Genetic Enhancement of Cereals Using Genomic Resources for Nutritional Food Security
Abstract
1. Introduction
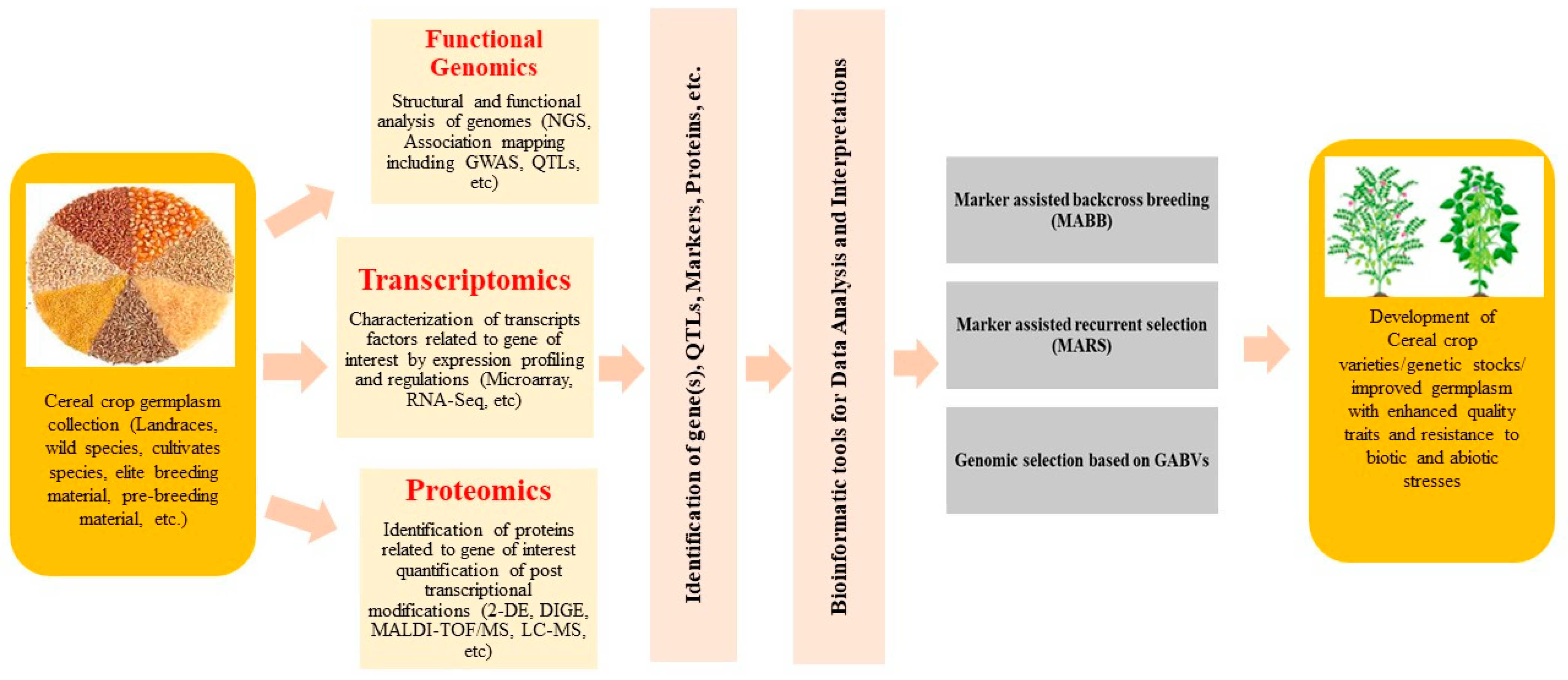
2. Genomic Resources
| Crop | Botanical Name | Genome Size (Mb) and Sequencing Method | Reference |
|---|---|---|---|
| Rice | Oryza sativa ssp. japonica (Nipponbare) Oryza sativa ssp. japonica (Nipponbare) Oryza sativa ssp. Indica |
| [22] [69] [70] |
| Maize | Zea mays (Palomero Toluqueno) (popcorn) Zea mays (B73) |
| [71] [24] |
| Sorghum | Sorghum bicolor (L.) Moench | 730 Mb; Sanger, Whole-Genome Sequencing (WGS) | [23,72] |
| Foxtail millet | Setaria italica | 515 Mb; Illumina, Whole-Genome Sequencing (WGS) | [73] |
| Bread wheat | Triticum aestivum | 17,000 Mb; 454, Whole-Genome Sequencing (WGS) | [74] |
| Barley | Hordeum vulgare | 5100 Mb; 454, Bacterial Artificial Chromosome (BAC-by-BAC) | [26,75] |
| Finger millet | Eleusine coracana | 1200 Mb; Illumina, Whole-Genome Sequencing (WGS) | [76,77] |
| Pearl millet | Cenchrus americanus | 1800 Mb; Illumina, Whole-Genome Sequencing (WGS) | [25] |
| Proso millet | Panicum miliaceum | 923 Mb; Illumina short-read coupled with Pac-Bio long-read sequencing | [78] |
| Barnyard millet | Echinochloa esculenta | 1.27 Mb; Illumina HiSeq platform | [79] |
3. Genomic Resources and Their Implications
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tasgin, E. Macronutrients and micronutrients in nutrition. Int. J. Innov. Res. Rev. 2017, 1, 10–15. [Google Scholar]
- Salse, J.; Feuillet, C. Comparative genomics of cereals. In Genomics-Assisted Crop Improvement; Springer Science+Business Media: New York, NY, USA, 2007; pp. 177–205. [Google Scholar]
- Lata, C.; Shivhare, R. Engineering cereal crops for enhanced abiotic stress tolerance. Proc. Indian Natl. Sci. Acad. 2021, 87, 63–83. [Google Scholar] [CrossRef]
- Sharma, M.; Kishore, A.; Roy, D.; Joshi, K. A comparison of the Indian diet with the EAT-Lancet reference diet. BMC Public Health 2020, 20, 812. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Chung, G. Genetic improvement of cereals and grain legumes. Genes 2020, 11, 1255. [Google Scholar] [CrossRef]
- Campbell, B.M.; Vermeulen, S.J.; Aggarwal, P.K.; Corner-Dollo, C.; Girvetz, E.; Loboguerrero, A.M.; Ramirez-Villegas, J.; Rosenstock, T.; Sebastian, L.; Thornton, P.K. Reducing risks to food security from climate change. Glob. Food Secur. 2016, 11, 34–43. [Google Scholar] [CrossRef]
- UNO United Nations Organization, Department of Economic and Social Affairs, Population Division. World Population Prospects 2019: Highlights. 2019. Available online: https://population.un.org/wpp/publications/files/wpp2019_highlights.pdf (accessed on 29 June 2023).
