Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Extraction and cDNA Synthesis
2.3. Primer Design
2.4. PCR and Quantitative Real-Time PCR Analysis
2.5. Stability Analysis of Candidate Reference Genes
3. Results
3.1. Primer Specificity and Amplification Efficiency of Candidate Reference Genes
3.2. Expression Analysis of Candidate Reference Genes under Biotic Stress and Hormone Treatment
3.3. Stability Analysis of the Candidate Reference Genes
3.4. Stability Analysis of the Candidate Reference Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.K.; Nair, R.M.; Lee, J.; Lee, S.-H. Genomic resources in mung bean for future breeding programs. Front. Plant Sci. 2015, 6, 626. [Google Scholar] [CrossRef]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.-K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mung bean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef] [PubMed]
- Rossman, D.R.; Rojas, A.; Jacobs, J.L.; Mukankusi, C.; Kelly, J.D.; Chilvers, M.I. Pathogenicity and virulence of soilborne oomycetes on Phaseolus vulgaris. Plant Dis. 2017, 101, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhang, Q.; Ding, P.; Yuan, X.; Wu, R.; Chen, J.; Xue, C.; Chen, X. First Report of root rot on mung bean caused by Pythium myriotylum in China. Plant Dis. 2021, 105, 233. [Google Scholar] [CrossRef]
- Yan, Q.; Hu, Y.; Zhang, Q.; Yuan, X.; Wu, R.; Chen, J.; Xue, C.; Chen, X. Occurrence of Root Rot Caused by Pythium aphanidermatum on mung bean (Vigna radiata) in China. Plant Dis. 2021, 105, 3764. [Google Scholar] [CrossRef]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Kharshandi, F.; Kayang, H. Antagonistic Potential of rhizobacterial isolates against fungal pathogens causing rhizome rot in turmeric. Arch. Microbiol. 2023, 205, 221. [Google Scholar] [CrossRef]
- Chenniappan, C.; Daniel, G.; Ponmurugan, P.; Narayanasamy, M. Association of multiple fungal pathogens to rhizome rot disease of turmeric. Arch. Phytopathol. Plant Prot. 2020, 53, 440–458. [Google Scholar] [CrossRef]
