
- Ňancucheo, I.; Johnson, D.B. Selective Removal of Transition Metals from Acidic Mine Waters by Novel Consortia of Acidophilic Sulfidogenic Bacteria. Microb. Biotechnol. 2012, 5, 34–44. [Google Scholar] [CrossRef]
- Peng, T.; Chen, L.; Wang, J.; Miao, J.; Shen, L.; Yu, R.; Gu, G.; Qiu, G.; Zeng, W. Dissolution and Passivation of Chalcopyrite during Bioleaching by Acidithiobacillus ferrivorans at Low Temperature. Minerals 2019, 9, 332. [Google Scholar] [CrossRef]
- He, Z.; Gao, F.; Zhao, J.; Hu, Y.; Qiu, G. Insights into the Dynamics of Bacterial Communities during Chalcopyrite Bioleaching. FEMS Microbiol. Ecol. 2010, 74, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Liljeqvist, M.; Rzhepishevska, O.I.; Dopson, M. Gene Identification and Substrate Regulation Provide Insights into Sulfur Accumulation during Bioleaching with the Psychrotolerant Acidophile Acidithiobacillus ferrivorans. Appl. Environ. Microbiol. 2013, 79, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, D.; Yang, J.; Wang, W.; Chen, P.; Zhang, S.; Yan, L. Acidithiobacillus thiooxidans and its Potential Application. Appl. Microbiol. Biotechnol. 2019, 103, 7819–7833. [Google Scholar] [CrossRef] [PubMed]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R.; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans Metabolism: From Genome Sequence to Industrial Applications. BMC Genom. 2008, 9, 597. [Google Scholar] [CrossRef]
- Jung, H.; Inaba, Y.; Banta, S. Genetic Engineering of the Acidophilic Chemolithoautotroph Acidithiobacillus ferrooxidans. Trends Biotechnol. 2021, 40, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, N.; Liu, X.; Zhou, Z.; Li, Q.; Fang, Y.; Fan, X.; Fu, X.; Liu, Y.; Yin, H. Characterization of the Acid Stress Response of Acidithiobacillus ferrooxidans ATCC 23270 Based on the Method of Microarray. J. Biol. Res. 2012, 17, 3–15. [Google Scholar]
- Zhang, S.; Yan, L.; Xing, W.; Chen, P.; Zhang, Y.; Wang, W. Acidithiobacillus ferrooxidans and its Potential Application. Extremophiles 2018, 22, 563–579. [Google Scholar] [CrossRef]
- Zhan, Y.; Yang, M.; Zhang, S.; Zhao, D.; Duan, J.; Wang, W.; Yan, L. Iron and Sulfur Oxidation Pathways of Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 2019, 35, 60. [Google Scholar] [CrossRef]
- Lee, Y.; Sethurajan, M.; van de Vossenberg, J.; Meers, E.; van Hullebusch, E.D. Recovery of Phosphorus from Municipal Wastewater Treatment Sludge through Bioleaching Using Acidithiobacillus thiooxidans. J. Environ. Manag. 2020, 270, 110818. [Google Scholar] [CrossRef]
- Naseri, T.; Bahaloo-Horeh, N.; Mousavi, S.M. Environmentally Friendly Recovery of Valuable Metals from Spent Coin Cells through Two-Step Bioleaching Using Acidithiobacillus thiooxidans. J. Environ. Manag. 2019, 235, 357–367. [Google Scholar] [CrossRef]
- Berlemont, R.; Gerday, C. Extremophiles. In Comprehensive Biotechnology; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 229–242. ISBN 978-0-08-088504-9. [Google Scholar]
- Plumb, J.J.; Muddle, R.; Franzmann, P.D. Effect of pH on Rates of Iron and Sulfur Oxidation by Bioleaching Organisms. Miner. Eng. 2008, 21, 76–82. [Google Scholar] [CrossRef]