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Massawe, F.; Mayes, S.; Cheng, A. Crop diversity: An unexploited treasure trove for food security. Trends Plant Sci. 2016, 21, 365–368. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Sood, M.; Jasrotia, M. Underutilized Crops and Their Value Addition; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2021; p. 11788. [Google Scholar]
- McKevith, B. Nutritional aspects of cereals. Nutr. Bull. 2004, 29, 111–142. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Gupta, M. Exploring the role of epigenetics in cereal and leguminous crops exposed to abiotic stress. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Transcriptional Regulation and Chromatin Remodeling in Plants; Springer: Cham, Switzerland, 2019; pp. 149–170. [Google Scholar]
- Eliazer Nelson, A.R.L.; Ravichandran, K.; Antony, U. The impact of the Green Revolution on indigenous crops of India. J. Ethn. Foods 2019, 6, 8. [Google Scholar] [CrossRef]
- Bouis, H.E.; Saltzman, A. Improving nutrition through biofortification: A review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Secur. 2017, 12, 49–58. [Google Scholar] [CrossRef]
- World Health Organization. Global Nutrition Report 2017: Nourishing the SDGs; Development Initiatives: Bristol, UK, 2017. [Google Scholar]
- Lyons, G.; Stangoulis, J.; Graham, R. High-selenium wheat: Biofortification for better health. Nutr. Res. Rev. 2003, 16, 45–60. [Google Scholar] [CrossRef]
- Buttriss, J. Nutrition and Food Processing; British Nutrition Foundation: London, UK, 1999. [Google Scholar]
- Henry, R.J. Biotechnology. In Cereals Processing Technology; Owens, G., Ed.; Woodhead Publishing: Abington, UK, 2001; pp. 53–76. [Google Scholar]
- Khush, G.S. Challenges for meeting the global food and nutrient needs in the new millenium. Proc. Nutr. Soc. 2001, 60, 15–26. [Google Scholar] [CrossRef]
- Lucca, P.; Hurrell, R.; Potrytkus, I. Fighting iron deficiency with iron-rich rice. J. Am. Coll. Nutr. 2002, 21, 184S–190S. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Zhang, H.; Zhao, Y.; Wang, X.; Rathore, A.; et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 2017, 35, 969–976. [Google Scholar] [CrossRef]
- Mayer, K.F.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Mochida, K.; Shinozaki, K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 2010, 51, 497–523. [Google Scholar] [CrossRef] [PubMed]
- Rakkammal, K.; Priya, A.; Pandian, S.; Maharajan, T.; Rathinapriya, P.; Satish, L.; Ceasar, S.A.; Sohn, S.-I.; Ramesh, M. Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview. Plants 2022, 11, 2852. [Google Scholar] [CrossRef] [PubMed]
- Salgotra, R.K.; Stewart, N.C. Functional markers for precision plant breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef] [PubMed]
- Pandian, S.; Rakkammal, K.; Rency, A.S.; Muthuramalingam, P.; Pandian, S.K.; Ramesh, M. Abiotic stress and applications of omics approaches to develop stress tolerance in agronomic crops. In Agronomic Crops: Volume 3: Stress Responses and Tolerance; 2020; pp. 557–578. [Google Scholar]
- Zhan, X.; Lu, Y.; Zhu, J.; Botella, J.R. Genome editing for plant research and crop improvement. J. Integr. Plant Biol. 2021, 63, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Salgotra, R.K.; Gupta, B.B.; Monika, S. Biotechnological interventions and their role in sustainable hill agriculture. J. Plant Sci. Res. 2015, 2, 1–8. [Google Scholar]
- Paes de Melo, B.; de Carpinetti, P.A.; Fraga, O.T.; Rodrigues-Silva, P.L.; Fioresi, V.S.; de Camargos, L.F.; da Silva Ferreira, M.F. Abiotic stresses in plants and their markers: A practice view of plant stress responses and programmed cell death mechanisms. Plants 2022, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Yaqoob, U.; Jan, N.; Raman, P.V.; Siddique, K.H.M.; John, R. Crosstalk between brassinosteroid signaling, ROS signaling and phenylpropanoid pathway during abiotic stress in plants: Does It exist? Plant Stress 2022, 4, 100075. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Stewart, C.N. Genetic augmentation of legume crops using genomic resources and genotyping platforms for nutritional food security. Plants 2022, 11, 1866. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Gupta, P.K.; Varshney, R.K. The Development and Use of Microsatellite Markers for Genetic Analysis and Plant Breeding with the Emphasis on Bread Wheat. Euphytica 2000, 113, 163–185. [Google Scholar] [CrossRef]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, 64062. [Google Scholar] [CrossRef]
