Evaluation of Candidate Reference Genes for Gene Expression Analysis in Wild Lamiophlomis rotata
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Screening of Candidate Internal Reference Genes and Designing of Primers
2.4. RT-qPCR Analysis of the Candidate Internal Reference Genes
2.5. Determination of the Correlation Coefficient of Primer Pairs
2.6. Stability Analysis of the Candidate Internal Reference Genes
2.7. Validation of the Validity of the Candidate Reference Genes
3. Results
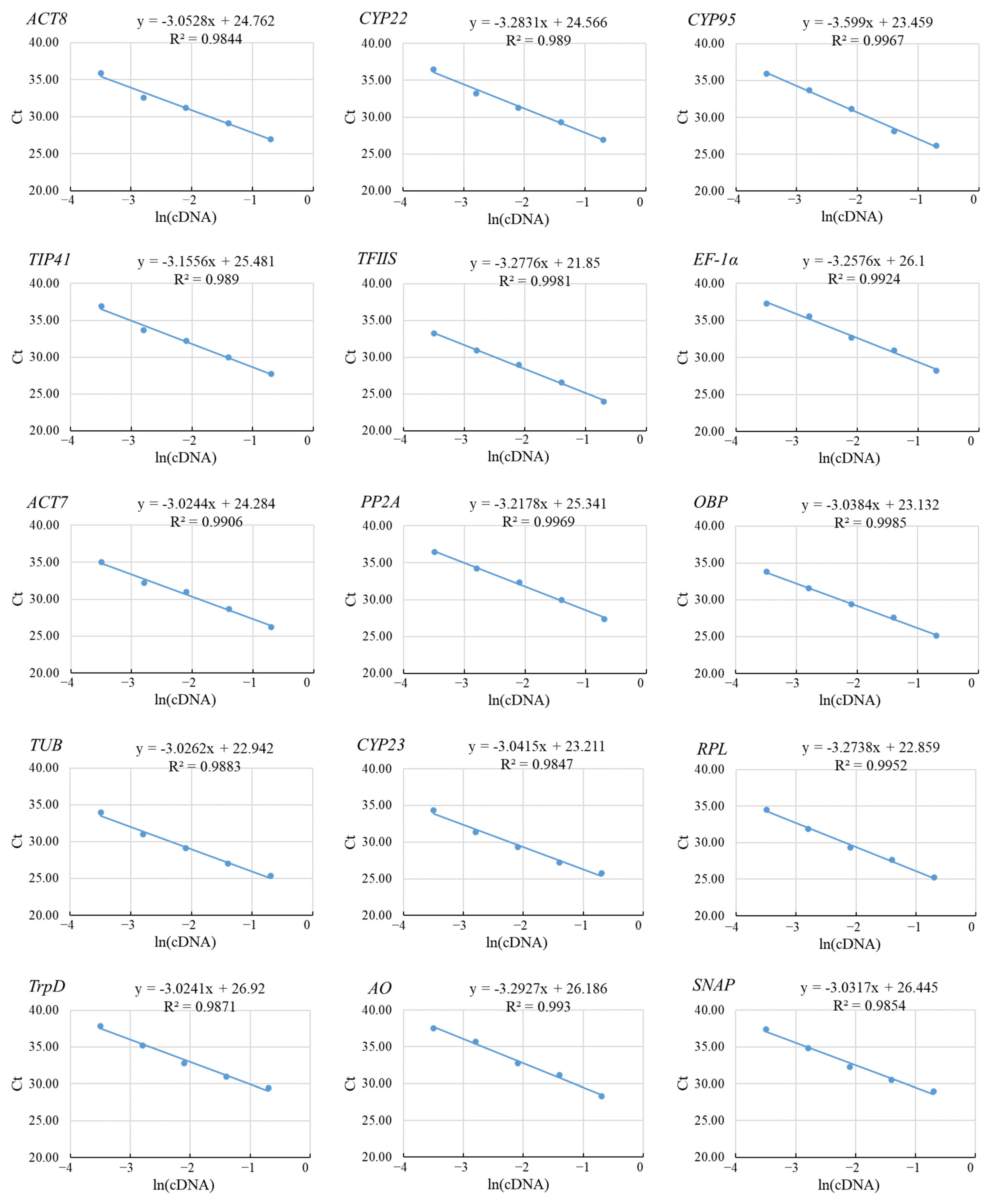
3.1. Primer Specificity and Correlation Coefficient of Primer Sets
3.2. Ct Values of the Candidate Reference Genes
3.3. Stability Analysis of the Candidate Reference Genes
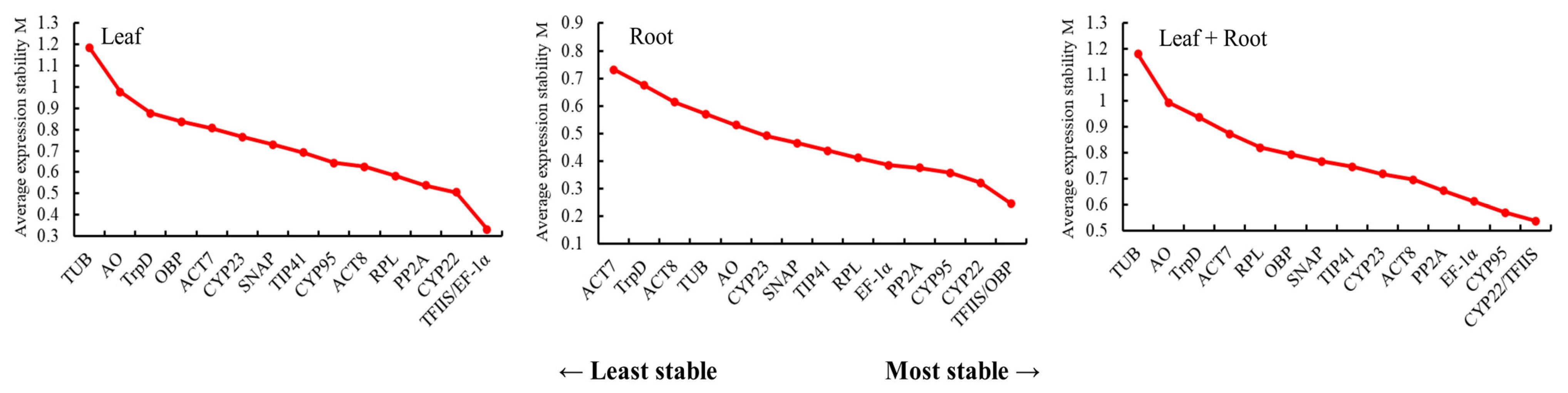
3.3.1. GeNorm Analysis of the Stability of the Candidate Reference Genes
3.3.2. NormFinder Analysis of the Stability of the Candidate Reference Genes
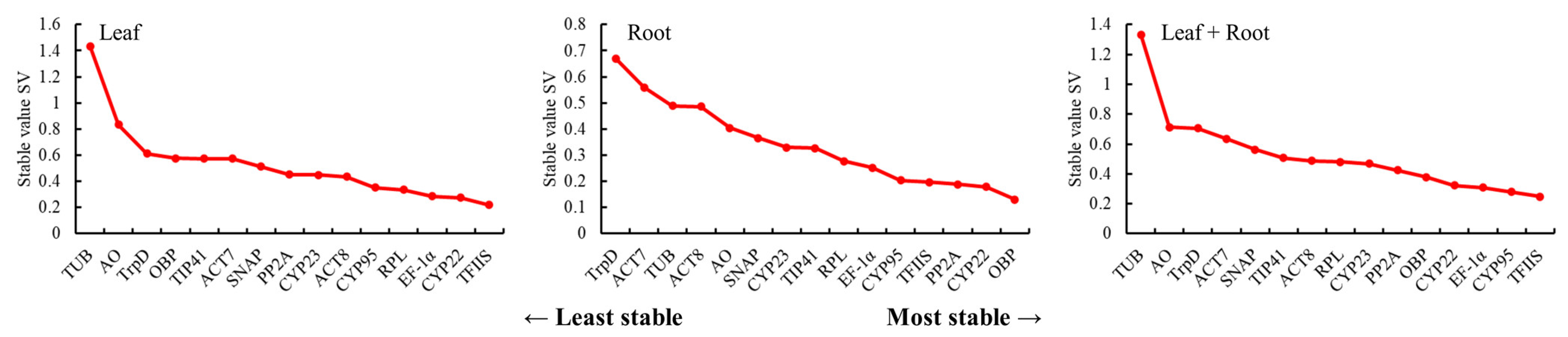
3.3.3. BestKeeper Analysis of the Stability of the Candidate Reference Genes
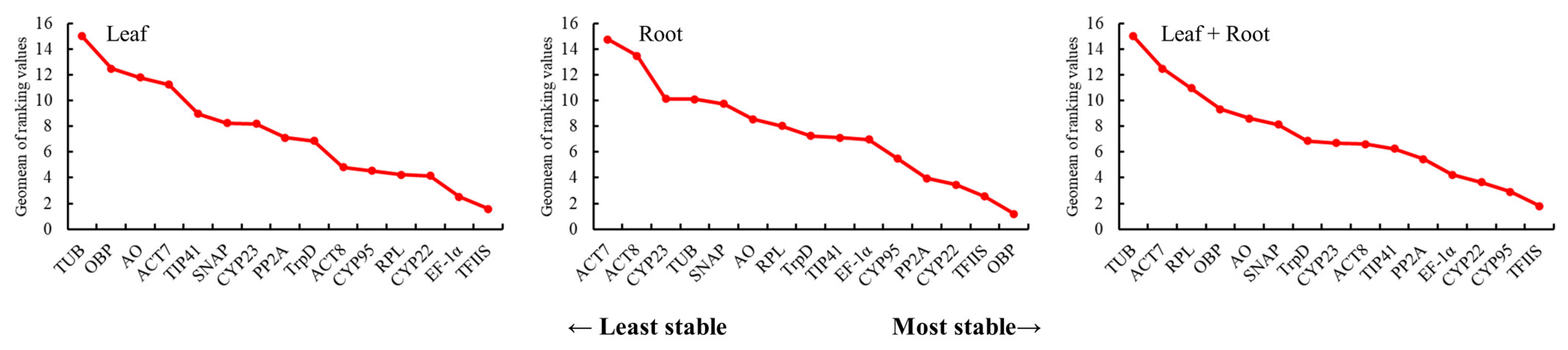
3.3.4. RefFinder Analysis of the Stability of the Candidate Reference Genes
3.4. Verification of the Stability of Internal Reference Genes
4. Discussion
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Li, Y.; Liang, X.; Zhou, X.; Wu, Z.; Yuan, L.; Wang, Y.; Li, Y. Selection of reference genes for qRT-PCR analysis in medicinal plant Glycyrrhiza under abiotic stresses and hormonal treatments. Plants 2020, 9, 1441. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, S.; Cao, Y.; Chen, R.; Wang, Z.; Cao, X. Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol. Biol. Rep. 2021, 48, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Zhang, B.; Yang, J.; Lan, J.; Yang, H.; Guo, K.; Li, J.; Chen, S. Validation of suitable reference genes by various algorithms for gene expression analysis in Isodon rubescens under different abiotic stresses. Sci. Rep. 2022, 12, 19599. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Z.; Bao, W.; Hu, H.; Chen, M.; Chai, T.; Wang, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol. 2019, 19, 498. [Google Scholar] [CrossRef]
- Kou, X.Y.; Zhang, L.; Yang, S.Z.; Li, G.H.; Ye, J.L. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Xu, L.; Xu, H.; Cao, Y.; Yang, P.; Feng, Y.; Tang, Y.; Yuan, S.; Ming, J. Validation of reference genes for quantitative real-time PCR during bicolor Tepal development in Asiatic hybrid lilies (Lilium spp.). Front. Plant Sci. 2017, 8, 669. [Google Scholar] [CrossRef]
