In Vitro Analysis of the Effect of SARS-CoV-2 Non-VOC and four Variants of Concern on MHC-Class-I Expression on Calu-3 and Caco-2 Cells †
Abstract
1. Introduction
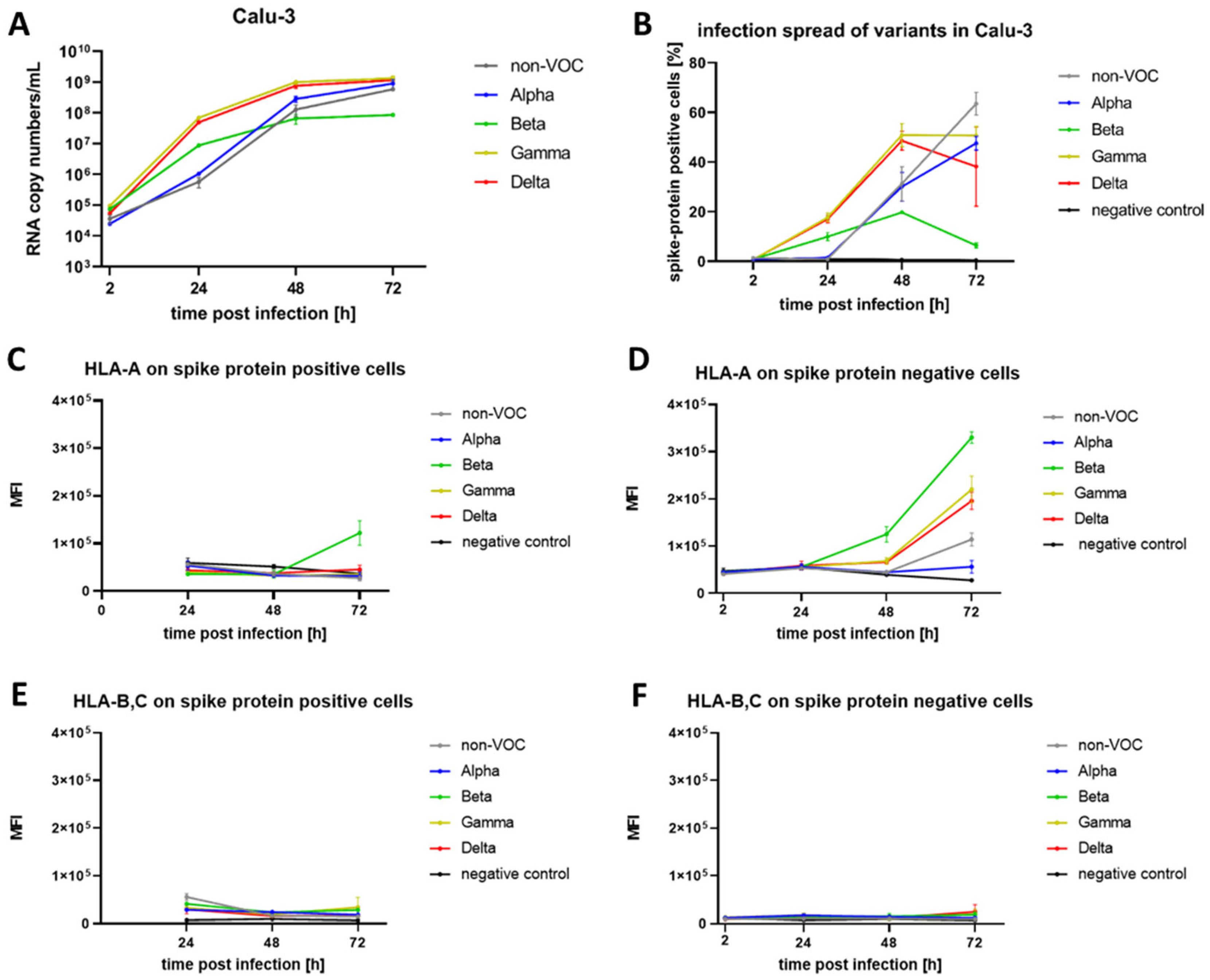
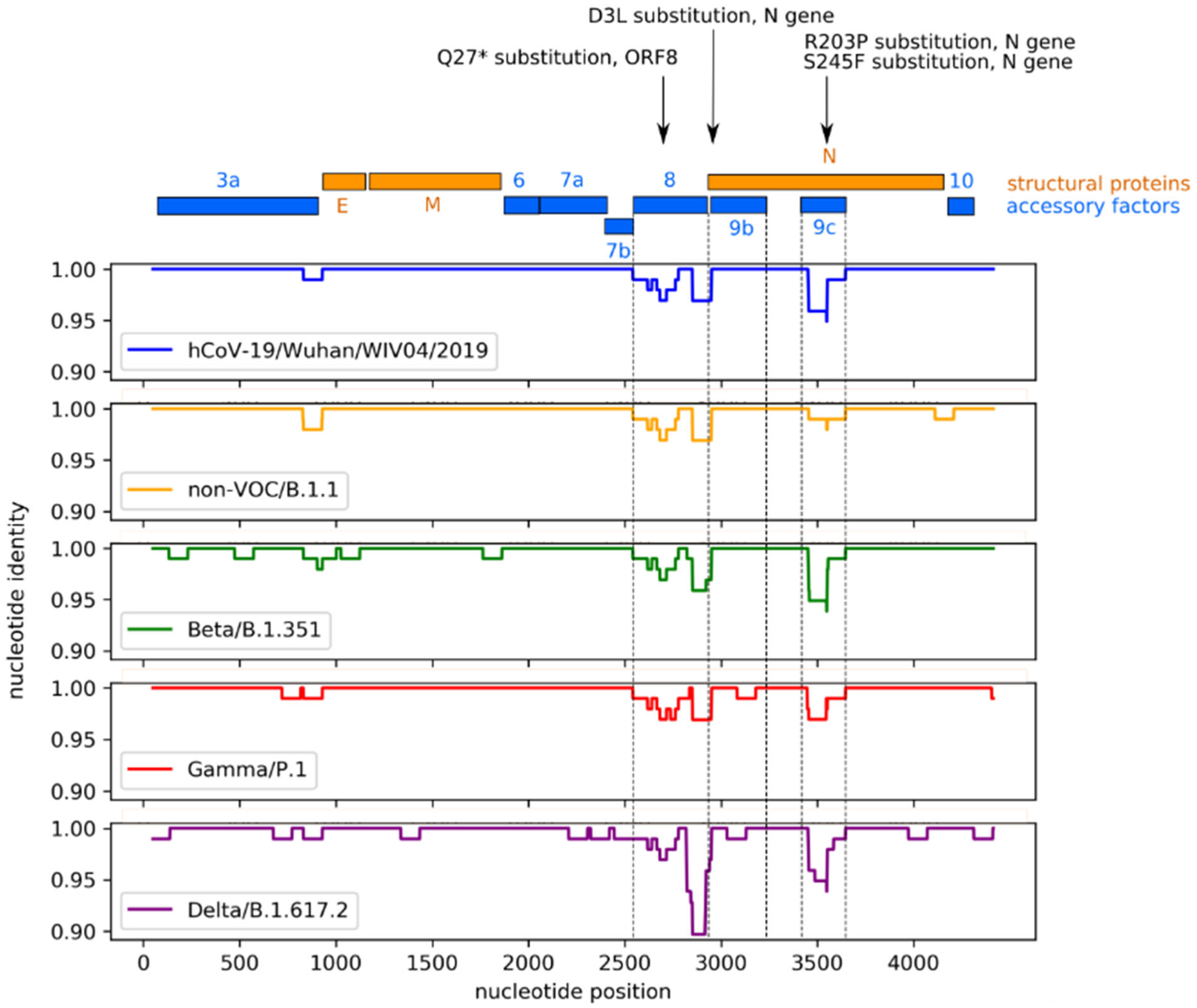
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Isolation and Propagation of Clinical SARS-CoV-2 Strains
4.3. SARS-CoV-2 Cell Culture Infection Experiments
4.4. Analysis of Extracellular Viral RNA by RT-qPCR
4.5. Flow Cytometry Preparation
4.6. ELISA Spike Protein
4.7. Immunohistochemistry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 20 June 2023).
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 20 March 2023).