- Sivprakash, K.R.; Prashanth, S.R.; Mohanty, B.P.; Parida, A. Genetic diversity of Black gram landraces as evaluated by AFLP markers. Curr. Sci. 2004, 86, 1411–1415. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Tautz, D. Hypervariability of simple sequences as a general source of polymorphic DNA markers. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, A.; Ausubel, F.M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993, 4, 403–410. [Google Scholar] [CrossRef]
- Paran, I.; Michelmore, R.W. Development of reliable PCR based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 1993, 85, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Lee, T.V.D.; Hornes, M.; Frijters, A.; Pot, J.; Paleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Gupta, P.K.; Roy, J.K.; Prasad, M. Single nucleotide polymorphisms: A new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr. Sci. 2001, 80, 524–535. [Google Scholar]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.V.P.R.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef]
- Pandey, M.K.; Roorkiwal, M.; Singh, V.; Ramalingam, A.; Kudapa, H.; Thudi, M.; Chitikineni, A.; Rathore, A.; Varshney, R.K. Emerging genomic tools for legume breeding: Current status and future prospects. Front. Plant Sci. 2016, 7, 455. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Zargar, S.M. Rediscovery of Genetic and Genomic Resources for Future Food Security; Springer: Singapore, 2020. [Google Scholar]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, R.; Duraiyan, J.; Kaliyappan, K.; Palanisamy, M. Microarray and its applications. J. Pharm. Bioallied Sci. 2012, 4, S310–S312. [Google Scholar] [PubMed]
- Russo, G.; Zegar, C.; Giordano, A. Advantages and limitations of microarray technology in human cancer. Oncogene 2003, 22, 6497–6507. [Google Scholar] [CrossRef] [PubMed]
- Loy, A.; Bodrossy, L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin. Chim. Acta 2006, 363, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Sabreena; Nazir, M.; Ganai, B.A.; Mir, R.A.; Zargar, S.M. Genetics and Genomics Resources of Millets: Availability, Advancements, and Applications. In Neglected and Underutilized Crops-Towards Nutritional Security and Sustainability; Springer: Singapore, 2021; pp. 153–166. [Google Scholar]
- Afzal, M.; Alghamdi, S.S.; Migdadi, H.H.; Khan, M.A.; Mirza, S.B.; El-Harty, E. Legume genomics and transcriptomics: From classic breeding to modern technologies. Saudi J. Biol. Sci. 2020, 27, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Kelley, J.M.; Gocayne, J.D.; Dubnick, M.; Polymeropoulos, M.H.; Xiao, H.; Merril, C.R.; Wu, A.; Olde, B.; Moreno, R.F.; et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 1991, 252, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Boguski, M.S.; Lowe, T.M.; Tolstoshev, C.M. dbEST–database for “expressed sequence tags”. Nat. Genet. 1993, 4, 332–333. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Wang, S.M. Decoding neuron transcriptome by SAGE. Encyclopedia of Neuroscience; Elsevier Ltd.: Amsterdam, The Netherlands, 2009; pp. 357–363. [Google Scholar]
- Martin, G.; Baurens, F.C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genom. 2016, 17, 1–12. [Google Scholar] [CrossRef]
- Ambrosino, L.; Colantuono, C.; Diretto, G.; Fiore, A.; Chiusano, M.L. Bioinformatics resources for plant abiotic stress responses: State of the art and opportunities in the fast evolving-omics era. Plants 2020, 9, 591. [Google Scholar] [CrossRef] [PubMed]
