Abstract
Castor (Ricinus communis) seeds are rich in a type of hydroxy fatty acid called ricinoleic acid, which is in high demand for the production of plant-based plastics, lubricants, and hydraulic oils. However, the high content of ricin, a toxic protein, in these seeds has restricted further expansion in the area of castor cultivation. Therefore, the development of ricin-free castor is needed. Genome editing technology, although successfully applied in several plant species, is still in the developing stages in castor and awaits the identification of an endogenous U6 promoter with robust function. Here, we searched for U6 small nuclear RNA (snRNA) genes in the castor genome. This led to the identification of six U6 snRNA genes. The promoters of these U6 snRNA genes were cloned, and their function was examined in castor cells using the particle delivery method. The results showed that a U6 promoter length of approximately 300 bp from the transcription start site was sufficient to activate gene expression. This study provides insights into the endogenous castor U6 promoter sequences and outlines a method for verifying the function of U6 promoters in plants using the particle delivery system.
1. Introduction
Castor (Ricinus communis) is an herbaceous plant of the Euphorbiaceae family that accumulates high amounts of hydroxy fatty acids, particularly ricinoleic acid, in its seeds [1]. Castor oil is composed of approximately 90% ricinoleic acid and is used as a lubricant and hydraulic oil owing to its high fluidity over a wide temperature range [1,2,3]. In addition, castor oil is almost the only feedstock for polyamide 11, a representative plant-derived engineered plastic of high commercial value [1,2,3,4]. Because of these properties, castor oil is in high demand, which continues to grow annually.
Castor has long been cultivated on a large scale in India, China, and Brazil as a non-food commercial crop and is increasingly being cultivated on poor soils in Mozambique, Ethiopia, and other countries [5]. In addition, because of its high heat and drought tolerance, castor can be grown on land unsuitable for other crops [6,7]. However, castor seeds contain ricin, an enzymatic protein that is highly toxic to humans and livestock because of its ability to inactivate eukaryotic ribosomes [8]. The presence of ricin in seeds is one of the main factors that limits the cultivation of castor, and the development of ricin-free castor genotypes is therefore needed.
CRISPR/Cas9, a genome editing technology, is the most promising method for producing ricin-free castor [9]. The application of the CRISPR/Cas9 system relies on (1) the availability of genome sequence information for accurate guide RNA (gRNA) design and (2) an established transformation method. The genome sequence of castor was released as a draft in 2010 [10], and chromosome-level genome assembly information was published in 2022 [11]. Although transformation methods for castor were recently reported [12], more suitable technologies for genome editing in castor are being established.
One of the problems with genome editing in plants is low editing efficiency [13]. Methods to improve editing efficiency have been extensively studied. Kor and colleagues recently summarized that gRNAs need to be vigorously and constantly expressed to improve genome editing efficiency, indicating that the promoter sequence used to express gRNAs has a significant impact on editing efficiency [14]. Furthermore, the authors reported that the promoter region of the U6 small nuclear (snRNA) gene was ideal for gRNA expression in various organisms [14]. Additional studies have shown that the use of endogenous U6 snRNA promoter sequences (henceforth U6 promoter) is necessary to achieve high editing efficiency [15,16,17,18]. In this study, we report the cloning of six U6 promoters from the castor genome and demonstrate their ability to express a transgene in castor cells. The development of a castor-optimized CRISPR/Cas9 system is essential for efficient genome editing in castor.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
To induce germination, castor seeds were placed on a wet paper towel and incubated for 72 h at 22 °C in the dark. Three-day-old seedlings were transferred to 1/5000a Wagner pots filled with cultured soil and grown under long-day conditions (16 h light/8 h dark) at 25 °C in a growth chamber (NK System, Osaka, Japan). One-tenth strength of Hoagland solution (1 L) was supplied to the plants once every 3 days. Experimental studies on plants, including the collection of plant material, were conducted in accordance with relevant institutional, national, and international guidelines and legislation.