- Yuan, W.; Wan, H.J.; Yang, Y.J. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants. Chin. Bull. Bot. 2012, 47, 427–436. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, X.; Sun, M.; Cong, H.; Qiao, F. Evaluation of reference genes for normalizing RT-qPCR in leaves and suspension cells of Cephalotaxus hainanensis under various stimuli. Plant Methods 2019, 15, 31. [Google Scholar] [CrossRef]
- Long, X.Y.; Wang, J.R.; Ouellet, T.; Rocheleau, H.; Wei, Y.M.; Pu, Z.E.; Jiang, Q.T.; Lan, X.J.; Zheng, Y.L. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010, 74, 307–311. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical de-termination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zhang, Y.; Liu, F.; Liu, T.; Chen, J.Y.; Fu, G.; Zheng, C.Y.; Su, D.D.; Wang, Y.N.; Zhou, H.K. Establishment of reference (housekeeping) genes via quantitative real-time PCR for investigation of the genomic basis of abiotic stress resistance in Psammochloa villosa (Poaceae). J. Plant Physiol. 2022, 268, 153575. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Li, Y.; Li, F.; Zheng, T.T.; Shi, L.; Zhang, Z.G.; Niu, T.M.; Wang, Q.Y.; Zhao, D.S.; Li, W.; Zhao, P. Lamiophlomis herba: A comprehensive overview of its chemical constituents, pharmacology, clinical applications, and quality control. Biomed. Pharmacother. 2021, 144, 112299. [Google Scholar] [CrossRef]
- Huang, X.J.; Wang, J.; Muhammad, A.; Tong, H.Y.; Wang, D.G.; Li, J.; Ihsan, A.; Yang, G.Z. Systems pharmacology-based dissection of mechanisms of Tibetan medicinal compound Ruteng as an effective treatment for collagen-induced arthritis rats. J. Ethnopharmacol. 2021, 272, 113953. [Google Scholar] [CrossRef]
- Xia, M.; Zhang, Y.; Wu, H.; Zhang, Q.; Liu, Q.; Li, G.; Zhao, T.; Liu, X.; Zheng, S.; Qian, Z.; et al. Forsythoside B attenuates neuro-inflammation and neuronal apoptosis by inhibition of NF-κB and p38-MAPK signaling pathways through activating Nrf2 post spinal cord injury. Int. Immunopharmacol. 2022, 111, 109120. [Google Scholar] [CrossRef]
- Ma, L.; Zhong, M.; Jiang, G.; Long, F.; Wu, W.; Jiang, Y. Elucidation of the active ingredients of Lamiophlomis herba against hemorrhage based on network pharmacology and tail snipping model in mice. Pharmazie 2020, 8, 381–384. [Google Scholar] [CrossRef]