- Gumulya, Y.; Boxall, N.; Khaleque, H.; Santala, V.; Carlson, R.; Kaksonen, A. In a Quest for Engineering Acidophiles for Biomining Applications: Challenges and Opportunities. Genes 2018, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Mykytczuk, N.C.S.; Trevors, J.T.; Ferroni, G.D.; Leduc, L.G. Cytoplasmic Membrane Fluidity and Fatty Acid Composition of Acidithiobacillus ferrooxidans in Response to PH Stress. Extremophiles 2010, 14, 427–441. [Google Scholar] [CrossRef] [PubMed]
- Rouchalova, D.; Rouchalova, K.; Janakova, I.; Cablik, V.; Janstova, S. Bioleaching of Iron, Copper, Lead, and Zinc from the Sludge Mining Sediment at Different Particle Sizes, pH, and Pulp Density Using Acidithiobacillus ferrooxidans. Minerals 2020, 10, 1013. [Google Scholar] [CrossRef]
- Kara, I.T.; Kremser, K.; Wagland, S.T.; Coulon, F. Bioleaching Metal-Bearing Wastes and by-Products for Resource Recovery: A Review. Environ. Chem. Lett. 2023, 1–22. [Google Scholar] [CrossRef]
- Soto, P.E.; Galleguillos, P.A.; Serón, M.A.; Zepeda, V.J.; Demergasso, C.S.; Pinilla, C. Parameters Influencing the Microbial Oxidation Activity in the Industrial Bioleaching Heap at Escondida Mine, Chile. Hydrometallurgy 2013, 133, 51–57. [Google Scholar] [CrossRef]
- Le Borgne, S.; Baquerizo, G. Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas. ChemEngineering 2019, 3, 72. [Google Scholar] [CrossRef]
- Jones, S.; Santini, J.M. Mechanisms of Bioleaching: Iron and Sulfur Oxidation by Acidophilic Microorganisms. Essays Biochem. 2023, 67, 685–699. [Google Scholar] [CrossRef]
- Schippers, A. Microorganisms Involved in Bioleaching and Nucleic Acid-Based Molecular Methods for their Identification and Quantification. In Microbial Processing of Metal. Sulfides; Springer: Dordrecht, The Netherlands, 2007; pp. 3–33. [Google Scholar]
- Wang, R.; Lin, J.-Q.; Liu, X.-M.; Pang, X.; Zhang, C.-J.; Yang, C.-L.; Gao, X.-Y.; Lin, C.-M.; Li, Y.-Q.; Li, Y.; et al. Sulfur Oxidation in the Acidophilic Autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 9, 3290. [Google Scholar] [CrossRef]
- Travisany, D.; Cortés, M.P.; Latorre, M.; Di Genova, A.; Budinich, M.; Bobadilla-Fazzini, R.A.; Parada, P.; González, M.; Maass, A. A New Genome of Acidithiobacillus thiooxidans Provides Insights into Adaptation to a Bioleaching Environment. Res. Microbiol. 2014, 165, 743–752. [Google Scholar] [CrossRef]
- Leathen, W.W.; Kinsel, N.A.; Braley, S.A. Ferrobacillus ferrooxidans: A Chemosynthetic Autotrophic Bacterium. J. Bacteriol. 1956, 72, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Christel, S.; Fridlund, J.; Buetti-Dinh, A.; Buck, M.; Watkin, E.L.; Dopson, M. RNA Transcript Sequencing Reveals Inorganic Sulfur Compound Oxidation Pathways in the Acidophile Acidithiobacillus ferrivorans. FEMS Microbiol. Lett. 2016, 363, fnw057. [Google Scholar] [CrossRef] [PubMed]
- Schippers, A.; Hedrich, S.; Vasters, J.; Drobe, M.; Sand, W.; Willscher, S. Biomining: Metal Recovery from Ores with Microorganisms; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–47. [Google Scholar]