- Unamba, C.I.; Nag, A.; Sharma, R.K. Next generation sequencing technologies: The doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 2015, 6, 1074. [Google Scholar] [CrossRef]
- Tofazzal Islam, T.I. CRISPR-Cas technology in modifying food crops. In CABI Reviews; CABI: Wallingford, UK, 2019; pp. 1–16. [Google Scholar]
- Katna, G.; Sood, V.K. Plant Genetic Resources, Traditional Knowledge and Their Use in Crop Improvement. In Plant Genetic Resources and Traditional Knowledge for Food Security; Springer: Singapore, 2015; pp. 23–38. [Google Scholar]
- Project, I.R.G.S. The Map-Based Sequence of the Rice Genome. Nature 2005, 436, 793–800. [Google Scholar]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Vielle-Calzada, J.P.; Martínez de la Vega, O.; Hernández-Guzmán, G.; Ibarra-Laclette, E.; Alvarez-Mejía, C.; Vega-Arreguín, J.C.; Jiménez-Moraila, B.; Fernández-Cortés, A.; Corona-Armenta, G.; Herrera-Estrella, L. The palomero genome suggests metal effects on domestication. Science 2009, 326, 1078. [Google Scholar] [CrossRef] [PubMed]
- Cooper, E.A.; Brenton, Z.W.; Flinn, B.S.; Jenkins, J.; Shu, S.; Flowers, D.; Luo, F.; Wang, Y.; Xia, P.; Barry, K.; et al. A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: Implications for the genetics of sugar metabolism. BMC Genom. 2019, 20, 420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (Setaria Italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Sato, K. History and future perspectives of barley genomics. DNA Res. 2020, 27, dsaa023. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, M.; Aluri, S.; Balachadran, M.T.; Sivarajan, S.R.; Patrignani, A.; Grüter, S.; Poveda, L.; Shimizu-Inatsugi, R.; Baeten, J.; Francoijs, K.J.; et al. Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid Crop. DNA Res. 2018, 25, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Aruna, Y.R.; Lohithaswa, H.C.; Mohanrao, A. Genome and transcriptome sequence of finger millet (Eleusine Coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom. 2017, 18, 465. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Ma, X.; Zhang, J.; Zhou, Y.; Liu, M.; Huang, L.; Sun, S.; Zhang, X.; Gao, X.; Zhan, W.; et al. Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 2019, 10, 464. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L.; et al. Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 2017, 8, 1031. [Google Scholar] [CrossRef]
- Jones, E.; Chu, W.C.; Ayele, M.; Ho, J.; Bruggeman, E.; Yourstone, K.; Rafalski, A.; Smith, O.S.; McMullen, M.D.; Bezawada, C.; et al. Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol. Breed. 2009, 24, 165–176. [Google Scholar] [CrossRef]
- Yang, Z.; Bai, Z.; Li, X.; Wang, P.; Wu, Q.; Yang, L.; Li, L.; Li, X. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 2012, 125, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nakazaki, T.; Chen, S.; Chen, W.; Saito, H.; Tsukiyama, T.; Okumoto, Y.; Xu, Z.; Tanisaka, T. Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 119, 85–91. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.; Robertson, M.; Ali, S.; Lenton, J.R.; Chandler, P.M. Isolation of gibberellin metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor. Appl. Genet. 2004, 109, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Raina, M.; Salgotra, R.K.; Pandotra, P.; Rathour, R.; Singh, K. Genetic enhancement for semi-dwarf and bacterial blight resistance with enhanced grain quality characteristics in traditional Basmati rice through marker-assisted selection. Comptes Rendus Biol. 2019, 342, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wang, L.; Gui, J.; Zhang, L.; Liu, Q.; Wang, J. Development and validation of a functional co-dominant SNP marker for the photoperiod thermo-sensitive genic male sterility pms3 (p/tms12-1) gene in rice. Breed. Sci. 2017, 67, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Szucs, P.; Yan, L.; Helguera, M.; Skinner, J.S.; Von Zitzewitz, J.; Hayes, P.M.; Dubcovsky, J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genom. 2005, 273, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Rabbi, S.H.A.; Kumar, A.; Mohajeri Naraghi, S.; Simsek, S.; Sapkota, S.; Solanki, S.; Alamri, M.S.; Elias, E.M.; Kianian, S.; Missaoui, A.; et al. Genome-wide association mapping for yield and related traits under drought stressed and non-stressed environments in wheat. Front. Genet. 2021, 12, 649988. [Google Scholar] [CrossRef]