- La, M.; Zhang, F.; Gao, S.; Liu, X.; Wu, Z.; Sun, L.; Tao, X.; Chen, W. Constituent analysis and quality control of Lamiophlomis rotata by LC-TOF/MS and HPLC-UV. J. Pharm. Biomed. Anal. 2015, 102, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jiang, S.; Dong, Q.; Dang, J.; Liu, Z.; Han, H.; Tao, Y.; Yue, H. Anti-rheumatoid arthritis effects of iridoid glucosides from Lamiophlomis rotata (Benth.) kudo on adjuvant-induced arthritis in rats by OPG/RANKL/NF-κB signaling pathways. J. Ethnopharmacol. 2021, 266, 13402. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.C.; Ma, H.P.; Hao, Y.; He, X.R.; Sun, A.J.; Jiang, W.; Li, M.X.; Jing, L.L.; He, L.; Ma, J.; et al. A new anti-fibrinolytic hemostatic com-pound 8-O-acetyl shanzhiside methylester extracted from Lamiophlomis rotata. J. Ethnopharmacol. 2016, 187, 232–238. [Google Scholar] [CrossRef]
- Zhu, B.; Gong, N.; Fan, H.; Peng, C.S.; Ding, X.J.; Jiang, Y.; Wang, Y.X. Lamiophlomis rotata, an orally available Tibetan herbal painkiller, specifically reduces pain hypersensitivity states through the activation of spinal glucagon-like peptide-1 receptors. Anesthesiology 2014, 121, 835–851. [Google Scholar] [CrossRef]
- Cui, Z.H.; Qin, S.S.; Zang, E.H.; Li, C.; Gao, L.; Li, Q.C.; Wang, Y.L.; Huang, X.Z.; Zhang, Z.Y.; Li, M.H. Traditional uses, phytochemistry, pharmacology and toxicology of Lamiophlomis rotata (Benth.) Kudo: A review. RSC Adv. 2020, 10, 11463–11474. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Li, Z.; Zhou, Y.; Shao, W.; Zhang, D. Active learning for efficient analysis of high-throughput nanopore data. Bioinformatics 2023, 39, 764. [Google Scholar] [CrossRef] [PubMed]
- Mursyidah, A.K.; Hafizzudin-Fedeli, M.; Nor Muhammad, N.A.; Latiff, A.; Firdaus-Raih, M.; Wan, K.L. Dissecting the biology of Rafflesia species: Current progress and future directions made possible with high-throughput sequencing data. Plant Cell Physiol. 2023, 6, 4. [Google Scholar] [CrossRef]
- Julca, I.; Tan, Q.W.; Mutwil, M. Toward kingdom-wide analyses of gene expression. Trends Plant Sci. 2023, 28, 235–249. [Google Scholar] [CrossRef]