- Akber, M.A.; Mubeen, M.; Sohail, M.A.; Khan, S.W.; Solanki, M.K.; Khalid, R.; Abbas, A.; Divvela, P.K.; Zhou, L. Global Distribution, Traditional and modern detection, diagnostic, and management approaches of Rhizoctonia solani associated with legume crops. Front. Microbiol. 2022, 13, 1091288. [Google Scholar] [CrossRef]
- Cao, W.; Zhang, H.; Zhou, Y.; Zhao, J.; Lu, S.; Wang, X.; Chen, X.; Yuan, L.; Guan, H.; Wang, G.; et al. Suppressing chlorophyll degradation by silencing OsNYC3 improves rice resistance to Rhizoctonia solani, the causal agent of sheath blight. Plant Biotechnol. J. 2022, 20, 335–349. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef] [PubMed]
- Arora, H.; Sharma, A.; Poczai, P.; Sharma, S.; Haron, F.F.; Gafur, A.; Sayyed, R.Z. Plant-derived protectants in combating soil-borne fungal infections in tomato and chilli. J. Fungi 2022, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Mageshwaran, V.; Gupta, R.; Singh, S.; Sahu, P.K.; Singh, U.B.; Chakdar, H.; Bagul, S.Y.; Paul, S.; Singh, H.V. Endophytic bacillus subtilis antagonize soil-borne fungal pathogens and suppress wilt complex disease in chickpea plants (Cicer arietinum L.). Front. Microbiol. 2022, 13, 994847. [Google Scholar] [CrossRef]
- Zhou, L.; Mideros, S.X.; Bao, L.; Hanlon, R.; Arredondo, F.D.; Tripathy, S.; Krampis, K.; Jerauld, A.; Evans, C.; St Martin, S.K.; et al. Infection and genotype remodel the entire soybean transcriptome. BMC Genom. 2009, 10, 49. [Google Scholar] [CrossRef]
- Bachmann, A.; Lavstsen, T. Analysis of var gene transcript patterns by quantitative real-time PCR. Methods Mol. Biol. 2022, 2470, 149–171. [Google Scholar] [CrossRef]
- Gao, K.; Khan, W.U.; Li, J.; Huang, S.; Yang, X.; Guo, T.; Guo, B.; Wu, R.; An, X. Identification and validation of reliable reference genes for gene expression studies in Koelreuteria paniculata. Genes 2022, 13, 714. [Google Scholar] [CrossRef]
- Tian, Y.; Chu, Z.; Wang, H.; Wang, G.; Wu, S.; Yang, Y. Selection and validation of reference genes for quantitative real-time PCR in Cymbidium sinense. Biotechniques 2022, 72, 51–59. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, J.; Wang, X.; Xiong, Y.; Xiong, Y.; Dong, Z.; Lei, X.; Yan, L.; Ma, X. Selection and validation of reference genes for qRT-PCR gene expression analysis in Kengyilia melanthera. Genes 2022, 13, 1445. [Google Scholar] [CrossRef]