2.2. Plasmid Construction
The RcU6 promoter fragments, with attB4 and attB1R sequences at their 5′ and 3′ ends, respectively, were amplified by PCR with specific primer sets (Table S1) and cloned into the pDONR P4-P1R entry vector (Thermo Fisher Scientific, Tokyo, Japan) using the Gateway BP recombination method (Thermo Fisher Scientific, Tokyo, Japan). The Venus-PTS1 sequence, with attB1 and attB2 sequences at its 5′ and 3′ ends, respectively, was amplified by PCR with specific primer sets (Table S1) and cloned into the pDONR221 entry vector (Thermo Fisher Scientific, Tokyo, Japan) using the Gateway BP recombination method (Thermo Fisher Scientific, Tokyo, Japan). To generate various RcU6 promoter constructs, Venus-PTS1 and the DNA fragments cloned into pDONR P4-P1R or pDONR221 were transferred into the R4pGWB401 destination vector [19] using the Gateway LR recombination reaction.
2.3. Transient Expression with the Particle Delivery System
A total of 10 mg of gold particles (1.0 μm diameter) coated with the plasmid (20 mg) were introduced into 25-day-old first or second castor true leaf cells using a Helios Gene Gun (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s instructions. During the bombardment, castor leaves were placed on wet filter paper, and helium gas was injected at a pressure of 200 psi. The bombarded samples were incubated in the dark at 22 °C for 16 h.
2.4. Confocal Microscopy
Castor leaf cells injected with the plasmid-coated gold particles were observed under a confocal laser scanning microscope (LSM510META; Carl Zeiss, Jena, Germany) [20]. Emission filters BP535-590 were used to detect signals from Venus-PTS1.
2.5. Nucleotide Acession Numbers
The nucleotide sequences of the six U6 promoters in this study are registered in DDBJ/GenBank/EMBL under accession numbers LC765373 for U6-1, LC765374 for U6-2, LC765375 for U6-3, LC765376 for U6-4, LC765377 for U6-5, and LC765378 for U6-6.
3. Results and Discussion
3.1. Castor U6 snRNA Gene Identification and Promoter Cloning
Endogenous promoters driving gRNA expression are essential for improving genome editing efficiency [15,16,17,18]. The upstream regions of endogenous U6 snRNA genes are most commonly used as promoters for genome editing [14]. The transcribed regions of U6 snRNA genes are highly conserved among species [21]. Therefore, U6 promoters have been searched for in various plant species and identified in their genomes based on the transcribed sequence of U6 snRNA genes in Arabidopsis thaliana. However, U6 snRNA genes reportedly exist as multicopies in many organisms and also as pseudogenes [22,23]. Therefore, functional verification of U6 snRNA genes is essential for identifying a functional promoter in the plant species of interest.
Based on the draft genome sequence of castor released in 2010 [10] and its chromosome-level genome assembly reported in 2022 [11], we identified eight U6 snRNA genes in the castor genome. These include RcU6-5 and RcU6-7 on chromosome 1, RcU6-3 and RcU6-4 on chromosome 6, RcU6-8 on chromosome 7, RcU6-1 and RcU6-2 on chromosome 8, and RcU6-6 on chromosome 10 (Table 1). All eight U6 genes showed high sequence similarity to the transcribed region sequence of the Arabidopsis U6 snRNA1 gene (Table 1). U6 snRNA genes possess an RNA polymerase III (POLIII) type 3 promoter, which in plants consists of a TATA box at −28 to −30 bp and a plant snRNA gene-specific element, upstream sequence element (USE), at approximately −70 bp [14]. Sequence analysis of a 100 bp sequence upstream of each of the eight U6 snRNA genes revealed the presence of a conserved TATA box and a USE in the promoter regions of RcU6-1 to RcU6-6 and that of Arabidopsis U6 snRNA1 (AtU6-1), but these elements were absent in the promoters of RcU6-7 and RcU6-8 (Figure 1). Since their promoter sequences do not contain the typical POLIII type 3 promoter elements, RcU6-7 and RcU6-8 could be pseudogenes and may not be effective for gRNA expression. Therefore, the promoters of six of the identified snRNA genes, RcU6-1 to RcU6-6, were considered for further analysis.

Table 1.