- Kapoor, B.; Kumar, A.; Kumar, P. Transcriptome repository of north-western himalayan endangered medicinal herbs: A paramount approach illuminating molecular perspective of phytoactive molecules and secondary metabolism. Mol. Genet. Genom. 2021, 296, 1177–1202. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Liu, L.; Jing, H.; Zuo, W.; Zeng, Y. Transcriptome analysis provides insights into Potentilla bifurca adaptation to high altitude. Life 2022, 12, 1337. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.O.; Lin, W.; Feng, E.; Ou, X. Transcriptome and metabolome response of eggplant against Ralstonia solanacearum infection. PeerJ 2023, 11, 14658. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Charron, J.B.; Jabaji, S. Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis E68 against Fusarium graminearum DAOMC 180378, the causal agent of Fusarium head blight. PLoS ONE 2023, 18, 0277983. [Google Scholar] [CrossRef]
- Saberi Riseh, R.; Skorik, Y.A.; Thakur, V.K.; Moradi Pour, M.; Tamanadar, E.; Noghabi, S.S. Encapsulation of plant biocontrol bacteria with alginate as a main polymer material. Int. J. Mol. Sci. 2021, 22, 11165. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Z.; Ding, Z.; Meng, H.; Shen, R.; Tang, H.; Liu, Y.G.; Chen, L. Public-transcriptome-database-assisted selection and validation of reliable reference genes for qRT-PCR in rice. Sci. China Life Sci. 2020, 63, 92–101. [Google Scholar] [CrossRef]
- Bai, X.; Chen, T.; Wu, Y.; Tang, M.; Xu, Z.-F. Selection and validation of reference genes for qRT-PCR analysis in the oil-rich tuber crop tiger nut (Cyperus esculentus) based on transcriptome data. Int. J. Mol. Sci. 2021, 22, 2569. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, K.C.G.; Desgagné-Penix, I.; Germain, H. Custom selected reference genes outperform pre-defined reference genes in transcriptomic analysis. BMC Genom. 2020, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, J.; Grégoire, J.C.; Vaillancourt, C. A simple method to assess group difference in RT-qPCR reference gene selection using GeNorm: The case of the placental sex. Sci. Rep. 2017, 7, 16923. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 31, 75–84. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zhu, G.; Liang, D.; He, B.; Wang, Y.; Cai, Y.; Zhang, Q. Reference gene selection for qPCR analysis in Schima superba under abiotic stress. Genes 2022, 13, 1887. [Google Scholar] [CrossRef]