- Chen, Z.; Halford, N.G.; Liu, C. Real-time quantitative PCR: Primer design, reference gene selection, calculations and statistics. Metabolites 2023, 13, 806. [Google Scholar] [CrossRef]
- Wang, W.; Hu, S.; Cao, Y.; Chen, R.; Wang, Z.; Cao, X. Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol. Biol. Rep. 2021, 48, 1115–1126. [Google Scholar] [CrossRef]
- Song, H.; Mao, W.; Duan, Z.; Que, Q.; Zhou, W.; Chen, X.; Li, P. Selection and validation of reference genes for measuring gene expression in Toona ciliata under different experimental conditions by quantitative real-time PCR analysis. BMC Plant Biol. 2020, 20, 450. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jin, H.; Li, X.; Yan, D.; Zhang, Y.; Li, M.; Gao, J.; Lu, B.; Chen, C.; Jiang, Y. Development of a novel real-time quantitative PCR method for detection of Ilyonectria robusta, the predominant species causing ginseng rusty root rot. Plant Dis. 2023, 107, 1680–1689. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Saxena, R.K.; Singh, V.K.; Krishnamurthy, L.; Varshney, R.K. Selection and validation of housekeeping genes as reference for gene expression studies in Pigeonpea (Cajanus cajan) under heat and salt stress conditions. Front. Plant Sci. 2015, 6, 1071. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Gao, M.; Liu, Y.; Ma, X.; Shuai, Q.; Gai, J.; Li, Y. Evaluation of reference genes for normalization of gene expression using quantitative RT-PCR under aluminum, cadmium, and heat stresses in soybean. PLoS ONE 2017, 12, e0168965. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xu, L.; Lai, H.; Chen, Y.; Yang, Z.; Huang, B. Identification and validation of reference genes for seashore paspalum response to abiotic stresses. Int. J. Mol. Sci. 2017, 18, 1322. [Google Scholar] [CrossRef]
- Chi, C.; Shen, Y.; Yin, L.; Ke, X.; Han, D.; Zuo, Y. Selection and validation of reference genes for gene expression analysis in Vigna angularis using quantitative real-time RT-PCR. PLoS ONE 2016, 11, e0168479. [Google Scholar] [CrossRef]
