Abstract
The Kazakhstan-Siberia Network for Spring Wheat Improvement (KASIB) was established in 2000, forming a collaboration between breeding and research programs through biannual yield trials. A core set of 142 genotypes from 15 breeding programs was selected, genotyped for 81 DNA functional markers and phenotyped for 10 agronomic traits at three sites in Kazakhstan (Karabalyk, Shortandy and Shagalaly) and one site in Russia (Omsk) in 2020–2022. The study aim was to identify markers demonstrating significant effects on agronomic traits. The average grain yield of individual trials varied from 118 to 569 g/m2. Grain yield was positively associated with the number of days to heading, plant height, number of grains per spike and 1000-kernel weight. Eight DNA markers demonstrated significant effects. The spring-type allele of the Vrn-A1 gene accelerated heading by two days (5.6%) and was present in 80% of the germplasm. The winter allele of the Vrn-A1 gene significantly increased grain yield by 2.7%. The late allele of the earliness marker per se, TaMOT1-D1, delayed development by 1.9% and increased yield by 4.5%. Translocation of 1B.1R was present in 21.8% of the material, which resulted in a 6.2% yield advantage compared to 1B.1B germplasm and a reduction in stem rust severity from 27.6 to 6.6%. The favorable allele of TaGS-D1 increased both kernel weight and yield by 2–3%. Four markers identified in ICARDA germplasm, ISBW2-GY (Kukri_c3243_1065, 3B), ISBW3-BM (TA004946-0577, 1B), ISBW10-SM2 (BS00076246_51, 5A), ISBW11-GY (wsnp_Ex_c12812_20324622, 4A), showed an improved yield in this study of 3–4%. The study recommends simultaneous validation and use of selected markers in KASIB’s network.
1. Introduction
Spring bread wheat is a major crop grown on over 16–17 M ha in North Kazakhstan and Western Siberia of Russia. This is a short-season crop sown in May and harvested in August–September. The production system includes other cereals (spring durum wheat, barley and oats), oil crops (sunflower, rapeseed, flax) and legumes (dry peas, lentil and chickpea). The spring wheat is produced entirely under rainfed conditions, and moisture stress is the key abiotic challenge limiting grain yield. Diseases (leaf and stem rusts, Septoria sp. and tan spot) and numerous insect pests occur regularly and limit production. The average grain yield in the region varies from 1.2 to 1.8 t/ha depending on the year [1]. Utilization of low-input production technologies and conventional tall cultivars is one of the reasons for the low yield. North Kazakhstan and Western Siberia are important for regional and global food security since most of the grain produced here is exported (www.fao.org/faostat, accessed on 1 September 2023). Therefore, raising the productivity and stability of wheat farming is an important task.
The wheat breeding framework in the region comprises primarily public breeding programs located at agricultural research stations, institutes and centers across the main agro-ecological regions [2]. The breeding system is based on crosses with subsequent selection using a modified pedigree method and yield testing at the advanced breeding stages. The development of new varieties takes 10–12 years, followed by 2–3 years of official registration trials and another 3–5 years for seed production, making it almost 20 years from the cross to the farmer’s field [2]. Very few programs in the region utilize modern genomic and phenomic tools. Multilocational trials at advanced breeding stages are also very limited. As a result, many released cultivars are susceptible to diseases, are tall and lodge in favorable environments, are not responsive to inputs and have limited adaptations. Several locally developed cultivars have been officially released but have not been grown at a large scale. In the meantime, there is competition from foreign, primarily European, cultivars with short stature, which are favored by farmers due to their being more technologically suitable and responding well to inputs. There is an obvious need for modernization of the breeding programs, including the utilization of genomic and phenomic tools.
DNA molecular markers have emerged as an important instrument for the selection of genotypes with desirable traits [3]. This constantly evolving technology offers high selection efficiency for various wheat traits, including resistance to biotic and abiotic stresses, adaptation, agronomic performance and grain quality. Different types of wheat molecular markers and their use in breeding have been summarized recently by Song et al. [4] and Sun et al. [5]. The research on the genetics and identification of molecular markers of spring wheat in North Kazakhstan and Western Siberia focused on grain elemental composition, leaf rust [6,7] and agronomic traits [8,9]. However, the outcomes of these studies have not been implemented in practical wheat breeding.
The Kazakhstan-Siberia Network for Spring Wheat Improvement (KASIB) was established in 2000, uniting more than 20 breeding and research programs. The network objective has been to exchange the germplasm and conduct cooperative multilocational testing to characterize the advanced breeding lines and new cultivars. Each participating institution submits 2–3 entries for a cooperative trial that lasts for 2 years. The resulting data are integrated and distributed among the network members. By 2018, more than 300 genotypes had been exchanged and tested in KASIB yield trials. KASIB’s core set, with 142 genotypes, was selected from this material. The set was genotyped for over 80 functional markers and phenotyped for three years at four sites. The study objective was to evaluate the effects of molecular markers on adaptation and productivity traits for utilization in practical breeding.