- Hedrich, S.; Johnson, D.B. Acidithiobacillus ferridurans sp. Nov., an Acidophilic Iron-, Sulfur- and Hydrogen-Metabolizing Chemolithotrophic Gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 4018–4025. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Lindstrom, E.B. Characterization of Thiobacillus caldus sp. nov., a Moderately Thermophilic Acidophile. Microbiology 1994, 140, 3451–3456. [Google Scholar] [CrossRef]
- Bryant, R.D.; McGroarty, K.M.; Costerton, J.W.; Laishley, E.J. Isolation and Characterization of a New Acidophilic Thiobacillus species (T. albertis). Can. J. Microbiol. 1983, 29, 1159–1170. [Google Scholar] [CrossRef]
- Wu, W.; Pang, X.; Lin, J.; Liu, X.; Wang, R.; Lin, J.; Chen, L. Discovery of a New Subgroup of Sulfur Dioxygenases and Characterization of Sulfur Dioxygenases in the Sulfur Metabolic Network of Acidithiobacillus caldus. PLoS ONE 2017, 12, e0183668. [Google Scholar] [CrossRef]
- Valenzuela, L.; Chi, A.; Beard, S.; Shabanowitz, J.; Hunt, D.F.; Jerez, C.A. Differential-Expression Proteomics for the Study of Sulfur Metabolism in the Chemolithoautotrophic Acidithiobacillus ferrooxidans. In Microbial Sulfur. Metabolism; Springer: Berlin/Heidelberg, Germnay, 2008; pp. 77–86. [Google Scholar]
- Suzuki, I.; Werkman, C.H. Glutathione and Sulfuroxidation by Thiobacillus thiooxidans. Proc. Natl. Acad. Sci. USA 1959, 45, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Kucera, J.; Pakostova, E.; Janiczek, O.; Mandl, M. Changes in Acidithiobacillus ferrooxidans Ability to Reduce Ferric Iron by Elemental Sulfur. Adv. Mat. Res. 2015, 1130, 97–100. [Google Scholar] [CrossRef]
- Rahman, P.K.S.M.; Gakpe, E. Production, Characterisation and Applications of Biosurfactants-Review. Biotechnology 2008, 7, 360–370. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Liu, J.; Qiu, G. Isolation and Characterization of Acidithiobacillus ferrooxidans Strain QXS-1 Capable of Unusual Ferrous Iron and Sulfur Utilization. Hydrometallurgy 2013, 136, 51–57. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Kim, E.-G.; Park, J.-R.; Ryu, Y.-H.; Moon, W.; Park, G.-H.; Ubaidillah, M.; Ryu, S.-N.; Kim, K.-M. Effect on Chemical and Physical Properties of Soil Each Peat Moss, Elemental Sulfur, and Sulfur-Oxidizing Bacteria. Plants 2021, 10, 1901. [Google Scholar] [CrossRef]
- Kanao, T.; Onishi, M.; Kajitani, Y.; Hashimoto, Y.; Toge, T.; Kikukawa, H.; Kamimura, K. Characterization of Tetrathionate Hydrolase from the Marine Acidophilic Sulfur-Oxidizing Bacterium, Acidithiobacillus thiooxidans Strain SH. Biosci. Biotechnol. Biochem. 2018, 82, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Silver, M.; Lundgren, D.G. Sulfur-Oxidizing Enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 1968, 46, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Sugio, T.; Mizunashi, W.; Inagaki, K.; Tano, T. Purification and Some Properties of Sulfur:Ferric Ion Oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 1987, 169, 4916–4922. [Google Scholar] [CrossRef]