- de Jesus Miranda, V.; Coelho, R.R.; Viana, A.A.B.; de Oliveira Neto, O.B.; Carneiro, R.M.D.G.; Rocha, T.L.; de Sa, M.F.G.; Fragoso, R.R. Validation of reference genes aiming accurate normalization of qPCR data in soybean upon nematode parasitism and insect attack. BMC Res. Notes 2013, 6, 196. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Qian, M.; Yang, N.; Zhu, C.; Xia, Y. Selection and validation of reference genes for real-time fluorescence quantitative PCR in mung beans. Plant Physiol. J. 2021, 57, 2203–2212. [Google Scholar] [CrossRef]
- Yang, J.; Yang, X.; Kuang, Z.; Li, B.; Lu, X.; Cao, X.; Kang, J. Selection of suitable reference genes for qRT-PCR expression analysis of Codonopsis pilosula under different experimental conditions. Mol. Biol. Rep. 2020, 47, 4169–4181. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Li, L.; Wang, K.; Zhao, M.; Li, S.; Jiang, Y.; Zhu, L.; Chen, J.; Wang, Y.; Sun, C.; Chen, P.; et al. Selection and validation of reference genes desirable for gene expression analysis by qRT-PCR in MeJA-treated ginseng hairy roots. PLoS ONE 2019, 14, e0226168. [Google Scholar] [CrossRef]
- Chen, X.; Mao, Y.; Huang, S.; Ni, J.; Lu, W.; Hou, J.; Wang, Y.; Zhao, W.; Li, M.; Wang, Q.; et al. Selection of suitable reference genes for quantitative real-time PCR in Sapium sebiferum. Front. Plant Sci. 2017, 8, 637. [Google Scholar] [CrossRef]
- Wang, G.; Tian, C.; Wang, Y.; Wan, F.; Hu, L.; Xiong, A.; Tian, J. Selection of reliable reference genes for quantitative RT-PCR in Garlic under salt stress. PeerJ 2019, 7, e7319. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Niu, J.; Quan, S. Identification of appropriate reference genes for RT-qPCR analysis in Juglans regia L. PLoS ONE 2018, 13, e0209424. [Google Scholar] [CrossRef]
- Li, T.; Yuan, W.; Qiu, S.; Shi, J. Selection of reference genes for gene expression analysis in Liriodendron hybrids’ somatic embryogenesis and germinative tissues. Sci. Rep. 2021, 11, 4957. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xiong, A.; Gao, Z.; Yu, X.; Li, M.; Hou, Y.; Sun, C.; Qu, S. Selection of suitable reference genes for RT-qPCR normalization under abiotic stresses and hormone stimulation in Persimmon (Diospyros kaki Thunb). PLoS ONE 2016, 11, e0160885. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, X.; Zhou, X.; Wu, Z.; Yuan, L.; Wang, Y.; Li, Y. Selection of reference genes for qRT-PCR analysis in medicinal plant Glycyrrhiza under abiotic stresses and hormonal treatments. Plants 2020, 9, 1441. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2011, 28, 343–357. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.-R.; Ahmad-Nizammuddin, N.-F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.-A. Comparative genome-wide analysis of WRKY, MADS-Box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef]
- Hou, Y.; Yu, X.; Chen, W.; Zhuang, W.; Wang, S.; Sun, C.; Cao, L.; Zhou, T.; Qu, S. MdWRKY75e enhances resistance to Alternaria alternata in Malus domestica. Hortic. Res. 2021, 8, 225. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-wide identification, characterization and expression analysis of the JAZ gene family in resistance to gray leaf spots in tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, B.; Xu, H.; Wu, J.; Xu, Z.; Wang, Y. The Phytophthora effector Avh94 manipulates host jasmonic acid signaling to promote infection. J. Integr. Plant Biol. 2022, 64, 2199–2210. [Google Scholar] [CrossRef]
- Dang, F.; Lin, J.; Li, Y.; Jiang, R.; Fang, Y.; Ding, F.; He, S.; Wang, Y. SlWRKY30 and SlWRKY81 synergistically modulate tomato immunity to Ralstonia solanacearum by directly regulating SlPR-STH2. Hortic. Res. 2023, 10, uhad050. [Google Scholar] [CrossRef] [PubMed]
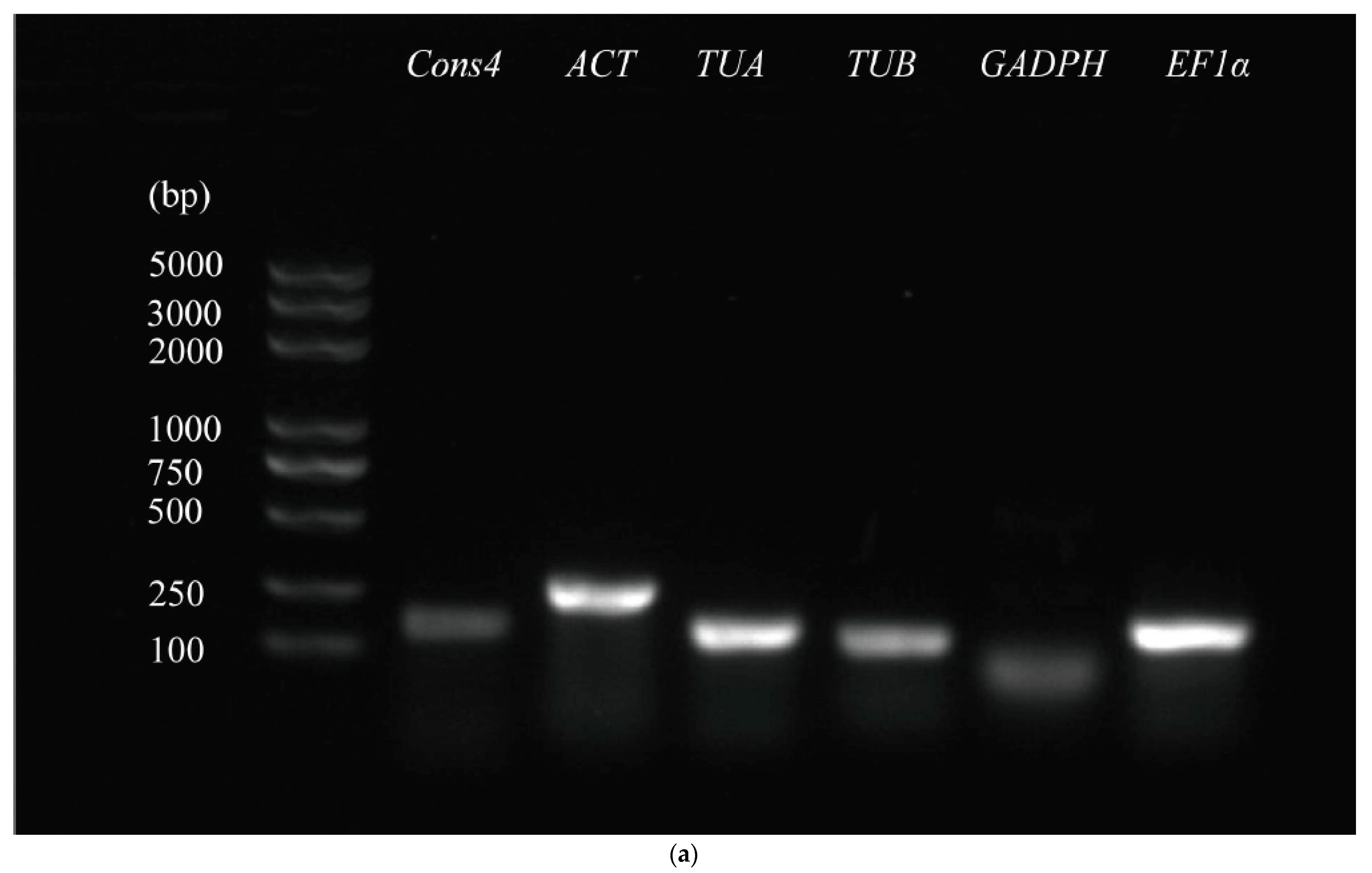
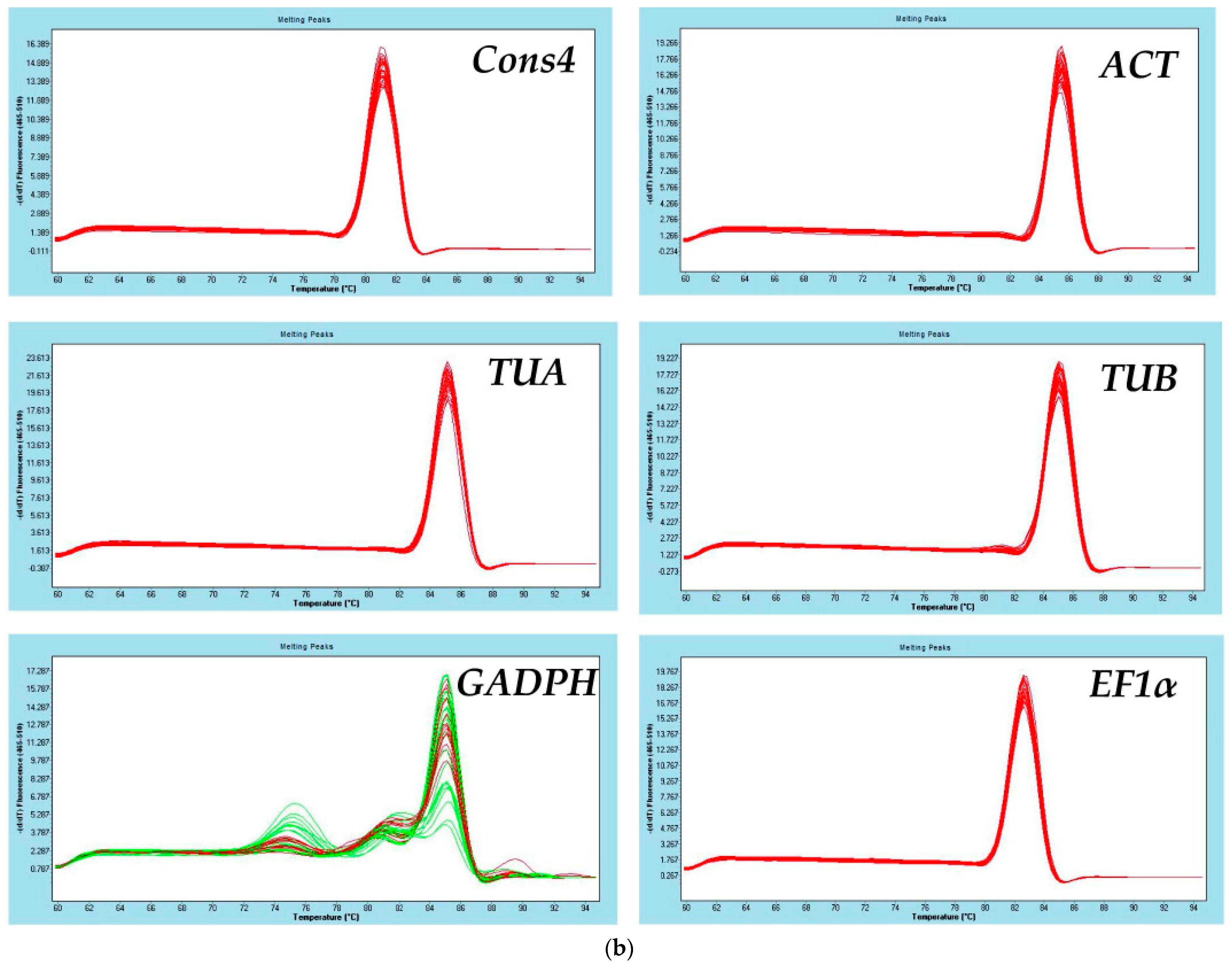
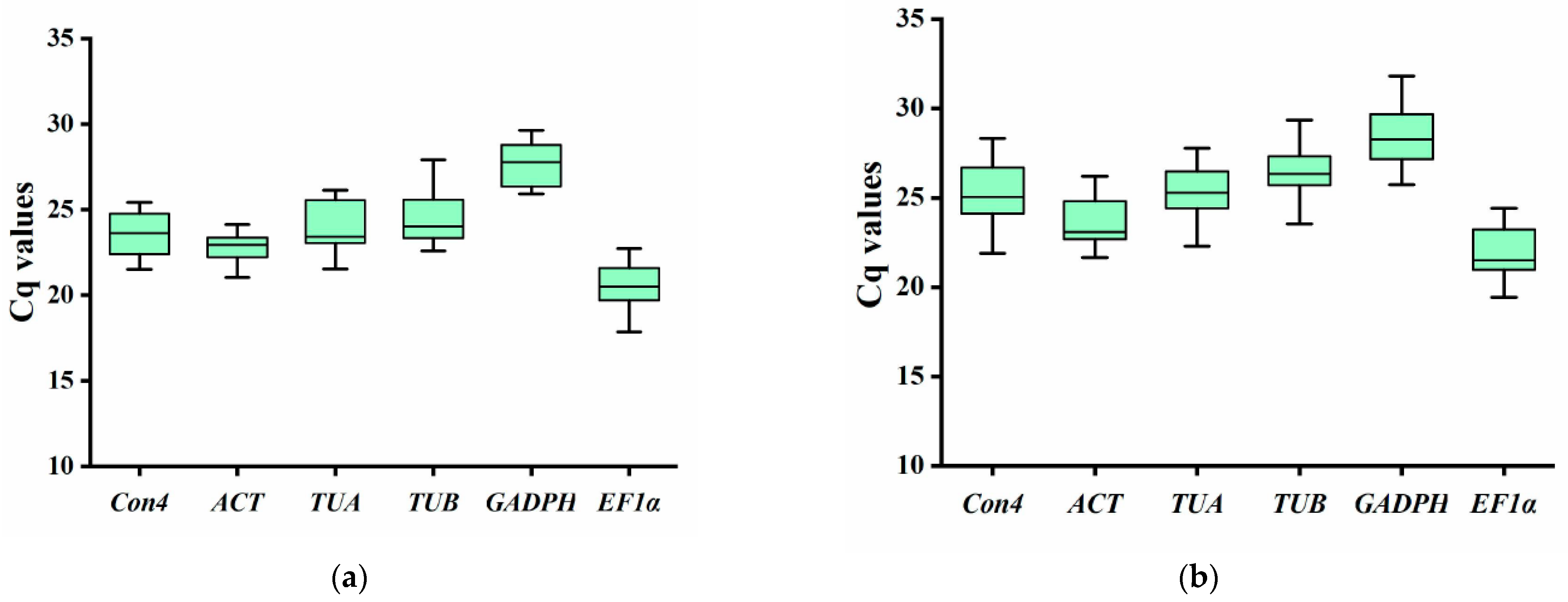



| Gene Name | Gene ID | Primer Sequence F/R (5′-3′) | Amplicon Length (bp) | Tm (°C) | Efficiency (%) | R2 |
|---|---|---|---|---|---|---|
| Cons4 | Vradi11g01010 | F:TCGCCAGATATTGCAGATAA | 156 | 81.24 | 104.43 | 0.996 |
| R:GGAGGAACAGTAGAAAGGGT | ||||||
| ACT | Vradi03g00210 | F:GGCGGTGTTCCCTAGCATTG | 246 | 85.59 | 95.96 | 0.999 |
| R:AGCGGTGCCTCGGTAAGAAG | ||||||
| TUA | Vradi08g19840 | F:GGTCAAATGCCAAGTGACAAAACAG | 150 | 85.21 | 97.392 | 0.998 |
| R:GTAAGGTCCAGTCCTAACCTCATCG | ||||||
| TUB | Vradi05g13910 | F:GCTTATGGATCTTGAACCTGGAA | 136 | 85.11 | 97.869 | 0.997 |