2. Materials and Methods
2.1. Material
A KASIB core set comprising 142 genotypes (Table S1) was selected from 325 KASIB accessions exchanged between cooperators from 2001 to 2018. The selection was made based on: (a) DNA diversity as represented by 63 random KASP-SNP markers, selection was made from different clusters developed by COREHUNTER software, v3; (b) geographic representation to include germplasm from all KASIB cooperators; and (c) agronomic performance including the five highest yielding genotypes from each KASIB trial. The germplasm represented eight breeding programs from Kazakhstan totaling 59 entries and nine programs from Russia with 83 entries. Geographic origin of the germplasm is presented in Table S1 and Figure 1. KASIB cooperators are located in diverse environments, varying in soil, climate and cropping systems. The distance from the westernmost site, Saratov to the easternmost, Barnaul is over 3000 km, while northernmost location, Tyumen is over 2000 km from the southernmost, Almaty. Most of the germplasm tested were advanced lines, while cultivars accounted for around 35%. The seeds were initially multiplied at Karabalyk Agricultural Experimental Station (AES) in 2018, distributed to three other sites and multiplied there in 2019, allowing replicated trials starting from 2020 with each site using their own seeds.
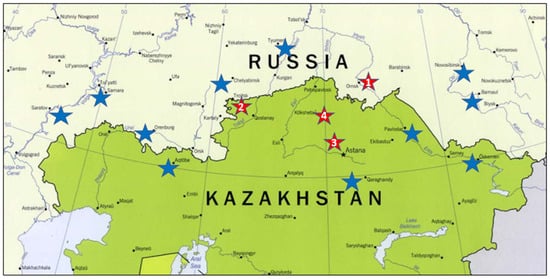
Figure 1.
Location of KASIB trials used in the study: 1. Omsk State Agrarian University, Omsk (55.026333 N, 73.310254 E); 2. Karabalyk Agricultural Experimental Station, Karabalyk, Kostanay reg. (53.845009 N, 62.122684 E); 3. A.I. Barayev Research and Production Centre for Grain Farming, Shortandy, Akmola reg. (51.636491 N, 71.021044 E); 4. North Kazakhstan Agricultural Experimental Station, Shagalaly, North Kazakhstan reg. (54.163557 N, 69.521307 E). Blue stars indicate KASIB sites not included in the study.
2.2. Yield Trials
The yield trails were conducted at four sites (Figure 1): Omsk State Agrarian University, named after P. Stolypin, (3 m2 plot; 2020 and 2022—4 reps; 2021—2 reps), Karabalyk AES (3 m2 plot; 2020 and 2022—2 reps; 2021—3 reps), A.I. Barayev Research and Production Centre for Grain Farming (3 m2 plot; 2020 and 2022—1 rep; 2021—2 reps), North Kazakhstan AES (5 m2 plot; 2021 and 2022—2 reps). The number of replications varied depending on individual program choices in a particular year to accommodate labor-intensive phenotyping. The experiments were laid out using a randomized complete block design. The following agronomic traits were recorded: number of days from emergence to heading and from heading to maturity, plant height (plot average), number of total and productive tillers per plant, spike length, number of grains per spike, 1000-kernel weight (TKW), grain weight per spike following the methodology described by Pask et al. [10] using 5 random plants per replication. Stem rust severity was evaluated as average per plot [10]. The trials were harvested by combine. ANOVAs for unbalanced design and associated analyses were conducted using R 4.3.0 using the sommer package. Best Linear Unbiased Predictions (BLUP) and Best Linear Unbiased Estimates (BLUE) for six target traits were calculated to account for unbalanced design as described by Smith et al. [11].
The preceding field was black fallow at all sites. Sowing took place at optimal dates between 15 and 25 of May. The seed rate varied from 300 to 400 seeds per m2 depending on location. Weeds were controlled with common herbicides. No disease or insect protection took place. The fertilizers were not applied following common production practices after fallow.
The soils across the study locations represented various types of chernozem: ordinary (Karabalyk), leached (Shortandy and Shagalaly) and meadow (Omsk) with pH within 6.5–7.5, average availability of phosphorus and high potassium. Nitrogen supply was above average. The weather conditions varied across the three years of study (Table 1), with 2020 being extremely dry and hot with the average temperature in July 1–2 °C higher than long-term values. Karabalyk site was exposed to moisture stress and heat in 2020 and 2022 as well. The growing conditions of 2022 were close to the long-term average in Omsk, Shortandy and Shagalaly. Among the diseases, only stem rust in Omsk in 2020 was observed, but it did not affect the yield performance. Overall, variable weather at four sites during three seasons allowed for a comprehensive evaluation of the germplasm.

Table 1.
The weather conditions of the experimental sites in 2020–2022 based on local meteo-stations.