- Suzuki, I. Oxidation of Elemental Sulfur by an Enzyme System of Thiobacillus thiooxidans. Biochim. Biophys. Acta (BBA)—General. Subj. 1965, 104, 359–371. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Liu, X.; Li, X.; Wen, Q.; Lin, J. Identification and Characterization of an ETHE1-like Sulfur Dioxygenase in Extremely Acidophilic Acidithiobacillus spp. Appl. Microbiol. Biotechnol. 2014, 98, 7511–7522. [Google Scholar] [CrossRef]
- Sattler, S.A.; Wang, X.; Lewis, K.M.; DeHan, P.J.; Park, C.-M.; Xin, Y.; Liu, H.; Xian, M.; Xun, L.; Kang, C. Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-β-Lactamase Superfamily. J. Biol. Chem. 2015, 290, 18914–18923. [Google Scholar] [CrossRef]
- Liu, H.; Xin, Y.; Xun, L. Distribution, Diversity, and Activities of Sulfur Dioxygenases in Heterotrophic Bacteria. Appl. Environ. Microbiol. 2014, 80, 1799–1806. [Google Scholar] [CrossRef]
- Jackson, M.R.; Melideo, S.L.; Jorns, M.S. Human Sulfide:Quinone Oxidoreductase Catalyzes the First Step in Hydrogen Sulfide Metabolism and Produces a Sulfane Sulfur Metabolite. Biochemistry 2012, 51, 6804–6815. [Google Scholar] [CrossRef]
- Grings, M.; Seminotti, B.; Karunanidhi, A.; Ghaloul-Gonzalez, L.; Mohsen, A.-W.; Wipf, P.; Palmfeldt, J.; Vockley, J.; Leipnitz, G. ETHE1 and MOCS1 Deficiencies: Disruption of Mitochondrial Bioenergetics, Dynamics, Redox Homeostasis and Endoplasmic Reticulum-Mitochondria Crosstalk in Patient Fibroblasts. Sci. Rep. 2019, 9, 12651. [Google Scholar] [CrossRef]
- Kabil, O.; Banerjee, R. Characterization of Patient Mutations in Human Persulfide Dioxygenase (ETHE1) Involved in H2S Catabolism. J. Biol. Chem. 2012, 287, 44561–44567. [Google Scholar] [CrossRef] [PubMed]
- Rohwerder, T.; Sand, W. The Sulfane Sulfur of Persulfides Is the Actual Substrate of the Sulfur-Oxidizing Enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 2003, 149, 1699–1710. [Google Scholar] [CrossRef]
- Guiliani, N.; Jerez, C.A. Molecular Cloning, Sequencing, and Expression of omp-40, the Gene Coding for the Major Outer Membrane Protein from the Acidophilic Bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 2000, 66, 2318–2324. [Google Scholar] [CrossRef]
- Ramírez, P.; Guiliani, N.; Valenzuela, L.; Beard, S.; Jerez, C.A. Differential Protein Expression during Growth of Acidithiobacillus ferrooxidans on Ferrous Iron, Sulfur Compounds, or Metal Sulfides. Appl. Environ. Microbiol. 2004, 70, 4491–4498. [Google Scholar] [CrossRef] [PubMed]
- Janosch, C.; Remonsellez, F.; Sand, W.; Vera, M. Sulfur Oxygenase Reductase (Sor) in the Moderately Thermoacidophilic Leaching Bacteria: Studies in Sulfobacillus thermosulfidooxidans and Acidithiobacillus caldus. Microorganisms 2015, 3, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-W.; Liu, Y.-Y.; Wu, J.-F.; She, Q.; Jiang, C.-Y.; Liu, S.-J. Novel Bacterial Sulfur Oxygenase Reductases from Bioreactors Treating Gold-Bearing Concentrates. Appl. Microbiol. Biotechnol. 2007, 74, 688–698. [Google Scholar] [CrossRef]