| R:GCCTTCGGTATAATGACCTTTCG | ||||||
| GADPH | Vradi0043s00410 | F:CGTTTTCACCCCTTTTCCG | 258 | 85.06 | 113.781 | 0.983 |
| R:CGTGATGCTTCCAGTGTCCG | ||||||
| EF1α | Vradi01g08330 | F:AGCGTGAAAGAGGAATTACCATCG | 153 | 82.65 | 98.095 | 0.999 |
| R:CAATAATAAGGACAGCACAATCAG |
| Gene Name | SA | MeJA | ETH | ABA | GA3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | |
| Cons4 | 0.024676533 | 1 | 0.222221876 | 3 | 0.384529941 | 5 | 0.259344484 | 3 | 0.069960504 | 1 |
| ACT | 0.024676533 | 2 | 0.109242042 | 1 | 0.122377254 | 1 | 0.333505004 | 4 | 0.413025931 | 5 |
| TUA | 0.037894549 | 3 | 0.147671416 | 2 | 0.339757665 | 3 | 0.211394671 | 2 | 0.176462268 | 2 |
| TUB | 0.377236807 | 4 | 0.253280028 | 4 | 0.287658317 | 2 | 0.547498395 | 5 | 0.379257235 | 4 |
| EF1α | 0.525706883 | 5 | 0.266094327 | 5 | 0.370284053 | 4 | 0.160752498 | 1 | 0.332945438 | 3 |
| Gene Name | Pythium myriotylum | Pythium aphanidermatum | Fusarium oxysporum | Rhizoctonia solani | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | Stability Value | Rank | |
| Cons4 | 0.590638821 | 5 | 0.309761038 | 3 | 0.444615042 | 5 | 0.60680042 | 5 | 0.231731888 | 4 |
| ACT | 0.202036918 | 1 | 0.387614988 | 5 | 0.251700828 | 4 | 0.530437025 | 4 | 0.299557287 | 5 |
| TUA | 0.445402607 | 4 | 0.337951776 | 4 | 0.235601773 | 3 | 0.113789254 | 1 | 0.247803221 | 3 |
| TUB | 0.274850505 | 3 | 0.184158256 | 2 | 0.076710811 | 1 | 0.316568757 | 3 | 0.285054343 | 2 |
| EF1α | 0.253857199 | 2 | 0.088874014 | 1 | 0.168786015 | 2 | 0.113789254 | 2 | 0.124083336 | 1 |
| Gene Name | SA | MeJA | ETH | ABA | GA3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | |
| Cons4 | 3.7 | 0.86 | 4 | 3.47 | 0.79 | 5 | 3.07 | 0.73 | 1 | 4.5 | 1.07 | 1 | 4.92 | 1.15 | 1 |
| ACT | 3.74 | 0.83 | 3 | 1.95 | 0.42 | 2 | 4.5 | 1.02 | 2 | 4.99 | 1.13 | 2 | 3.67 | 0.82 | 2 |
| TUA | 3.38 | 0.77 | 2 | 2.52 | 0.56 | 3 | 5.32 | 1.26 | 4 | 6.49 | 1.55 | 3 | 5.73 | 1.35 | 3 |
| TUB | 2.04 | 0.48 | 1 | 1.37 | 0.31 | 1 | 4.74 | 1.17 | 3 | 7.36 | 1.85 | 5 | 5.06 | 1.21 | 5 |
| EF1α | 6.37 | 1.26 | 5 | 4.09 | 0.78 | 4 | 7.71 | 1.55 | 5 | 8.18 | 1.67 | 4 | 7.27 | 1.45 | 4 |