2.3. Genotyping
DNA extraction was conducted from lyophilized tissue, followed by quantification, quality control and DNA purification as described in CIMMYT wheat molecular genetics: Laboratory Protocols and Applications to Wheat Breeding [12]. Kompetitive Allele Specific PCR (KASP) markers for 81 different loci were used (Table S3) with analysis performed by Biosearch Technologies (Teddington, Middlesex, UK). The primer sequences, amplification conditions and detailed genotyping procedures of some genes are described in [13], while for some genes this information was provided by respective research groups. The markers selection was based on a standard set of markers available at CIMMYT and ICARDA for wheat at the time of analysis in 2019. KASP markers for which one of the alleles was represented at a frequency higher than 80% were not considered for evaluation of marker–trait association. For several markers, especially disease and insect resistance, phenotyping was not conducted. Eventually, only 17 markers on 12 chromosomes were selected for evaluation in the study: development rate—5 loci, disease resistance—1 locus, plant height—1 locus, yield and yield components—10 loci. The marker–trait associations were identified by comparing the average values of BLUPs or original data with alternative marker alleles using the respective standard errors.
3. Results
3.1. Grain Yield and Agronomic Traits across the Study Sites and Years
The average values of grain yield and other agronomic traits for the four sites across the years are presented in Figure 2 and yearly data in Table S3. The Omsk environment was the most favorable for spring wheat compared to the three sites in Kazakhstan. The average grain yield in Omsk was 478 g/m2, compared to 178–249 g/m2 at other sites. The plant height in Omsk was 88.3 cm for three years, or 17.4–45.2% taller than in other sites. Among yield components, spike fertility, reflected by number of grains per spike, was 35.6 compared to 24.1–26.8 at the sites in Kazakhstan. However, the Omsk trials were marginally superior to those of the North Kazakhstan station for 1000-kernel weight: 40.8 vs. 39.1 g. The lowest TKW was in Karabalyk—31.5 g. ANOVA results demonstrated highly significant effects of sites, years, genotypes and the interactions genotypes - sites, sites x years for all traits (height, spike length, number of grains per spike, grain weight per spike, TKW and grain yield) (Table S4). However, the interaction genotypes - years was only significant for TKW.
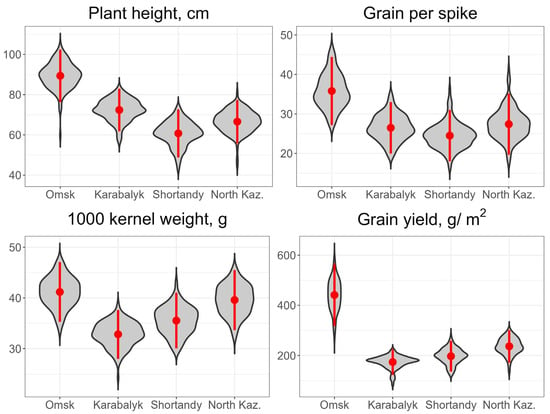
Figure 2.
Best Linear Unbiased Estimates of agronomic traits at the four experimental sites in 2020–2022. Mean and confidence interval of the mean (95%) are indicated by a red point and red bar, respectively, within each violin plot.
Biplot analysis of 11 trials using average yearly values of 6 traits demonstrated the differences between the years and sites (Figure 3). Three seasons in Omsk were quite similar and formed one compact group. Similarly, the three seasons at Karabalyk are also grouped. Shortandy and Shagalaly define two other groups in the same quadrat of the biplot, suggesting similar performances of the spring wheat panel at these two locations. The expression of all traits was the highest in Omsk, as indicated by the length and direction of the arrows. The combinations of agronomic traits in Karabalyk in 2021 and Shagalaly in 2022 were quite unique, setting these sites apart from each other and from other trials. Overall, the 12 sites-years combinations were sufficiently diverse to allow the comprehensive evaluation of KASIB core material for adaptation and agronomic traits.
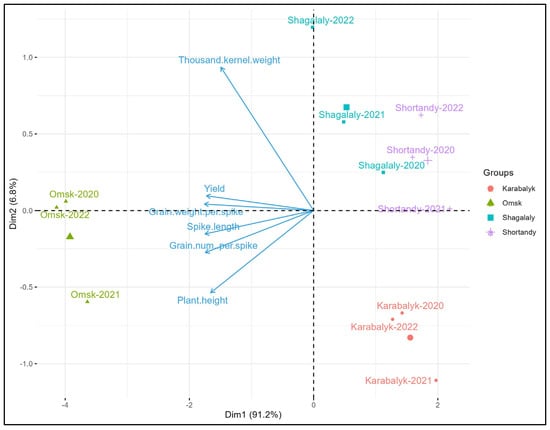
Figure 3.
Biplot analysis of 12 trials using average yearly values of variety BLUEs.
3.2. Association of Grain Yield with Adaptation Traits and Yield Components
Pearson coefficients of correlations were calculated between grain yield and yield component traits across all 142 genotypes for each site and year separately (Table 2) using BLUEs. Plant height had a significant positive correlation in all nine trials. Taller germplasm tended to provide consistently higher yields, especially in Shagalaly and Shortandy. The number of grains per spike had a higher association with the grain yield compared to TKW, considering individual locations. The respective coefficients of correlation were significant in 10 trials out of 11, compared to 6 for TKW. The grain weight per spike, as an integral trait, had as high of an association with grain yield as did the number of grains per spike. The correlations computed for average values for each year across sites are much higher compared to individual trials, and all five traits demonstrate high and positive association with grain yield. This means that all agronomic traits included in this study are important in defining spring wheat productivity in the target environments.