- You, X.-Y.; Guo, X.; Zheng, H.-J.; Zhang, M.-J.; Liu, L.-J.; Zhu, Y.-Q.; Zhu, B.; Wang, S.-Y.; Zhao, G.-P.; Poetsch, A.; et al. Unraveling the Acidithiobacillus caldus Complete Genome and Its Central Metabolisms for Carbon Assimilation. J. Genet. Genom. 2011, 38, 243–252. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Holmes, D.S. Comparative Genome Analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into Their Metabolism and Ecophysiology. Hydrometallurgy 2008, 94, 180–184. [Google Scholar] [CrossRef]
- Chen, L.; Ren, Y.; Lin, J.; Liu, X.; Pang, X.; Lin, J. Acidithiobacillus caldus Sulfur Oxidation Model Based on Transcriptome Analysis between the Wild Type and Sulfur Oxygenase Reductase Defective Mutant. PLoS ONE 2012, 7, e39470. [Google Scholar] [CrossRef]
- Yin, H.; Zhang, X.; Li, X.; He, Z.; Liang, Y.; Guo, X.; Hu, Q.; Xiao, Y.; Cong, J.; Ma, L.; et al. Whole-Genome Sequencing Reveals Novel Insights into Sulfur Oxidation in the Extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 2014, 14, 179. [Google Scholar] [CrossRef]
- Liu, L.-J.; Stockdreher, Y.; Koch, T.; Sun, S.-T.; Fan, Z.; Josten, M.; Sahl, H.-G.; Wang, Q.; Luo, Y.-M.; Liu, S.-J.; et al. Thiosulfate Transfer Mediated by DsrE/TusA Homologs from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina. J. Biol. Chem. 2014, 289, 26949–26959. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C. Cytoplasmic Sulfur Trafficking in Sulfur-Oxidizing Prokaryotes. IUBMB Life 2015, 67, 268–274. [Google Scholar] [CrossRef]
- Ehrenfeld, N.; Levicán, G.J.; Parada, P. Heterodisulfide Reductase from Acidithiobacilli is a Key Component Involved in Metabolism of Reduced Inorganic Sulfur Compounds. Adv. Mat. Res. 2013, 825, 194–197. [Google Scholar] [CrossRef]
- Camacho, D.; Frazao, R.; Fouillen, A.; Nanci, A.; Lang, B.F.; Apte, S.C.; Baron, C.; Warren, L.A. New Insights into Acidithiobacillus thiooxidans Sulfur Metabolism Through Coupled Gene Expression, Solution Chemistry, Microscopy, and Spectroscopy Analyses. Front. Microbiol. 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.; Dahl, C. A Novel Bacterial Sulfur Oxidation Pathway Provides a New Link between the Cycles of Organic and Inorganic Sulfur Compounds. ISME J. 2018, 12, 2479–2491. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Koch, T.; Steffens, L.; Finkensieper, J.; Zigann, R.; Cronan, J.E.; Dahl, C. Lipoate-Binding Proteins and Specific Lipoate-Protein Ligases in Microbial Sulfur Oxidation Reveal an Atypical Role for an Old Cofactor. Elife 2018, 7, e37439. [Google Scholar] [CrossRef]
- Tabita, R.; Silver, M.; Lundgren, D.G. The Rhodanese Enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 1969, 47, 1141–1145. [Google Scholar] [CrossRef]
- Gardner, M.N.; Rawlings, D.E. Production of Rhodanese by Bacteria Present in Bio-Oxidation Plants Used to Recover Gold from Arsenopyrite Concentrates. J. Appl. Microbiol. 2000, 89, 185–190. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Bardischewsky, F.; Rother, D.; Quentmeier, A.; Fischer, J. Prokaryotic Sulfur Oxidation. Curr. Opin. Microbiol. 2005, 8, 253–259. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Quentmeier, A.; Bardischewsky, F.; Rother, D.; Kraft, R.; Kostka, S.; Prinz, H. Novel Genes Coding for Lithotrophic Sulfur Oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 2000, 182, 4677–4687. [Google Scholar] [CrossRef] [PubMed]