- Tomar, V.; Singh, D.; Dhillon, G.S.; Singh, R.P.; Poland, J.; Joshi, A.K.; Singh, P.K.; Bhati, P.K.; Kumar, S.; Rahman, M.; et al. New QTLs for spot blotch disease resistance in wheat (Triticum aestivum L.) using genome-wide association mapping. Front. Genet. 2021, 11, 613217. [Google Scholar] [CrossRef]
- He, X.; Juliana, P.; Kabir, M.R.; Roy, K.K.; Islam, R.; Marza, F.; Peterson, G.; Singh, G.P.; Chawade, A.; Joshi, A.K.; et al. Screening and mapping for head blast resistance in a panel of CIMMYT and South Asian bread wheat germplasm. Front. Genet. 2021, 12, 679162. [Google Scholar] [CrossRef]
- Pradhan, A.K.; Kumar, S.; Singh, A.K.; Budhlakoti, N.; Mishra, D.C.; Chauhan, D.; Mittal, S.; Grover, M.; Kumar, S.; Gangwar, O.P.; et al. Identification of QTLs/defense genes effective at seedling stage against prevailing races of wheat stripe rust in India. Front. Genet. 2020, 11, 572975. [Google Scholar] [CrossRef]
- Kokhmetova, A.; Sehgal, D.; Ali, S.; Atishova, M.; Kumarbayeva, M.; Leonova, I.; Dreisigacker, S. Genome-wide association study of tan spot resistance in a hexaploid wheat collection from Kazakhstan. Front. Genet. 2021, 11, 581214. [Google Scholar] [CrossRef]
- Xu, F.; Chen, S.; Yang, X.; Zhou, S.; Wang, J.; Zhang, Z.; Huang, Y.; Song, M.; Zhang, J.; Zhan, K.; et al. Genome-wide association study on root traits under different growing environments in wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 646712. [Google Scholar] [CrossRef]
- Sehgal, D.; Mondal, S.; Crespo-Herrera, L.; Velu, G.; Juliana, P.; Huerta-Espino, J.; Shrestha, S.; Poland, J.; Singh, R.; Dreisigacker, S. Haplotype-based, genome-wide association study reveals stable genomic regions for grain yield in CIMMYT spring bread wheat. Front. Genet. 2020, 11, 589490. [Google Scholar] [CrossRef]
- Cao, J.; Shang, Y.; Xu, D.; Xu, K.; Cheng, X.; Pan, X.; Liu, X.; Liu, M.; Gao, C.; Yan, S.; et al. Identification and validation of new stable QTLs for grain weight and size by multiple mapping models in common wheat. Front. Genet. 2020, 11, 584859. [Google Scholar] [CrossRef]
- Adhikari, A.; Basnet, B.R.; Crossa, J.; Dreisigacker, S.; Camarillo, F.; Bhati, P.K.; Jarquin, D.; Manes, Y.; Ibrahim, A.M. Genome-wide association mapping and genomic prediction of anther extrusion in CIMMYT hybrid wheat breeding program via modeling pedigree, genomic relationship, and interaction with the environment. Front. Genet. 2020, 11, 586687. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, J.; Mavi, G.S.; Dhillon, G.S.; Sharma, A.; Singh, R.; Devi, U.; Chhuneja, P. Pyramiding of high grain weight with stripe rust and leaf rust resistance in elite Indian wheat cultivar using a combination of marker assisted and phenotypic selection. Front. Genet. 2020, 11, 593426. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q.; Tang, M.; Zhang, X.; Pan, Y.; Yang, X.; Gao, G.; Lv, R.; Tao, W.; Jiang, L.; et al. QTL mapping and identification of candidate genes for heat tolerance at the flowering stage in rice. Front. Genet. 2021, 11, 621871. [Google Scholar] [CrossRef]
- Lee, C.M.; Park, H.S.; Baek, M.K.; Jeong, O.Y.; Seo, J.; Kim, S.M. QTL mapping and improvement of pre-harvest sprouting resistance using japonica weedy rice. Front. Plant Sci. 2023, 14, 1194058. [Google Scholar] [CrossRef]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef]
- Kim, K.W.; Nawade, B.; Nam, J.; Chu, S.H.; Ha, J.; Park, Y.J. Development of an inclusive 580K SNP array and its application for genomic selection and genome-wide association studies in rice. Front. Plant Sci. 2022, 13, 1036177. [Google Scholar] [CrossRef] [PubMed]
- Reyes, V.P.; Kitony, J.K.; Nishiuchi, S.; Makihara, D.; Doi, K. Utilization of genotyping-by-sequencing (GBS) for rice pre-breeding and improvement: A review. Life 2022, 12, 1752. [Google Scholar] [CrossRef] [PubMed]