Table 2.
Pearson coefficients of correlation between the BLUEs values for grain yield and agronomic traits across study sites, 2020–2022.
The rate of development or earliness measured by the number of days from emergence to heading is a key character for short-season wheat like in North Kazakhstan and Siberia. There is a clear positive association between the number of days to heading (within a range of 35–45 days) and grain yield (Figure 4). However, the later-heading and -maturing genotypes demonstrated lower yield, perhaps due to exposure to unfavorable conditions during maturity. The number of days from heading to maturity had a significant negative correlation with the number of days to heading (0.59) and an insignificant one with the grain yield.
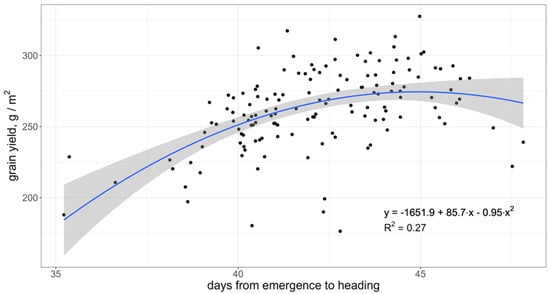
Figure 4.
Relationship between the number of days to heading and grain yield computed from BLUE of the 142 accessions for the set of target environments. The grey area represents 95% confidence interval of the regression.
The adaptation traits and yield components studied in this experiment had various degrees of association with the grain yield, which also depended on the site and year. This provides an optimal basis for evaluation of the effects of molecular markers on these traits.
3.3. Effects of Molecular Markers on Agronomic Traits
The Rht8 allele of the Ribonuclease H-Like 1 gene [14] was present in most germplasm (78%) but did not reduce the height compared to the group of germplasm possessing the recessive allele (Table 3). Five markers linked to the genes controlling the rate of development, i.e., the number of days from emergence to heading, were assessed in the KASIB core set. The spring allele of the Vrn-A1 gene accelerated heading by two days (5.6%), with a significant difference in average values across all trials and for each individual trial. This allele was present in 80% of the germplasm. The presence of the winter allele of the Vrn-A1 gene significantly increased grain yield by 2.7%. The second marker with a significant effect on heading date was TaMOT1-D1, which accelerated development by 1.9% across all trials and was significant in five trials out of six. The presence of the allele which extends the growing period also significantly increased yield by 4.5%.

Table 3.
Effects of molecular markers on agronomic traits, BLUPs, 2020–2023.
Two markers with effects on 1000-kernel weight and grain yield were evaluated: alleles at GS5-2334-SNP did not result in significant differences in either of the traits, while TaGS-D1 significantly increased grain weight and yield by 2.1% and 3.2%, respectively. Marker ISBW10-SM2-QTL, with an effect on tillering capacity, proved its effect under KASIB conditions, significantly increasing the number of total and productive tillers per plant by 7.8 and 4.3%, respectively. However, it did not have a significant effect on yield. Two markers linked to genes affecting yield performance under drought conditions (TaCwi-4A and Dreb1) did not prove effective in the current study. They had no effect even in the trials with low yield caused by moisture stress.
For grain yield, the presence of the respective alleles of the markers ISBW3-BM-QTL, ISBW11-GY-QTL and ISBW2-GY-QTL increased the productivity by 3.5–4.5% across all sites and were also effective in most individual trials. Marker ISBW11-GY-QTL increased grain yield in Omsk in three years by 7.4%.
Translocation 1B.1R was present in 21.8% of the material studied. The groups of material possessing 1B.1R had a 6.2% yield advantage compared to the 1B.1B group across all 11 trials. However, its positive effect on yield in Omsk was 12.3% during three years, in North Kazakhstan—7.5%, in Shortandy—6.2%, while no advantage was found under Karabalyk conditions. The presence of 1B.1R translocation substantially reduced stem rust severity from 27.6 to 6.6%. Another marker of stem rust resistance, IWAB8036, did not significantly affect the pathogen severity.
3.4. Markers Distribution across the Breeding Programs
The allelic frequencies of eight effective markers described above were estimated in 322 cultivars and breeding lines originating from 15 breeding programs in Kazakhstan and Russia (Table 4). Allele vrn-A1, associated with winter growth habit, is considered favorable for productivity due to a positive association between the number of days to heading and grain yield (Figure 4). The frequency of this allele varied from zero in Aktobe, Chelyabinsk and Novosibirsk to 95% in the East Kazakhstan breeding program. Its presence in Kazakh germplasm was almost four times more frequent than in Russian material. The frequency of 1B.1R translocation was below 5% at six breeding programs in Kazakhstan, while it exceeded 20% at five programs in Russia. Its frequency in the germplasm from Kurgan Seeds company and Omsk Agricultural Research Center was 57.1%. Overall, the average frequency of markers associated with grain yield and its components exceeded 60%. For some favorable alleles, the frequency was 100%: TaGS-D1b and ISBW2-GY (T) in East Kazakhstan AES germplasm; ISBW10-SM2 (G) in material from Karagandy AES, Kazakh Grain Center and Chelyabinsk Agricultural Research Institute (ARI).