- Qu, R.; Miao, Y.; Cui, Y.; Cao, Y.; Zhou, Y.; Tang, X.; Yang, J.; Wang, F. Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Yin, D.; Zhang, M.; Gao, Z.; Tuluhong, M.; Li, X.; Li, J.; Li, B.; Cui, G. Validation of appropriate reference genes for qRT–PCR normalization in oat (Avena sativa L.) under UV-B and high-light stresses. Int. J. Mol. Sci. 2022, 23, 11187. [Google Scholar] [CrossRef]
- Akbarabadi, A.; Ismaili, A.; Kahrizi, D.; Nazarian Firouzabadi, F. Validation of expression stability of reference genes in response to herbicide stress in wild oat (Avena ludoviciana). Cell Mol. Biol. 2018, 4, 113–118. [Google Scholar] [CrossRef]
- Hossain, M.S.; Ahmed, R.; Haque, M.S.; Alam, M.M.; Islam, M.S. Identification and validation of reference genes for real-time quantitative RT-PCR analysis in jute. BMC Mol. Biol. 2019, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, C.; Zhang, J.; Chen, F.; Gong, Y.; Li, Y.; Su, Y.; Wei, Y.; Zhao, Y. Selection of the reference gene for expression normalization in Papaver somniferum, L. under abiotic stress and hormone treatment. Genes 2020, 11, 124. [Google Scholar] [CrossRef]
- Liu, C.; Wang, L.; Sun, Y.; Zhao, X.; Chen, T.; Su, X.; Guo, H.; Wang, Q.; Xi, X.; Ding, Y.; et al. Probe synthesis reveals eukaryotic translation elongation factor[1-Alpha]1 as the anti-pancreatic cancer target of BE-43547A. Angew. Chem. Int. Ed. Engl. 2022, 61, e202206953. [Google Scholar] [CrossRef]
- Kristensen, R.; Torp, M.; Kosiak, B.; Holst-Jensen, A. Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycol. Res. 2005, 109, 173–186. [Google Scholar] [CrossRef]
- Tatsuka, M.; Mitsui, H.; Wada, M.; Nagata, A.; Nojima, H.; Okayama, H. Elongation factor-1α gene determines susceptibility to transformation. Nature 1992, 359, 333–336. [Google Scholar] [CrossRef]
- Hammond, G.R.V.; Pacheco, J. Oxysterol binding protein: Tether, transporter... and Flux Capacitor? Trends Cell Biol. 2019, 29, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, F.; Liu, Z.; Feng, X.; Li, Y.; Zhu, P. Inactivation of BoORP3a, an oxysterol-binding protein, causes a low wax phenotype in ornamental kale. Hortic. Res. 2022, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Shivhare, R.; Kaur, A.; Bansal, S.; Sonah, H.; Deshmukh, R.; Giri, J.; Lata, C.; Ram, H. Reference gene identification for gene expression analysis in rice under different metal stress. J. Biotechnol. 2021, 332, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.W.; Wang, D.R.; Wang, X.; You, C.X.; Wang, X.F. Genome-wide identification and stress response analysis of cyclophilin gene family in apple (Malus × domestica). BMC Genom. 