- Levicán, G.; Bruscella, P.; Guacunano, M.; Inostroza, C.; Bonnefoy, V.; Holmes, D.S.; Jedlicki, E. Characterization of the PetI and Res. Operons of Acidithiobacillus ferrooxidans. J. Bacteriol. 2002, 184, 1498–1501. [Google Scholar] [CrossRef] [PubMed]
- Moya-Beltrán, A.; Gajdosik, M.; Rojas-Villalobos, C.; Beard, S.; Mandl, M.; Silva-García, D.; Johnson, D.B.; Ramirez, P.; Quatrini, R.; Kucera, J. Influence of Mobile Genetic Elements and Insertion Sequences in Long- and Short-Term Adaptive Processes of Acidithiobacillus ferrooxidans Strains. Sci. Rep. 2023, 13, 10876. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-F.; Fu, L.-J.; Lin, J.-Q.; Pang, X.; Liu, X.-M.; Wang, R.; Wang, Z.-B.; Lin, J.-Q.; Chen, L.-X. The Σ54-Dependent Two-Component System Regulating Sulfur Oxidization (Sox) System in Acidithiobacillus caldus and Some Chemolithotrophic Bacteria. Appl. Microbiol. Biotechnol. 2017, 101, 2079–2092. [Google Scholar] [CrossRef]
- Suhadolnik, M.L.S.; Costa, P.S.; Castro, G.M.; Lobo, F.P.; Nascimento, A.M.A. Comprehensive Insights into Arsenic- and Iron-Redox Genes, Their Taxonomy and Associated Environmental Drivers Deciphered by a Meta-Analysis. Environ. Int. 2021, 146, 106234. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.H.; Bandeiras, T.M.; Urich, T.; Teixeira, M.; Gomes, C.M.; Kletzin, A. Coupling of the Pathway of Sulphur Oxidation to Dioxygen Reduction: Characterization of a Novel Membrane-Bound Thiosulphate:Quinone Oxidoreductase. Mol. Microbiol. 2004, 53, 1147–1160. [Google Scholar] [CrossRef]
- Wang, Z.-B.; Li, Y.-Q.; Lin, J.-Q.; Pang, X.; Liu, X.-M.; Liu, B.-Q.; Wang, R.; Zhang, C.-J.; Wu, Y.; Lin, J.-Q.; et al. The Two-Component System RsrS-RsrR Regulates the Tetrathionate Intermediate Pathway for Thiosulfate Oxidation in Acidithiobacillus caldus. Front. Microbiol. 2016, 7, 1755. [Google Scholar] [CrossRef]
- Rzhepishevska, O.I.; Valdés, J.; Marcinkeviciene, L.; Gallardo, C.A.; Meskys, R.; Bonnefoy, V.; Holmes, D.S.; Dopson, M. Regulation of a Novel Acidithiobacillus caldus Gene Cluster Involved in Metabolism of Reduced Inorganic Sulfur Compounds. Appl. Environ. Microbiol. 2007, 73, 7367–7372. [Google Scholar] [CrossRef]
- van Zyl, L.J.; van Munster, J.M.; Rawlings, D.E. Construction of arsB and tetH Mutants of the Sulfur-Oxidizing Bacterium Acidithiobacillus caldus by Marker Exchange. Appl. Environ. Microbiol. 2008, 74, 5686–5694. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, X.; Wang, H.; Li, X.; Lin, J. Construction and Characterization of TetH Overexpression and Knockout Strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 2014, 196, 2255–2264. [Google Scholar] [CrossRef]
- Kanao, T.; Kamimura, K.; Sugio, T. Identification of a Gene Encoding a Tetrathionate Hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 2007, 132, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Beard, S.; Paradela, A.; Albar, J.P.; Jerez, C.A. Growth of Acidithiobacillus ferrooxidans ATCC 23270 in Thiosulfate Under Oxygen-Limiting Conditions Generates Extracellular Sulfur Globules by Means of a Secreted Tetrathionate Hydrolase. Front. Microbiol. 2011, 2, 79. [Google Scholar] [CrossRef] [PubMed]