- Bartholomé, J.; Prakash, P.T.; Cobb, J.N. Genomic Prediction: Progress and Perspectives for Rice Rice Improvement. In Genomic Prediction of Complex Traits: Methods and Protocols; Humana: New York, NY, USA, 2022; pp. 569–617. [Google Scholar]
- Subedi, S.R.; Sandhu, N.; Singh, V.K.; Sinha, P.; Kumar, S.; Singh, S.P.; Ghimire, S.K.; Pandey, M.; Yadaw, R.B.; Varshney, R.K.; et al. Genome-wide association study reveals significant genomic regions for improving yield, adaptability of rice under dry direct seeded cultivation condition. BMC Genom. 2019, 20, 471. [Google Scholar] [CrossRef]
- Daware, A.; Malik, A.; Srivastava, R.; Das, D.; Ellur, R.K.; Singh, A.K.; Tyagi, A.K.; Parida, S.K. Rice Pangenome Genotyping Array: An efficient genotyping solution for pangenome-based accelerated genetic improvement in rice. Plant J. 2023, 113, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Cheng, P.; Cheng, Y.; Feng, Y.; Huang, D.; Huang, T.; Song, X.; Ying, J. QTL-Seq identified a major QTL for grain length and weight in rice using near isogenic F2 population. Rice Sci. 2018, 25, 121–131. [Google Scholar]
- Zhao, M.; Ma, Z.; Wang, L.; Tang, Z.; Mao, T.; Liang, C.; Gao, H.; Zhang, L.; He, N.; Fu, L.; et al. SNP-based QTL mapping for panicle traits in the japonica super rice cultivar Liaoxing 1. Crop J. 2020, 8, 769–780. [Google Scholar] [CrossRef]
- Ranganatha, H.M.; Lohithaswa, H.C.; Pandravada, A. Mapping and validation of major quantitative trait loci for resistance to northern corn leaf blight along with the determination of the relationship between resistances to multiple foliar pathogens of maize (Zea mays L.). Front. Genet. 2021, 11, 548407. [Google Scholar] [CrossRef]
- Adewale, S.A.; Badu-Apraku, B.; Akinwale, R.O.; Paterne, A.A.; Gedil, M.; Garcia-Oliveira, A.L. Genome-wide association study of Striga resistance in early maturing white tropical maize inbred lines. BMC Plant Biol. 2020, 20, 203. [Google Scholar] [CrossRef]
- Zebire, D.; Menkir, A.; Adetimirin, V.; Mengesha, W.; Meseka, S.; Gedil, M. Identifying suitable tester for evaluating Striga resistant lines using DArTseq markers and agronomic traits. PLoS ONE 2021, 16, e0253481. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cao, Y.; Wang, Y.; Ding, Y. Development of the maize 5.5 K loci panel for genomic prediction through genotyping by target sequencing. Front. Plant Sci. 2022, 13, 4544. [Google Scholar] [CrossRef]
- Yu, G.; Cui, Y.; Jiao, Y.; Zhou, K.; Wang, X.; Yang, W.; Xu, Y.; Yang, K.; Zhang, X.; Li, P.; et al. Comparison of sequencing-based and array-based genotyping platforms for genomic prediction of maize hybrid performance. Crop J. 2023, 11, 490–498. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, R.; Zhao, Y.; Yao, J.; Li, W.; Yang, Z.; Sun, F.; Yang, X. Identifying QTL and candidate genes for prolificacy in maize. Crop J. 2023, 11, 531–539. [Google Scholar] [CrossRef]
- Gupta, M.; Choudhary, M.; Singh, A.; Sheoran, S.; Singla, D.; Rakshit, S. Meta-QTL analysis for mining of candidate genes and constitutive gene network development for fungal disease resistance in maize (Zea mays L.). Crop J. 2023, 11, 511–522. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Crossa, J.; Mugo, S.; Atlin, G.N.; Tarekegne, A.; Meisel, B.; Sehabiague, P.; Vivek, B.S.; Oikeh, S.; et al. Improving maize grain yield under drought stress and non-stress environments in sub-Saharan Africa using marker-assisted recurrent selection. Crop Sci. 2016, 56, 344–353. [Google Scholar] [CrossRef]
- Ellis, M.; Spielmeyer, W.; Gale, K.; Rebetzke, J.; Richards, A. ‘‘Perfect’’ markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor. Appl. Genet. 2002, 105, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Mikołajczak, K.; Ogrodowicz, P.; Surma, M.; Adamski, T.; Kuczyńska, A. Introgression of LTP2 gene through marker assisted backcross in barley (Hordeum vulgare L.). Electron. J. Biotechnol. 2016, 24, 9–11. [Google Scholar] [CrossRef][Green Version]