Summary of U6 snRNA genes identified in the genome sequence of castor (ASM1957865v1) using the transcribed region of the Arabidopsis U6-1 snRNA gene as the query sequence.
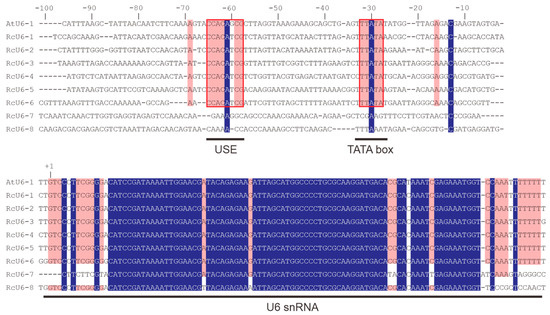
Figure 1.
Alignment of eight castor U6 and Arabidopsis U6-1 genes. The sequence of the transcribed region of the U6 genes and approximately 100 bp upstream of their transcription start sites was aligned. USE, TATA box, and U6 snRNA sequences are underlined. The conserved USE and TATA box sequences are outlined in red. Bases shaded in purple indicate those common to all DNA sequences described. Bases shaded in pink indicate those common to AtU6-1, RcU6-1, RcU6-2, RcU6-3, RcU6-4, RcU6-5 and RcU6-6.
3.2. Functional Analysis of the Rc U6 Gene Promoters
Heterologous U6 promoters have been reported to be functional in several plant species [24,25]. However, in several studies, heterologous U6 promoters were shown to reduce genome editing efficiency compared with endogenous U6 promoters [15,16,17,18], indicating that functional verification of promoters in native species is a better approach than testing their activity in heterologous systems. Castor has long been cultivated as a useful crop. However, the transformation of castor has been reported relatively recently [12], and a transient expression system using protoplasts for the verification of RcU6 promoter function has not yet been established. Therefore, we carried out functional verification of the RcU6 promoters in castor cells using the particle delivery system, which allows the transient expression of genes in a consistent manner across various plant species [26,27]. POLIII is known to be involved in the transcription of 5S rRNA, tRNAs, and small RNAs [14]. However, POLIII has also been reported to transcribe mRNAs, such as those encoding proteins [15,28]. Therefore, we performed the functional verification of RcU6 promoters using the fluorescent protein Venus because it is easy to investigate U6 promoter activity as a fluorescent signal. The peroxisomal targeting signal 1, PTS1 [29], was ligated to the C-terminus of Venus [30] to facilitate the observation of Venus as a peroxisome-localized fluorescent signal. The Venus-PTS1 fusion and each of the six RcU6 promoters were further cloned into the R4pGWB401 plasmid [19,31] to generate the RcU6-1pro:Venus-PTS1, RcU6-2pro:Venus-PTS1, RcU6-3pro:Venus-PTS1, RcU6-4pro:Venus-PTS1, RcU6-5pro:Venus-PTS1, and RcU6-6pro:Venus-PTS1 constructs. The DNA of each plasmid was introduced into castor leaf cells using a gene gun [20]. The introduction of plasmid DNAs without the U6 promoter resulted in no Venus fluorescence in peroxisomes (Figure 2A). However, the introduction of the RcU6-1pro:Venus-PTS1 plasmid resulted in Venus fluorescence in peroxisomes, which appeared as spherical spots in castor leaf cells. This indicated that RcU6-1 promoter could transcribe Venus-PTS1. Similarly, the introduction of the other five RcU6 promoters driving the Venus-PTS1 fusion also produced fluorescence signals in peroxisomes (Figure 2C–G). The results show that the six newly cloned RcU6 promoters are transcriptionally active in castor cells. Judging from the fluorescence pattern of Venus-PTS1, it can be concluded that there is no difference in the six U6 promoter activities. These results also suggest that the Venus gene was correctly transcribed and translated, and the Venus protein was transported to peroxisomes, indicating that the U6 promoter is functional when cloned upstream of the Venus gene. Thus, this study provides a simple and robust technique for verifying promoter function in plant species for which transformation techniques are not yet established.