Table 4.
Distribution of eight molecular markers of agronomic traits in the germplasm from KASIB breeding programs.
The important question was if the observed differences in the distribution of effective markers are related to agro-ecological conditions of the breeding programs or reflect the parentage, crossing and selection strategy employed by the breeders. At the time of the development of this germplasm, the breeders were not aware of the molecular markers and used conventional selection. The biplot location of the breeding programs according to the markers’ frequency (Figure 5) does not indicate the effect of agro-ecology as such. The breeding fields of the Omsk Agric. Research Center and University are only 5 km apart, but the markers’ frequency patterns are different. The same situation was found with Karabalyk AES and Fiton Breeding Company, and Kurgan ARI and Kurgan Seed Company. They are located close to each other, but the markers’ composition is different. There was a very close similarity between the breeding programs of Omsk Agrarian University and Kurgan Seeds, as well as Kazakh Grain Center and Karagandy ARI. East Kazakhstan AES stands apart from all other programs.
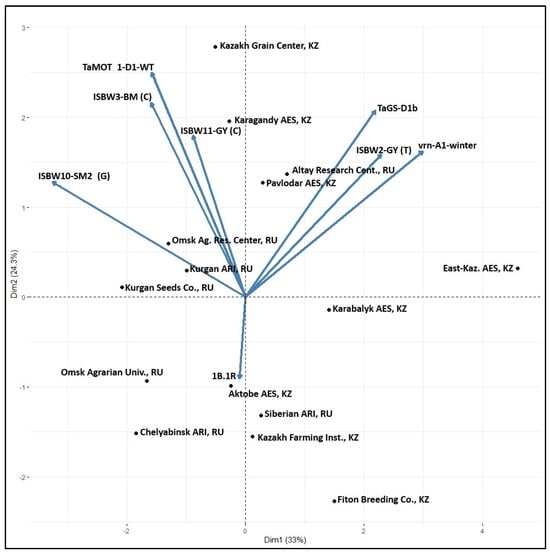
Figure 5.
Biplot distribution of spring wheat breeding programs at 15 institutions according to the frequency of alleles at eight markers of agronomic traits (Table 4).
3.5. Identification of Superior Germplasm
A multilocational trial with 12 sites–years allowed the identification of superior high yielding germplasm using BLUEs. Due to the effect of maturity on grain yield, the highest yielding genotypes were identified separately for early (total of 24 genotypes), intermediate (96 genotypes) and late (22 genotypes) groups (Table 5 and Table S5). Four of the five highest yielding entries in the early group originated from the program in Russia including Siberian ARI in Novosibirsk (entries 27 and 29), Kurgan Seed (entry 127) and Samara ARI (entry 38). Lut. 166-SP94 was bred at Kazakh Farming Institute in Almaty, the most southern KASIB site. None of these lines possessed 1B.1R translocation and all carried the spring allele at the Vrn-A1 locus. The presence of yield-related alleles varied depending on genotypes from four to six.

Table 5.
Grain yield, number of days to heading, TKW and presence of the effective markers in the top yielding germplasm.
The top yielding cultivar in the intermediate group, Saratov-75, originated from South-East ARI in the Volga region. It did not possess 1B.1R translocation or the winter allele of the Vrn-A1 locus. There are only five genotypes originating from Kazakhstan in the top twenty high yielding lines in this maturity group. The best Kazakh genotype, Lut. 1764, was ranked tenth. Five of the eight top-yielding genotypes possessed 1B.1R translocation. Four of the ten top-yielding genotypes possessed all five favorable alleles of markers TaGS-D1b, ISBW10-SM2 (G), ISBW2-GY (T), ISBW3-BM (C), ISBW11-GY (C). This is, perhaps, another indirect indication of their efficiency.
The highest yielding cultivar in the late group and the whole set was Zauljbinka from East-Kazakhstan ARI, which possessed both 1B.1R translocation and the vrn-A1 allele. The second and the third highest yielding genotypes in this group originated from Altay ARI. Cultivar Tobolskaya, which possessed all five favorable yield-related alleles, and it demonstrated good performance in KASIB regular trials and wide distribution in the production.
Considering the entire set of the best 30 genotypes, none of them possessed all eight favorable alleles. However, three breeding lines (146-Lut. 186/04-61, 49-Lut. 1764, 50-Lutescens 1082) combined seven of the eight favorable alleles.