- Hernandez, J.; Steffenson, B.J.; Filichkin, T.; Fisk, S.P.; Helgerson, L.; Meints, B.; Vining, K.J.; Marshall, D.; del Blanco, A.; Chen, X.; et al. Introgression of rpg4/Rpg5 into barley germplasm provides insights into the genetics of resistance to Puccinia graminis f. sp. tritici race ttksk and resources for developing resistant cultivars. Phytopathology 2019, 109, 1018–1028. [Google Scholar] [CrossRef]
- Tommasini, L.; Yahiaoui, N.; Srichumpa, P.; Keller, B. Development of functional markers specific for seven Pm3 resistance alleles and their validation in the bread wheat gene pool. Theor. Appl. Genet. 2006, 114, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Periyannan, S.; Bansal, U.; Bariana, H.; Deal, K.; Luo, M.; Dvorak, J.; Lagudah, E. Identification of a robust molecular marker for the detection of the stem rust resistance gene Sr45 in common wheat. Theor. Appl. Genet. 2014, 127, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Krattinger, S.G.; Herrera-Foessel, S.; Singh, R.P.; Huerta-Espino, J.; Spielmeyer, W.; Brown-Guedira, G.; Selter, L.L.; Keller, B. Gene specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 2009, 119, 889–898. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, Y.; He, Z.; Wu, Y.P.; Xiao, Y.G.; Ma, C.X.; Xia, X.C. Characterization of phytoene synthase 1 gene (Psy1) located on common wheat chromosome 7A and development of a functional marker. Theor. Appl. Genet. 2008, 116, 213–221. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, X.; He, Z.; Ma, W.; Appels, R.; Pena, R.J.; Xia, X.C. Characterization of low molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (T. aestivum L.). Theor. Appl. Genet. 2009, 118, 525–539. [Google Scholar] [CrossRef]
- Ramkumar, G.; Sivaranjani, A.; Pandey, M.K.; Sakthivel, K.; Shobha, N.R.; Sudarshan, I.; Prasad, G.S.V.; Neeraja, C.N.; Sundaram, R.M.; Viraktamath, B.C.; et al. Development of a PCR-based SNP marker system for effective selection of kernel length and kernel elongation in rice. Mol. Breed. 2010, 26, 735–740. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, S.; Yang, G.; Zha, W.; Cai, H.; Li, S.; Chen, Z.; Liu, K.; Xu, H.; You, A. A perfect functional marker for the gene of intermediate amylose content Wx-in in rice (Oryza sativa L.). Crop Breed. Appl. Biotechnol. 2018, 18, 103–109. [Google Scholar] [CrossRef]
- Thornsberry, J.M.; Goodman, M.M.; Doebley, J.; Kresovich, S.; Nielsen, D.; Buckler, E.S. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Gen. 2001, 28, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, R.; Hossain, F.; Muthusamy, V.; Baveja, A.; Mehta, B.K.; Zunjare, R.U. Development and validation of breeder-friendly functional markers of sugary1 gene encoding starch-debranching enzyme affecting kernel sweetness in maize (Zea mays). Crop Pasture Sci. 2019, 70, 868–875. [Google Scholar] [CrossRef]
- Dhutmal, R.R.; Mundhe, A.G.; More, A.W. Molecular marker techniques: A Review. Int. J. Curr. Microbiol. App. Sci. 2018, 6, 816–825. [Google Scholar]
- Amom, T.; Nongdam, P. The use of molecular marker methods in plants: A review. Int. J. Curr. Res. Rev. 2017, 9, 1–7. [Google Scholar]
- Joseph, M.; Gopalakrishnan, S.; Sharma, R.K.; Singh, V.P.; Singh, A.K.; Singh, N.K.; Mohapatra, T. Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breed. 2004, 13, 377–387. [Google Scholar] [CrossRef]
- Yugander, A.; Sundaram, R.M.; Singh, K.; Ladhalakshmi, D.; Subba Rao, L.V.; Madhav, M.S.; Badri, J.; Prasad, M.S.; Laha, G.S. Incorporation of the novel bacterial blight resistance gene Xa38 into the genetic background of elite rice variety ‘Improved Samba Mahsuri’. PLoS ONE 2018, 13, e0198260. [Google Scholar] [CrossRef]
- Ellur, R.K.; Khanna, A.; Gopala Krishnan, S.; Bhowmick, P.K.; Vinod, K.K.; Nagarajan, M.; Mondal, K.K.; Singh, N.K.; Singh, K.; Prabhu, K.V.; et al. Marker-aided incorporation of Xa38, a novel bacterial blight resistance gene, in PB1121 and comparison of its resistance spectrum with xa13 + Xa21. Sci. Rep. 2016, 6, 29188. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.; Varier, M.; Prakash, A.; Prasad, D. Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 2018, 8, 2413. [Google Scholar] [CrossRef]