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021, 21, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.S.; Marwal, R.; Radhakrishnan, V.S.; Ponnusamy, K.; Jolly, B.; Bhoyar, R.C.; Sardana, V.; Naushin, S.; Rophina, M.; Mellan, T.A.; et al. Genomic characterization and Epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science 2021, 374, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 96, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 5 March 2023).
- Yoo, J.-S.; Sasaki, M.; Cho, S.X.; Kasuga, Y.; Zhu, B.; Ouda, R.; Orba, Y.; de Figueiredo, P.; Sawa, H.; Kobayashi, K.S. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021, 12, 6602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zang, T.M.; Stevenson, E.M.; Lei, X.; Copertino, D.C.; Mota, T.M.; Boucau, J.; Garcia-Beltran, W.F.; Jones, R.B.; Bieniasz, P.D. Inhibition of major histocompatibility complex-I antigen presentation by sarbecovirus ORF7a proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2209042119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef] [PubMed]
- Holloway, G.; Fleming, F.E.; Coulson, B.S. MHC class I expression in intestinal cells is reduced by rotavirus infection and increased in bystander cells lacking rotavirus antigen. Sci. Rep. 2018, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R. Locus-specific constitutive and cytokine-induced HLA class I gene expression. J. Immunol. 2003, 170, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Mautner, L.; Baillie, C.-K.; Herold, H.M.; Volkwein, W.; Guertler, P.; Eberle, U.; Ackermann, N.; Sing, A.; Pavlovic, M.; Goerlich, O.; et al. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2020, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Di Canzio, J.; Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 1998, 221, 35–41. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahlmann, N.A.; Mautner, L.; Hoyos, M.; Sallard, E.; Berger, C.; Dangel, A.; Jönsson, F.; Fischer, J.C.; Kreppel, F.; Zhang, W.; et al. In Vitro Analysis of the Effect of SARS-CoV-2 Non-VOC and four Variants of Concern on MHC-Class-I Expression on Calu-3 and Caco-2 Cells. Genes 2023, 14, 1348. https://doi.org/10.3390/genes14071348
Bahlmann NA, Mautner L, Hoyos M, Sallard E, Berger C, Dangel A, Jönsson F, Fischer JC, Kreppel F, Zhang W, et al. In Vitro Analysis of the Effect of SARS-CoV-2 Non-VOC and four Variants of Concern on MHC-Class-I Expression on Calu-3 and Caco-2 Cells. Genes. 2023; 14(7):1348. https://doi.org/10.3390/genes14071348
Chicago/Turabian StyleBahlmann, Nora A., Lena Mautner, Mona Hoyos, Erwan Sallard, Carola Berger, Alexandra Dangel, Franziska Jönsson, Johannes C. Fischer, Florian Kreppel, Wenli Zhang, and et al. 2023. "In Vitro Analysis of the Effect of SARS-CoV-2 Non-VOC and four Variants of Concern on MHC-Class-I Expression on Calu-3 and Caco-2 Cells" Genes 14, no. 7: 1348. https://doi.org/10.3390/genes14071348
APA StyleBahlmann, N. A., Mautner, L., Hoyos, M., Sallard, E., Berger, C., Dangel, A., Jönsson, F., Fischer, J. C., Kreppel, F., Zhang, W., Esposito, I., Bölke, E., Baiker, A., & Ehrhardt, A. (2023). In Vitro Analysis of the Effect of SARS-CoV-2 Non-VOC and four Variants of Concern on MHC-Class-I Expression on Calu-3 and Caco-2 Cells. Genes, 14(7), 1348. https://doi.org/10.3390/genes14071348