4. Discussion
This is the first study in the KASIB region to assess the practical value of using a set of well-known molecular markers with respect to agronomic traits in a large set of diverse spring wheat germplasm using multi-locational trials with four sites and three seasons. The material used in the study represented 15 breeding programs, which cover over 80% of all spring wheat areas in Kazakhstan and Russia. Substantial phenotyping efforts allowed detailed characterization of the germplasm in contrasting conditions with significant genotype x environment interactions. Evaluation of the effects of molecular markers on agronomic traits led to the following general observations: (a) 17 out of 81 KASP markers evaluated in the germplasm were informative, the latter being excluded due to their unbalanced distribution or lack of phenotyping data; (b) eight markers (or 50%) of the set of informative markers proved to have a significant effect on the target traits; and (c) the effects of favorable alleles on agronomic traits was estimated to be in the range of 2–5%, but in combination, they can contribute to better performance; (d) the markers with significant effects in this study were developed outside of the KASIB region, demonstrating their universal nature.
The gene Vrn-A1 has a large effect on vernalization requirements, diverse allelic variation [15] and contributes to genotype x environment interaction [16]. A previous study of 148 spring bread wheat cultivars in Western Siberia showed the presence of the vrn-A1 allele at a frequency of 8% [17], whereas in this experiment it was also 8% in Russian material, but 35% in Kazakhstan germplasm. However, there were no previous reports on the effect of this allele on grain yield in the region.
TaMOT-1-D1 is a marker related to the Eps-D1 gene which accounts for the variation in flowering time when vernalization and photoperiod requirements are satisfied [18]. Askhadullin et al. [19] mention the importance of this gene for earliness in spring wheat cultivars from the Volga region. However, no data on the frequency or effects of this gene on earliness or yield in the KASIB region were available. The current study demonstrated that the late allele of this gene is widely spread in Kazakh and Russian germplasm and positively contributed to number of days to heading and grain yield.
The history of 1B.1R translocation and its use in wheat breeding is well described [20]. The translocation contributed to a successful breeding program at CIMMYT and was incorporated in global wheat germplasm [21]. The translocation contributed to yield potential [22] but also negatively affected the grain’s technological quality [23]. Korobkova et al. [24] documented the presence of 1B.1R translocation in over 50% of 66 modern winter wheat varieties from the Krasnodar breeding program. The frequency of 1B.1R translocation was over 25% in spring wheat stem rust resistance material studied in North Kazakhstan and Russia, as reported by Shamanin et al. [25]. The authors also mentioned a positive effect of 1B.1R translocation on grain yield. The current study proves the value of this translocation in the region with its pleiotropic effect on grain yield and stem rust resistance through the presence of the Sr31 gene, affective against a wide range of pathotypes, including Ug99 [25].
The gene TaGS-D1 on chromosome 7DS in wheat was described as ortholog to the rice gene OsGS3 that plays a principal role in controlling grain weight and length in rice [26]. The study on European winter wheat did not show an effect of this marker on grain weight [27]; neither did a study of CIMMYT spring wheat [28]. Shan et al. [29] demonstrated its positive effect on yield components. So far, the gene’s effect had not been studied in Kazakhstan and Russia. Its frequency in the KASIB core set was around 70% and it increased both TKW and yield by 2–3%.
ICARDA identified several markers affecting yield and its components through association mapping in Sudan and Morocco. These markers include ISBW2-GY (Kukri_c3243_1065, 3B), ISBW3-BM (TA004946-0577, 1B), ISBW10-SM2 (BS00076246_51, 5A), ISBW11-GY (wsnp_Ex_c12812_20324622, 4A) [30,31]. Despite the differences in agro-ecological conditions of North Africa and the KASIB region, these markers turned out to be effective in the current study, improving yield by 3–4%.
The distribution of a subset of eight effective markers in a large set of material across 15 breeding programs revealed large differences in their frequencies. These differences originate from the practical availability of diverse germplasm sources, from unconscious selection for optimal agronomic types, grain yield and resistance to biotic and abiotic stresses. However, the breeding programs’ choices of parents and selection strategy also contribute to this genetic diversity. Superior high-yielding genotypes were identified in all three maturity groups. Their multilocational characterization is supplemented by genetic composition for eight effective markers selected in this study. These superior genotypes can be used as parents for crossing programs.
What are the next steps in the utilization of molecular markers-assisted selection in practical breeding in KASIB institutions? The eight effective markers, including three with pleiotropic effects, identified in the current study have to be supplemented by SNP markers identified through GWAS on this diversity panel of spring wheat adapted to the region. Several studies have been completed, and results need to be converted to KASP markers for efficient use [12,13,14,15,16]. The KASIB core set GWAS for over 20 traits is underway and will contribute potentially useful markers. The simultaneous validation and use of existing and new markers need to include as many KASIB network sites as possible. Precision phenotyping and field trials will require the use of modern experimental design and statistical tools. Genotyping platforms in the region or efficient outsourcing would be an essential part of the markers’ use framework. Training of the breeders on the utilization of molecular markers, experimental design and phenotyping would be an important component of the successful application of genomic technologies in breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes15010086/s1, Table S1. The list of materials used in the study; Table S2. The list of KASP markers used in the study; Table S3. Grain yield and agronomic traits at three experimental sites in 2020–2022; Table S4. ANOVA results for grain yield and agronomic traits at three experimental sites in 2020–2022; Table S5. Grain yield, number of days to heading, TKW and presence of effective molecular markers in KASIB set.