- Griesbeck, C.; Hauska, G.; Schütz, M. Biological Sulfide Oxidation: Sulfide-Quinone Reductase (SQR), the Primary Reaction. Recent. Res. Dev. Microbiol. 2000, 4, 179–203. [Google Scholar]
- Griesbeck, C.; Schütz, M.; Schödl, T.; Bathe, S.; Nausch, L.; Mederer, N.; Vielreicher, M.; Hauska, G. Mechanism of Sulfide-Quinone Reductase Investigated Using Site-Directed Mutagenesis and Sulfur Analysis. Biochemistry 2002, 41, 11552–11565. [Google Scholar] [CrossRef]
- Cherney, M.M.; Zhang, Y.; Solomonson, M.; Weiner, J.H.; James, M.N.G. Crystal Structure of Sulfide:Quinone Oxidoreductase from Acidithiobacillus ferrooxidans: Insights into Sulfidotrophic Respiration and Detoxification. J. Mol. Biol. 2010, 398, 292–305. [Google Scholar] [CrossRef]
- Zhang, Y.; Weiner, J.H. Characterization of the Kinetics and Electron Paramagnetic Resonance Spectroscopic Properties of Acidithiobacillus ferrooxidans Sulfide:Quinone Oxidoreductase (SQR). Arch. Biochem. Biophys. 2014, 564, 110–119. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Zhao, D.; Ni, Y.; Wang, W.; Yan, L. Complete Genome Sequence of Acidithiobacillus ferrooxidans YNTRS-40, a Strain of the Ferrous Iron- and Sulfur-Oxidizing Acidophile. Microorganisms 2019, 8, 2. [Google Scholar] [CrossRef]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Extending the Models for Iron and Sulfur Oxidation in the Extreme Acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009, 10, 394. [Google Scholar] [CrossRef]
- Kupka, D.; Liljeqvist, M.; Nurmi, P.; Puhakka, J.A.; Tuovinen, O.H.; Dopson, M. Oxidation of Elemental Sulfur, Tetrathionate and Ferrous Iron by the Psychrotolerant Acidithiobacillus strain SS3. Res. Microbiol. 2009, 160, 767–774. [Google Scholar] [CrossRef]
- Hao, L.; Liu, X.; Wang, H.; Lin, J.; Pang, X.; Lin, J. Detection and Validation of a Small Broad-Host-Range Plasmid PBBR1MCS-2 for Use in Genetic Manipulation of the Extremely Acidophilic Acidithiobacillus sp. J. Microbiol. Methods 2012, 90, 309–314. [Google Scholar] [CrossRef]
- Chen, L.; Lin, J.; Li, B.; Lin, J.; Liu, X. Method Development for Electrotransformation of Acidithiobacillus caldus. J. Microbiol. Biotechnol. 2010, 20, 39–44. [Google Scholar] [CrossRef]
- Meng, J.; Wang, H.; Liu, X.; Lin, J.; Pang, X.; Lin, J. Construction of Small Plasmid Vectors for Use in Genetic Improvement of the Extremely Acidophilic Acidithiobacillus caldus. Microbiol. Res. 2013, 168, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lin, J.; Liu, X.; Pang, X.; Lin, H.; Lin, J. Transposition of IS Elements Induced by Electroporation of Suicide Plasmid in Acidithiobacillus caldus. Enzyme Microb. Technol. 2013, 53, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Mahadevan, R. Genetic Engineering of Acidithiobacillus ferridurans with CRISPR-Cas9/DCas9 Systems. bioRxiv 2022. [Google Scholar] [CrossRef]