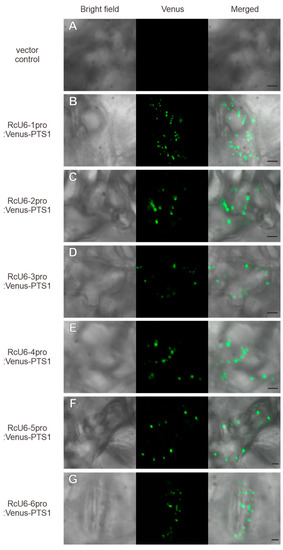
Figure 2.
Functional verification of RcU6 promoters. The vector without the U6 promoter (A), RcU6-1pro:Venus-PTS1 (B), RcU6-2pro:Venus-PTS1 (C), RcU6-3pro:Venus-PTS1 (D), RcU6-4pro:Venus-PTS1 (E), RcU6-5pro:Venus-PTS1 (F), and RcU6-6pro:Venus-PTS1 (G) constructs were individually introduced into castor leaf cells using the particle delivery method. Scale bars in the merged pictures represent 5 µm.
3.3. Truncation of Rc U6 Promoters
The ability of U6 promoters to express gRNAs has been well studied. In Arabidopsis, an approximately 100 bp sequence upstream of the transcription start site of the Rc-1 gene has been shown to function well for driving gene expression. Genome editing in plants differs from that in animals; since it is difficult to inject gRNA and Cas9 nuclease into egg cells in plants, unlike in animals, the CRISPR/Cas9 cassette is introduced into the plant genome [9,13,32]. Therefore, shortening the size of the transgene helps in increasing transformation efficiency, and the most effective way of shortening the size of the CRISPR/Cas9 cassette is by truncating the U6 promoter. Therefore, we attempted to truncate the RcU6 promoters to approximately 500 bp from the transcription start size. The truncated promoters, namely, RcU6-1pro(533), RcU6-2pro(504), RcU6-3pro(590), RcU6-4pro(596), RcU6-5pro(550), and RcU6-6pro(611), were cloned into the R4pGWB401 vector along with the Venus-PTS1 fusion, and the resulting plasmids were transiently expressed in castor leaf cells by particle bombardment. The introduction of plasmid DNA without the inserted promoter resulted in no Venus fluorescence in peroxisomes (Figure 3A). However, peroxisome-localized Venus fluorescence was observed in castor leaf cells transformed with RcU6-1pro(533):Venus-PTS1, RcU6-2pro(504):Venus-PTS1, RcU6-3pro(590):Venus-PTS1, RcU6-4pro(596):Venus-PTS1, RcU6-5pro(550):Venus-PTS1, and RcU6-6pro (611):Venus-PTS1 constructs (Figure 3B–G). These results show that U6 promoters truncated to approximately 500 bp in length are active in castor cells. In some crops, endogenous promoters truncated to approximately 300 bp have been reported to show activity [15,17,33]. Therefore, we truncated the RcU6 promoters to approximately 300 bp from the transcription start site, and named them RcU6-1pro(307), RcU6-2pro(289), RcU6-3pro(315), RcU6-4pro(332), RcU6-5pro(260), and RcU6-6pro(289). The introduction of plasmid DNA without the inserted promoter resulted in no Venus fluorescence in peroxisomes (Figure 4A). However, fluorescence was detected in the peroxisomes of castor leaf cells transformed with RcU6-1pro(307):Venus-PTS1, RcU6-2pro(289):Venus-PTS1, RcU6-3pro(315):Venus-PTS1, RcU6-4pro(332):Venus-PTS1, RcU6-5pro(260):Venus-PTS1, and RcU6-6pro (289):Venus-PTS1 constructs (Figure 4B–G). These results demonstrate that the RcU6 promoters, truncated to approximately 300 bp in length, are also transcriptionally active.
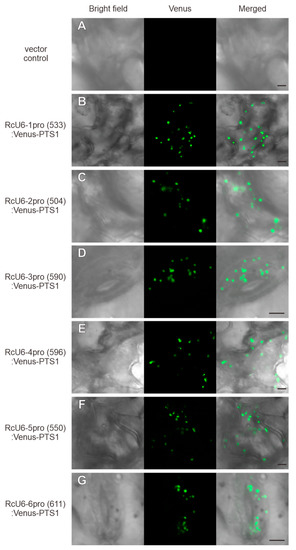
Figure 3.