Author Contributions
Conceptualization, A.M., V.S. and V.C.; methodology, S.D., A.R., C.B. and L.G.; software, C.B. and L.G.; formal analysis, A.M., S.S., C.B. and L.G.; investigation, A.B., Y.D., T.S., E.F., V.C., S.S. and R.Z.; resources, V.C. and R.Z.; data curation, A.M., S.S., C.B. and L.G.; writing—original draft preparation, A.M., V.S., A.R. and S.D.; writing—review and editing, A.M., L.G., C.B. and V.S.; visualization, T.S.; supervision, A.M., V.C., T.S. and V.S.; funding acquisition, T.S., V.S. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
KASIB Network activities in the Russian Federation have been supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement No. 075-15-2021-534 of 28 May 2021). Statistical processing of the experimental data of the KASIB program has been supported by the Russian Science Foundation (Agreement No. 22-16-20008 of 23 March 2022). Phenotyping of the germplasm at three sites in Kazakhstan was supported by the Ministry of Agriculture Targeted-Funding Program BR10765056 awarded to Kazakh Agrotechnical University, named after S. Seifullin, titled “Development of the highly productive varieties and hybrids of grain crops based on the achievements of biotechnology, genetics, physiology, biochemistry of plants for their sustainable production in various soil and climatic zones Kazakhstan”. KASIB genotyping was supported by CRP WHEAT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available from the corresponding author upon request.
Acknowledgments
Breeding programs participating in the KASIB network and sharing the germplasm and data are greatly appreciated.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karatayev, M.; Clarke, M.; Salnikov, V.; Bekseitova, R.; Nizamova, M. Monitoring climate change, drought conditions and wheat production in Eurasia: The case study of Kazakhstan. Heliyon 2022, 8, e08660. [Google Scholar] [CrossRef] [PubMed]
- Goncharov, N.P. Scientific support to plant breeding and seed production in Siberia in the XXI century. Vavilov J. Genet. Breed. 2021, 25, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Babar, H.; Akpınar, B.A.; Alaux, M.; Algharib, A.M.; Sehgal, D.; Ali, Z.; Aradottir, G.I.; Batley, J.; Bellec, A.; Bentley, A.R.; et al. Capturing wheat phenotypes at the genome level. Front. Plant. Sci. 2022, 851079. [Google Scholar]
- Song, L.; Wang, R.; Yang, X.; Zhang, A.; Liu, D. Molecular markers and their applications in marker-assisted selection (MAS) in bread wheat (Triticum aestivum L.). Agriculture 2023, 13, 642. [Google Scholar] [CrossRef]
- Sun, C.; Hu, H.; Cheng, Y.; Yang, X.; Qiao, Q.; Wang, C.; Zhang, L.; Chen, D.; Zhao, S.; Dong, Z.; et al. Genomics-assisted breeding: The next-generation wheat breeding era. Plant Breed. 2023, 142, 259–268. [Google Scholar] [CrossRef]
- Genievskaya, Y.; Abugalieva, S.; Rsaliyev, A.; Yskakova, G.; Turuspekov, Y. QTL mapping for seedling and adult plant resistance to leaf and stem rusts in Pamyati Azieva × Paragon mapping population of bread wheat. Agronomy 2020, 10, 1285. [Google Scholar] [CrossRef]
- Kelbin, V.N.; Skolotneva, E.S.; Shamanin, V.P.; Salina, E.A. Diversity of stem rust resistance in modern Siberian bread wheat (Triticum aestivum) germplasm. Plant Breed. 2022, 141, 194–203. [Google Scholar] [CrossRef]
- Bhatta, M.; Shamanin, V.P.; Shepelev, S.S.; Baenziger, P.S.; Pozherukova, V.; Pototskaya, I.; Morgounov, A.I. Marker-trait associations for enhancing agronomic performance, disease resistance, and grain quality in synthetic and bread wheat accessions in western Siberia. G3 Genes Genomes Genet. 2019, 9, 4209–4222. [Google Scholar] [CrossRef]
- Amalova, A.; Abugalieva, S.; Babkenov, A.; Babkenova, S.; Turuspekov, Y. Genome-wide association study of yield components in spring wheat collection harvested under two water regimes in Northern Kazakhstan. Peer J. 2021, 9, e11857. [Google Scholar] [CrossRef]
- Pask, A.J.D.; Pietragalla, J.; Mullan, D.M.; Reynolds, M.P. Physiological Breeding II: A Field Guide to Wheat Phenotyping; CIMMYT: Mexico City, Mexico, 2012. [Google Scholar]
- Smith, A.B.; Cullis, B.R.; Thompson, R. The analysis of crop cultivar breeding and evaluation trials: An overview of current mixed model approaches. J. Agric. Sci. 2005, 143, 449–462. [Google Scholar] [CrossRef]