- Krier, J.B.; Kalia, S.S.; Green, R.C. Genomic Sequencing in Clinical Practice: Applications, Challenges, and Opportunities. Dialogues Clin. Neurosci. 2016, 18, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Fariq, A.; Blazier, J.C.; Yasmin, A.; Gentry, T.J.; Deng, Y. Whole Genome Sequence Analysis Reveals High Genetic Variation of Newly Isolated Acidithiobacillus ferrooxidans IO-2C. Sci. Rep. 2019, 9, 13049. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.S.; Schaperdoth, I.; Macalady, J.L. Biogeography of Sulfur-Oxidizing Acidithiobacillus Populations in Extremely Acidic Cave Biofilms. ISME J. 2016, 10, 2879–2891. [Google Scholar] [CrossRef]
- Gu, H.; Liang, S.; Zhao, J. Novel Sequencing and Genomic Technologies Revolutionized Rice Genomic Study and Breeding. Agronomy 2022, 12, 218. [Google Scholar] [CrossRef]
- Feng, S.; Qiu, Y.; Huang, Z.; Yin, Y.; Zhang, H.; Zhu, D.; Tong, Y.; Yang, H. The Adaptation Mechanisms of Acidithiobacillus caldus CCTCC M 2018054 to Extreme Acid Stress: Bioleaching Performance, Physiology, and Transcriptomics. Environ. Res. 2021, 199, 111341. [Google Scholar] [CrossRef]
- Yin, Z.; Feng, S.; Tong, Y.; Yang, H. Adaptive Mechanism of Acidithiobacillus thiooxidans CCTCC M 2012104 under Stress during Bioleaching of Low-Grade Chalcopyrite Based on Physiological and Comparative Transcriptomic Analysis. J. Ind. Microbiol. Biotechnol. 2019, 46, 1643–1656. [Google Scholar] [CrossRef]
- Li, M.; Wen, J. Recent Progress in the Application of Omics Technologies in the Study of Bio-Mining Microorganisms from Extreme Environments. Microb. Cell. Fact. 2021, 20, 178. [Google Scholar] [CrossRef] [PubMed]

| A. ferrooxidans | A. ferrivorans | A. ferriphilus | A. ferridurans | A. ferrianus | A. thiooxidans | A. caldus | A. sulfuriphilus | A. albertensis | |
|---|---|---|---|---|---|---|---|---|---|
| Reference genome | ASM1346280v1 | NEW_PRJEB5721 | ASM2084402v1 | ASM396665v1 | ASM1037809v1 | ASM966247v1 | ASM869422v1 | ASM372122v1 | ASM193165v1 |
| Strain | YNTRS-40 | PRJEB5721 | GT2 | JCM 18981 | MG | ATCC 19377 | MTH-04 | CJ-2 | DSM 14366 |
| Deposited genomes | 64 | 12 | 12 | 9 | 1 | 26 | 24 | 1 | 2 |
| Completed genomes | 5 | 3 | 2 | 1 | 0 | 1 | 4 | 0 | 0 |
| Total genes | 3542 | 3781 | 2633 | 3173 | 3467 | 3707 | 2995 | 3083 | 3909 |
| Hypothetical proteins | 38.18% | 43.67% | 34.26% | 36.53% | 39.72% | 45.75% | 41.17% | 38.11% | 43.34% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibáñez, A.; Garrido-Chamorro, S.; Coque, J.J.R.; Barreiro, C. From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus. Genes 2023, 14, 1772. https://doi.org/10.3390/genes14091772
Ibáñez A, Garrido-Chamorro S, Coque JJR, Barreiro C. From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus. Genes. 2023; 14(9):1772. https://doi.org/10.3390/genes14091772
Chicago/Turabian StyleIbáñez, Ana, Sonia Garrido-Chamorro, Juan J. R. Coque, and Carlos Barreiro. 2023. "From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus" Genes 14, no. 9: 1772. https://doi.org/10.3390/genes14091772
APA StyleIbáñez, A., Garrido-Chamorro, S., Coque, J. J. R., & Barreiro, C. (2023). From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus. Genes, 14(9), 1772. https://doi.org/10.3390/genes14091772