- Abdul Fiyaz, R.; Shivani, D.; Chaithanya, K.; Mounika, K.; Chiranjeevi, M.; Laha, G.S.; Viraktamath, B.C.; Subba Rao, L.V.; Sundaram, R.M. Genetic improvement of rice for bacterial blight resistance: Present status and future prospects. Rice Sci. 2022, 29, 118–132. [Google Scholar] [CrossRef]
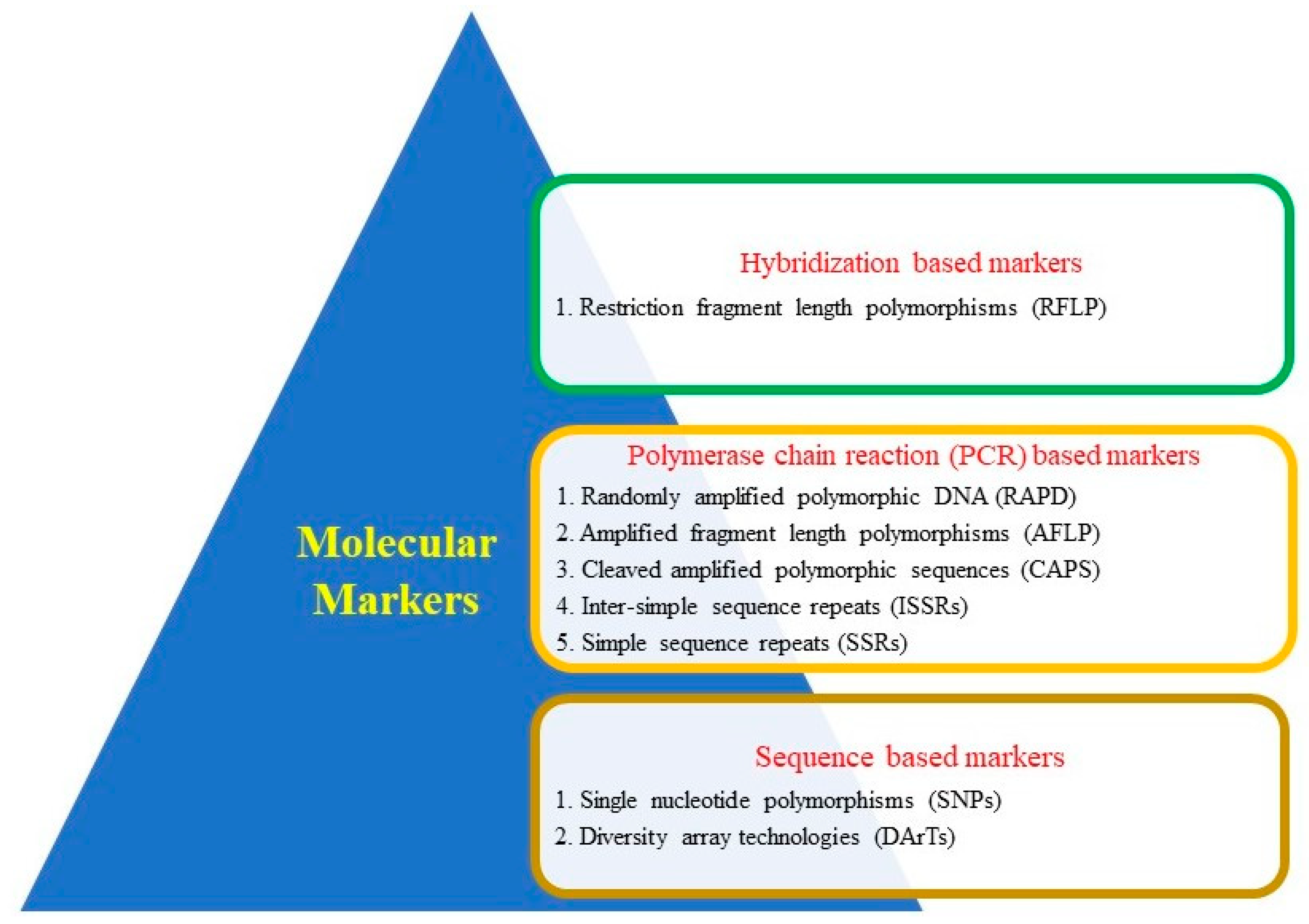

| Crop | Molecular Breeding Approaches | Traits Improved | References |
|---|---|---|---|
| Wheat | QTL mapping and GWAS using Infinium 90 K SNP assay | Drought tolerance | [87] |
| GWAS using GBS | Spot blotch resistance | [88] | |
| GWAS using Illumina Infinium 15 K BeadChip | Head blast resistance | [89] | |
| GWAS using 35 K axiom® arrays | Stripe rust resistance | [90] | |
| GWAS using DArTseq technology | Tan spot resistance | [91] | |
| GWAS using wheat 660 K SNP array | Identification of chromosomal regions of root traits | [92] | |
| Haplotype-based GWAS and genotyping by sequencing | Grain yield | [93] | |
| QTL mapping (RIL population derived from specific locus amplified fragment sequencing (SLAF-seq)) | Grain weight and size | [94] | |
| GWAS using 20 K Infinitum iSelect SNP array and genomic prediction of anther extrusion in hybrid breeding | Helps in rapid breeding for a particular trait | [95] | |
| Marker-assisted selection | Pyramided genes of high grain weight, stripe rust, and leaf rust resistance | [96] | |
| Rice | QTL mapping | Heat tolerance | [97] |
| QTL mapping | Pre-harvest sprouting resistance | [98] | |
| SNP genotyping array RiceSNP50 | Functional genomics and molecular breeding | [99] | |
| 580 K SNP array | Genomic selection and GWAS | [100] | |
| Genotyping-by-sequencing | Pre-breeding and improvement | [101] | |
| Genomic prediction | Rice improvement | [102] | |
| GWAS | Adaptability to dry direct-seeded rice (DDSR) system | [103] | |
| Rice pangenome genotyping array (RPGA) SNP genotyping | Genetic improvement | [104] | |
| QTL-seq using NIL-F2 | Grain length and weight | [103] | |
| SNP-based QTL mapping | Panicle traits | [104] | |
| Maize | QTL mapping | Northern corn leaf blight resistance | [105] |
| GWAS | Striga resistance | [106] | |
| DArT seq SNP genotyping | Striga resistance hybrid breeding | [107] | |
| 5.5 K SNPs using genotyping by target sequencing (GBTS) | Genomic prediction | [108] | |
| GBS, 40 K SNP array and target sequence capture | Genomic prediction | [109] | |
| QTL mapping | Prolificacy trait | [110] | |
| Meta-QTL analysis | Fungal disease resistance | [111] | |
| Marker-assisted recurrent selection (MARS) | Grain yield | [112] | |
| GWAS | Ear rot resistance | [113] | |
| QTL/genomic region identification | Biotic stress resistance | [114] | |
| Barley | LTP2 through marker-assisted recurrent selection (MARS) | Semi-dwarf | [115] |
| Introgression of rpg4/Rpg5 gene through marker-assisted recurrent selection (MABB) | Stem rust disease resistance | [116] | |
| Pearl millet | Introgression of qRSg1 and qRSg4 genes through marker-assisted selection (MAS) | Downy mildew disease resistance | [117] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhary, N.; Salgotra, R.K.; Chauhan, B.S. Genetic Enhancement of Cereals Using Genomic Resources for Nutritional Food Security. Genes 2023, 14, 1770. https://doi.org/10.3390/genes14091770
Chaudhary N, Salgotra RK, Chauhan BS. Genetic Enhancement of Cereals Using Genomic Resources for Nutritional Food Security. Genes. 2023; 14(9):1770. https://doi.org/10.3390/genes14091770
Chicago/Turabian StyleChaudhary, Neeraj, Romesh Kumar Salgotra, and Bhagirath Singh Chauhan. 2023. "Genetic Enhancement of Cereals Using Genomic Resources for Nutritional Food Security" Genes 14, no. 9: 1770. https://doi.org/10.3390/genes14091770
APA StyleChaudhary, N., Salgotra, R. K., & Chauhan, B. S. (2023). Genetic Enhancement of Cereals Using Genomic Resources for Nutritional Food Security. Genes, 14(9), 1770. https://doi.org/10.3390/genes14091770