Functional verification of RcU6 promoters truncated to approximately 500 bp. The vector without the U6 promoter (A), RcU6-1pro(533):Venus-PTS1 (B), RcU6-2pro(504):Venus-PTS1 (C), RcU6-3pro(590):Venus-PTS1 (D), RcU6-4pro(596):Venus-PTS1 (E), RcU6-5pro(550):Venus-PTS1 (F), and RcU6-6pro(611):Venus-PTS1 (G) were separately introduced into castor leaf cells by particle bombardment. The numbers in brackets indicate the length of the promoter. Scale bars in the merged pictures represent 5 µm.
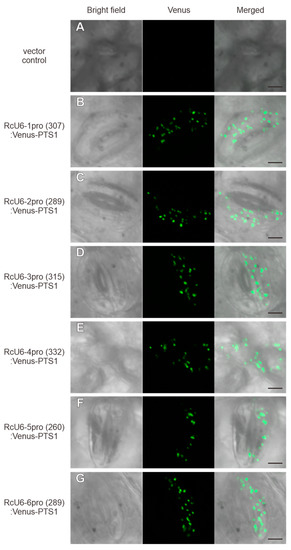
Figure 4.
Functional verification of RcU6 promoters truncated to approximately 300 bp. The vector without the U6 promoter (A), RcU6-1pro(307):Venus-PTS1 (B), RcU6-2pro(289):Venus-PTS1 (C), RcU6-3pro(315):Venus-PTS1 (D), RcU6-4pro(332):Venus-PTS1 (E), RcU6-5pro(260):Venus-PTS1 (F), and RcU6-6pro (289):Venus-PTS1 (G) were separately introduced into castor leaf cells by particle bombardment. The numbers in brackets indicate the length of the promoter. Scale bars in the merged pictures represent 5 µm.
4. Conclusions
Our study demonstrates that six RcU6 promoters truncated to approximately 300 bp are functional in castor cells. These results improve the potential of genome editing technology in castor, an important commercial crop, and provide a strategy for the construction of castor-optimized CRISPR/Cas9 cassettes in future studies. Moreover, our results show that transient expression analysis of U6 promoters using the particle delivery method is an effective approach for the functional verification of promoters in plant species for which transformation methods have not yet been established, thus facilitating the generation of genetically modified crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14071327/s1, Table S1: List of primers used in this study.
Author Contributions
Conceptualization, M.K. and S.M.; methodology, M.K. and S.M.; validation, M.K. and S.M.; formal analysis, M.K. and K.H.; investigation, M.K. and S.M.; writing—original draft preparation, M.K.; writing—review and editing, S.M.; supervision, S.M.; project administration, M.K.; funding acquisition, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI, Grant Number 19K22438 (M.K.).