2022, 23, 806. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, Q.-J.; Sun, G.-Q.; Zheng, K.; Yao, Z.-P.; Han, Y.-H.; Wang, L.-P.; Duan, Y.-J.; Yu, D.-Q.; Qu, Y.-Y. Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 2019, 20, 349. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ahmad, A.; Kushwaha, N.; Shokeen, N.; Negi, S.; Gautam, K.; Singh, A.; Tiwari, P.; Garg, R.; Agarwal, R.; et al. Selection of ideal reference genes for gene expression analysis in COVID-19 and Mucormycosis. Microbiol. Spectr. 2022, 10, e0165622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Wang, J.; Hu, T.; Tong, Y.; Zhou, J.; Song, Y.; Gao, W.; Huang, L. Overexpression and RNA interference of TwDXR regulate the accumulation of terpenoid active ingredients in Tripterygium wilfordii. Biotechnol. Lett. 2018, 40, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Henriquez, M.A.; Soliman, A.; Li, G.; Hannoufa, A.; Ayele, B.T.; Daayf, F. Molecular cloning, functional characterization and expression of potato (Solanum tuberosum) 1-deoxy-d-xylulose 5-phosphate synthase 1 (StDXS1) in response to Phytophthora infestans. Plant Sci. 2016, 243, 71–83. [Google Scholar] [CrossRef]
- Hao, G.; Shi, R.; Tao, R.; Fang, Q.; Jiang, X.; Ji, H.; Feng, L.; Huang, L. Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol. Biochem. 2013, 70, 21–32. [Google Scholar] [CrossRef]
- Tong, Y.; Su, P.; Zhao, Y.; Zhang, M.; Wang, X.; Liu, Y.; Zhang, X.; Gao, W.; Huang, L. Molecular cloning and characterization of DXS and DXR genes in the terpenoid biosynthetic pathway of Tripterygium wilfordii. Int. J. Mol. Sci. 2015, 16, 25516–25535. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, F.; Li, S.; Wang, Y.; Zhou, C.; Shi, M.; Wang, J.; Chen, Y.; Wang, Y.; Wang, H.; et al. Molecular cloning and characterization of two 1-deoxy-D-xylulose-5-phosphate synthase genes involved in tanshinone biosynthesis in Salvia miltiorrhiza. Mol. Breed. 2016, 36, 124. [Google Scholar] [CrossRef]
- Kim, Y.B.; Kim, S.M.; Sathasivam, R.; Kim, Y.K.; Park, S.U.; Kim, S.U. Overexpression of Ginkgo biloba Hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase 2 gene (GbHDR2) in Nicotiana tabacum cv. Xanthi. 3 Biotech. 2021, 11, 337. [Google Scholar] [CrossRef] [PubMed]




| Sample Name | Sampling Location | Altitude | East Longitude | North Latitude |