| Gene Name | Pythium myriotylum | Pythium aphanidermatum | Fusarium oxysporum | Rhizoctonia solani | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | CV | SD | Rank | |
| Cons4 | 5.38 | 1.35 | 5 | 4.64 | 1.09 | 4 | 4.55 | 1.2 | 5 | 5.53 | 1.45 | 4 | 5.89 | 1.44 | 5 |
| ACT | 2.45 | 0.56 | 2 | 2.67 | 0.6 | 1 | 2.52 | 0.62 | 2 | 6.58 | 1.66 | 5 | 4.48 | 1.04 | 1 |
| TUA | 3.49 | 0.86 | 3 | 5.02 | 1.21 | 5 | 2.3 | 0.6 | 1 | 4.55 | 1.2 | 3 | 5.72 | 1.41 | 3 |
| TUB | 1.72 | 0.45 | 1 | 3.86 | 0.97 | 3 | 2.44 | 0.65 | 3 | 3.25 | 0.92 | 1 | 5.55 | 1.42 | 4 |
| EF1α | 4.73 | 1.04 | 4 | 3.56 | 0.73 | 2 | 4.22 | 0.97 | 4 | 4.94 | 1.12 | 2 | 5.7 | 1.22 | 2 |
| Rank | Hormone Induction | Biotic Stress | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SA | MeJA | ETH | ABA | GA3 | Pythium myriotylum | Pythium aphanidermatum | Fusarium oxysporum | Rhizoctonia solani | Total | |
| 1 | TUA | ACT | TUB | TUA | TUA | TUA | EF1α | ACT | EF1α | TUA |
| 2 | TUB | TUB | ACT | EF1α | TUB | EF1α | Cons4 | TUA | TUA | EF1α |
| 3 | ACT | TUA | EF1α | TUB | ACT | TUB | TUB | EF1α | TUB | ACT |
| 4 | EF1α | EF1α | TUA | Cons4 | EF1α | ACT | TUA | Cons4 | Cons4 | TUB |
| 5 | Cons4 | Cons4 | Cons4 | ACT | Cons4 | Cons4 | ACT | TUB | ACT | Cons4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, H.; Wu, T.; Zheng, Y.; Wang, R.; Xue, D.; Yan, Q.; Yuan, X.; Chen, X. Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR. Genes 2023, 14, 1739. https://doi.org/10.3390/genes14091739
Zhou Y, Liu H, Wu T, Zheng Y, Wang R, Xue D, Yan Q, Yuan X, Chen X. Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR. Genes. 2023; 14(9):1739. https://doi.org/10.3390/genes14091739
Chicago/Turabian StyleZhou, Yanyan, Huan Liu, Ting Wu, Yu Zheng, Ruimin Wang, Dong Xue, Qiang Yan, Xingxing Yuan, and Xin Chen. 2023. "Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR" Genes 14, no. 9: 1739. https://doi.org/10.3390/genes14091739
APA StyleZhou, Y., Liu, H., Wu, T., Zheng, Y., Wang, R., Xue, D., Yan, Q., Yuan, X., & Chen, X. (2023). Screening of Reference Genes under Biotic Stress and Hormone Treatment of Mung Bean (Vigna radiata) by Quantitative Real-Time PCR. Genes, 14(9), 1739. https://doi.org/10.3390/genes14091739