Institutional Review Board Statement
The study was approved by the Safety Committee for Genetic Recombination Experiments of the National Institutes of Natural Sciences, National Institute for Basic Biology (approval numbers: B14-004-A and B14-005-A) and was carried out properly in accordance with the safety management regulations for genetic recombination experiments.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
We thank Tsuyoshi Nakagawa (Shimane University) for providing experimental materials, and the staff at the Model Organisms Facility and Trans-Omics Facility at the NIBB Trans-Scale Biology Center for technical support. We also acknowledge Kenji Komazawa (Itoh Oil Chemicals Co., Ltd.) for the joint research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Patel, V.R.; Dumancas, G.G.; Kasi Viswanath, L.C.; Maples, R.; Subong, B.J.J. Castor Oil: Properties, Uses, and Optimization of Processing Parameters in Commercial Production. Lipid Insights 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Nitbani, F.O.; Tjitda, P.J.P.; Wogo, H.E.; Detha, A.I.R. Preparation of Ricinoleic Acid from Castor Oil: A Review. J. Oleo Sci. 2022, 71, 781–793. [Google Scholar] [CrossRef]
- Kunduru, K.R.; Basu, A.; Haim Zada, M.; Domb, A.J. Castor Oil-Based Biodegradable Polyesters. Biomacromolecules 2015, 16, 2572–2587. [Google Scholar] [CrossRef]
- Chakraborty, I.; Chatterjee, K. Polymers and Composites Derived from Castor Oil as Sustainable Materials and Degradable Biomaterials: Current Status and Emerging Trends. Biomacromolecules 2020, 21, 4639–4662. [Google Scholar] [CrossRef]
- Timko, J.A.; Amsalu, A.; Acheampong, E.; Teferi, M.K. Local Perceptions about the Effects of Jatropha (Jatropha curcas) and Castor (Ricinus communis) Plantations on Households in Ghana and Ethiopia. Sustain. Sci. Pract. Policy 2014, 6, 7224–7241. [Google Scholar] [CrossRef]
- Xu, W.; Chen, Z.; Ahmed, N.; Han, B.; Cui, Q.; Liu, A. Genome-Wide Identification, Evolutionary Analysis, and Stress Responses of the GRAS Gene Family in Castor Beans. Int. J. Mol. Sci. 2016, 17, 1004. [Google Scholar] [CrossRef]
- Han, B.; Fu, L.; Zhang, D.; He, X.; Chen, Q.; Peng, M.; Zhang, J. Interspecies and Intraspecies Analysis of Trehalose Contents and the Biosynthesis Pathway Gene Family Reveals Crucial Roles of Trehalose in Osmotic-Stress Tolerance in Cassava. Int. J. Mol. Sci. 2016, 17, 1077. [Google Scholar] [CrossRef]
- Polito, L.; Bortolotti, M.; Battelli, M.G.; Calafato, G.; Bolognesi, A. Ricin: An Ancient Story for a Timeless Plant Toxin. Toxins 2019, 11, 324. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G.; et al. Draft Genome Sequence of the Oilseed Species Ricinus communis. Nat. Biotechnol. 2010, 28, 951–956. [Google Scholar] [CrossRef]
- Lu, J.; Pan, C.; Fan, W.; Liu, W.; Zhao, H.; Li, D.; Wang, S.; Hu, L.; He, B.; Qian, K.; et al. A Chromosome-Level Genome Assembly of Wild Castor Provides New Insights into Its Adaptive Evolution in Tropical Desert. Genom. Proteom. Bioinform. 2022, 20, 42–59. [Google Scholar] [CrossRef]
- Alexandrov, O.S.; Petrov, N.R.; Varlamova, N.V.; Khaliluev, M.R. An Optimized Protocol for In Vitro Indirect Shoot Organogenesis of Impala Bronzovaya and Zanzibar Green Ricinus communis L. Varieties. Horticulturae 2021, 7, 105. [Google Scholar] [CrossRef]
- Cable, J.; Ronald, P.C.; Voytas, D.; Zhang, F.; Levy, A.A.; Takatsuka, A.; Arimura, S.-I.; Jacobsen, S.E.; Toki, S.; Toda, E.; et al. Plant Genome Engineering from Lab to Field-a Keystone Symposia Report. Ann. N. Y. Acad. Sci. 2021, 1506, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Kor, S.D.; Chowdhury, N.; Keot, A.K.; Yogendra, K.; Chikkaputtaiah, C.; Sudhakar Reddy, P. RNA Pol III Promoters-Key Players in Precisely Targeted Plant Genome Editing. Front. Genet. 2022, 13, 989199. [Google Scholar] [CrossRef]