- Dreisigacker, S.; Sehgal, D.; Reyes Jaimez, A.E. CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding; CIMMYT: Mexico City, Mexico, 2016; pp. 1–154. [Google Scholar]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Chai, L.; Xin, M.; Dong, C.; Chen, Z.; Zhai, H.; Zhuang, J.; Cheng, X.; Wang, N.; Geng, J.; Wang, X.; et al. A natural variation in Ribonuclease H-like gene underlies Rht8 to confer “Green Revolution” trait in wheat. Mol. Plant 2022, 15, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Sherman, J.D.; Yan, L.; Talbert, L.; Dubcovsky, J. A PCR marker for growth habit in common wheat based on allelic variation at the VRN-A1 gene. Crop. Sci. 2004, 44, 1832–1838. [Google Scholar] [CrossRef]
- Alipour, H.; Abdi, H.; Rahimi, Y.; Bihamta, M.R. Genotype-by-year interaction for grain yield of Iranian wheat cultivars and its interpretation using Vrn and Ppd functional markers and environmental covariables. Cereal Res. Commun. 2021, 49, 681–690. [Google Scholar] [CrossRef]
- Efremova, T.T.; Chumanova, E.V.; Trubacheeva, N.V.; Arbuzova, V.S.; Belan, I.A.; Pershina, L.A. Prevalence of VRN1 locus alleles among spring common wheat cultivars cultivated in Western Siberia. Russ. J. Genet. 2016, 52, 146–153. [Google Scholar] [CrossRef]
- Zikhali, M.; Wingen, L.U.; Griffiths, S. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J. Exp. Botany 2016, 67, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Askhadullin, D.F.; Askhadullin, D.F.; Vasilova, N.Z.; Brykova, A.N. Earliness of spring bread wheat accessions under the conditions of the Middle Volga Region. Procd. Appl. Botany Genet. Breed. 2022, 183, 66–75. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Garkava-Gustavsson, L.; Åhman, I. A systematic review of rye (Secale cereale L.) as a source of resistance to pathogens and pests in wheat (Triticum aestivum L.). Hereditas 2017, 154, 14. [Google Scholar] [CrossRef]
- Rajaram, S. Prospects and promise of wheat breeding in the 21st century. Euphytica 2001, 119, 3–15. [Google Scholar] [CrossRef]
- Villareal, R.L.; Rajaram, S.; Mujeeb-Kazi, A.; Del Toro, E. The effect of chromosome 1B/1R translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed. 1991, 106, 77–81. [Google Scholar] [CrossRef]
- Moskal, K.; Kowalik, S.; Podyma, W.; Łapiński, B.; Boczkowska, M. The pros and cons of rye chromatin introgression into wheat genome. Agronomy 2021, 11, 456. [Google Scholar] [CrossRef]
- Korobkova, V.A.; Bespalova, L.A.; Yanovsky, A.S.; Chernook, A.G.; Kroupin, P.Y.; Arkhipov, A.V.; Yurkina, A.I.; Nazarova, L.A.; Mudrova, A.A.; Voropaeva, A.D.; et al. Permanent spreading of 1RS.1AL and 1RS.1BL translocations in modern wheat breeding. Plants 2023, 12, 1205. [Google Scholar] [CrossRef] [PubMed]
- Shamanin, V.; Salina, E.; Wanyera, R.; Zelenskiy, Y.; Olivera, P.; Morgounov, A. Genetic diversity of spring wheat from Kazakhstan and Russia for resistance to stem rust Ug99. Euphytica 2016, 212, 287–296. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Y.-L.; Gao, L.-F.; Zhao, G.-Y.; Zhuo, R.-H.; Zhang, B.-S.; Jia, J.-Z. TaCK6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Zanke, C.; Ling, J.; Plieske, J.; Kollers, S.; Ebmeyer, E.; Korzun, V.; Argillier, O.; Stiewe, G.; Hinze, M.; Neumann, F.; et al. Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front. Plant Sci. 2015, 6, 644. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, D.; Mondal, S.; Guzman, C.G.; Franco, C.; Singh, R.; Dreisigacker, S. Validation of candidate gene-based markers and identification of novel loci for thousand-grain weight in spring bread wheat. Front. Plant Sci. 2019, 10, 01189. [Google Scholar] [CrossRef] [PubMed]
- Shan, D.; Ali, M.; Shahid, M.; Arif, A.; Waheed, M.Q.; Xia, X.; Trethowan, R.; Tester, M.; Poland, J.; Ogbonnaya, F.C.; et al. Genetic networks underlying salinity tolerance in wheat uncovered with genome-wide analyses and selective sweeps. Theor. Appl. Genet. 2022, 135, 2925–2941. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, W.; Suleiman, S.; Tahir, I.; Sanchez-Garcia, M.; Jighly, A.; Hagras, A.; Thabet, S.; Baum, M. Heat-tolerant QTLs associated with grain yield and its components in spring bread wheat under heat-stressed environments of Sudan and Egypt. Crop. Sci. 2019, 59, 199–211. [Google Scholar] [CrossRef]
- Alemu, A.; Suliman, S.; Hagras, A.; Thabet, S.; Al-Abdallat, A.; Abdelmula, A.A.; Tadesse, W. Multi-model genome-wide association and genomic prediction analysis of 16 agronomic, physiological and quality related traits in ICARDA spring wheat. Euphytica 2021, 217, 205. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).