- Long, L.; Guo, D.-D.; Gao, W.; Yang, W.-W.; Hou, L.-P.; Ma, X.-N.; Miao, Y.-C.; Botella, J.R.; Song, C.-P. Optimization of CRISPR/Cas9 Genome Editing in Cotton by Improved SgRNA Expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Massel, K.; Lam, Y.; Hintzsche, J.; Lester, N.; Botella, J.R.; Godwin, I.D. Endogenous U6 Promoters Improve CRISPR/Cas9 Editing Efficiencies in Sorghum Bicolor and Show Potential for Applications in Other Cereals. Plant Cell Rep. 2022, 41, 489–492. [Google Scholar] [CrossRef]
- Johansen, I.E.; Liu, Y.; Jørgensen, B.; Bennett, E.P.; Andreasson, E.; Nielsen, K.L.; Blennow, A.; Petersen, B.L. High Efficacy Full Allelic CRISPR/Cas9 Gene Editing in Tetraploid Potato. Sci. Rep. 2019, 9, 17715. [Google Scholar] [CrossRef]
- Riu, Y.-S.; Kim, G.H.; Chung, K.W.; Kong, S.-G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants 2023, 12, 878. [Google Scholar] [CrossRef]
- Nakagawa, T.; Nakamura, S.; Tanaka, K.; Kawamukai, M.; Suzuki, T.; Nakamura, K.; Kimura, T.; Ishiguro, S. Development of R4 Gateway Binary Vectors (R4pGWB) Enabling High-Throughput Promoter Swapping for Plant Research. Biosci. Biotechnol. Biochem. 2008, 72, 624–629. [Google Scholar] [CrossRef]
- Kanai, M.; Hayashi, M.; Kondo, M.; Nishimura, M. The Plastidic DEAD-Box RNA Helicase 22, HS3, Is Essential for Plastid Functions Both in Seed Development and in Seedling Growth. Plant Cell Physiol. 2013, 54, 1431–1440. [Google Scholar] [CrossRef]
- Didychuk, A.L.; Butcher, S.E.; Brow, D.A. The Life of U6 Small Nuclear RNA, from Cradle to Grave. RNA 2018, 24, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.E.; Caputi, M.; Beemon, K.L. Packaging and Reverse Transcription of SnRNAs by Retroviruses May Generate Pseudogenes. RNA 2004, 10, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Guerineau, F.; Waugh, R. The U6 Small Nuclear RNA Gene Family of Potato. Plant Mol. Biol. 1993, 22, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Wilson, F.M.; Harrison, K.; Armitage, A.D.; Simkin, A.J.; Harrison, R.J. CRISPR/Cas9-Mediated Mutagenesis of Phytoene Desaturase in Diploid and Octoploid Strawberry. Plant Methods 2019, 15, 45. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Liu, Z. Efficient Genome Editing of Wild Strawberry Genes, Vector Development and Validation. Plant Biotechnol. J. 2018, 16, 1868–1877. [Google Scholar] [CrossRef]
- Lorence, A.; Verpoorte, R. Gene Transfer and Expression in Plants. Methods Mol. Biol. 2004, 267, 329–350. [Google Scholar]
- Taylor, N.J.; Fauquet, C.M. Microparticle Bombardment as a Tool in Plant Science and Agricultural Biotechnology. DNA Cell Biol. 2002, 21, 963–977. [Google Scholar] [CrossRef]
- Gunnery, S.; Mathews, M.B. Functional mRNA Can Be Generated by RNA Polymerase III. Mol. Cell. Biol. 1995, 15, 3597–3607. [Google Scholar] [CrossRef]
- Hayashi, M.; Aoki, M.; Kato, A.; Kondo, M.; Nishimura, M. Transport of Chimeric Proteins That Contain a Carboxy-Terminal Targeting Signal into Plant Microbodies. Plant J. 1996, 10, 225–234. [Google Scholar] [CrossRef]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A Variant of Yellow Fluorescent Protein with Fast and Efficient Maturation for Cell-Biological Applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Nakamura, S.; Mano, S.; Tanaka, Y.; Ohnishi, M.; Nakamori, C.; Araki, M.; Niwa, T.; Nishimura, M.; Kaminaka, H.; Nakagawa, T.; et al. Gateway Binary Vectors with the Bialaphos Resistance Gene, bar, as a Selection Marker for Plant Transformation. Biosci. Biotechnol. Biochem. 2010, 74, 1315–1319. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef] [PubMed]
- Bernard, G.; Gagneul, D.; Alves Dos Santos, H.; Etienne, A.; Hilbert, J.-L.; Rambaud, C. Efficient Genome Editing Using CRISPR/Cas9 Technology in Chicory. Int. J. Mol. Sci. 2019, 20, 1155. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).