|---|---|---|---|---|
| GL | Guoluo county, Qinghai, China | 3750 m | 100°14′38″ | 34°29′10″ |
| XW | Xiewu county, Qinghai, China | 3860 m | 97°21′3″ | 33°7′46″ |
| YS | Yushu county, Qinghai, China | 3880 m | 97°1′23″ | 32°51′4″ |
| ZD | Zaduo county, Qinghai, China | 4208 m | 95°10′48″ | 32°52′12″ |
| CD | Chengduo county, Qinghai, China | 4270 m | 97°27′16″ | 33°18′2″ |
| Gene Name | Gene Transcript Genbank No. | Primer Sequence | Product Length (bp) | Primer Efficiency |
|---|---|---|---|---|
| ACT8 | OQ471970 | F: ACTCACTTGCGGTCCAGTTATCC R: ATAACAGCTCCAGGGACTTCCAC | 107 | 1.13 |
| CYP22 | OQ471972 | F: CTTCGACATCACCATCGGAAAC R: TTCCTGTATTCGCCTGTGCAGT | 117 | 1.02 |
| CYP95 | OQ471971 | F: GCAACGGTCTCTCCTCCAAGA R: CTCACAGGGCTTCGACTTGGT | 86 | 0.90 |
| TIP41 | OQ471973 | F: GGGAAGACTGCCAGGATCAAAT R: AGCCAGAAGCGCAAGAGAAGAT | 175 | 1.07 |
| TFIIS | OQ471974 | F: GAGTTTGAGCCACGCTCGATT R: TCTTGCACCTCCCACAAGTGA | 163 | 1.02 |
| EF-1α | OQ471975 | F: ACTGGGACTTCTCAGGCTGATTG R: CTGGCCTTGGAGTACTTTGGTGT | 188 | 1.03 |
| ACT7 | OQ471976 | F: CACCACCCGAAAGAAAGTACAGTG R: AGGACCCGATTCATCATACTCTCC | 110 | 1.14 |
| PP2A | OQ471977 | F: GCTCATGTGTTGCTCCCTCCT R: GCTGCCAGCCTCTTCACAAGT | 155 | 1.05 |
| OBP | OQ471978 | F: GGAACCTCTTCCTGGCACAGA R: AACCAGACTTGGCGAGGTCAC | 197 | 1.13 |
| TUB | OQ471979 | F: CAAACTCGCCGTGAACCTCAT R: CGTCCCACATTTGCTGGGTAA | 134 | 1.14 |
| CYP23 | OQ471980 | F: TGCGCCCTGTGCAATTCTATC R: AACTGTCTTTGGCGCGACACT | 132 | 1.13 |
| RPL | OQ471981 | F: GAAACCCGCTGTCGTTAAACC R: CTATCATCGCGCTTTCCTTCC | 151 | 1.02 |
| TrpD | OQ471982 | F: CTGAGGCTGAGGCTTCTCTTGA R: CACCACCAGTCCCAACAATGTC | 192 | 1.14 |
| AO | OQ471983 | F: GTCGCCTACAAGCCAAATAGGG R: GACGACATCCATGTGCATACCA | 99 | 1.01 |
| SNAP | OQ471984 | F: GTATGAAGACGCTGCCGATTTG R: GCATTAGCTGCTTCATGCTTGC | 141 | 1.14 |
| Gene Name | Gene Transcript Genbank No. | Primer Sequence (5′–3′) (Forward/Reverse) | Amplicon Length (bp) |
|---|---|---|---|
| DXS | OQ471985 | F: GAAGGGGAGAGGGTGGCTCTAT R: GAGCACCATCCAACGGCTTAC | 136 |
| HDR | OQ471986 | F: CTTGCCGGAGACCAGAATATC R: GCCTTGGCGTTAAACTCAGAC | 111 |
| DXR | OQ471987 | F: CGAGCAGAACTTGTCACATCG R: CTGCAAACTAGCCGCGTAATC | 84 |
| Reference Gene | Leaf | Root | Leaf + Root | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CV Value | SD Value | Rank | CV Value | SD Value | Rank | CV Value | SD Value | Rank | |
| TrpD | 2.84 | 0.92 | 1 | 1.58 | 0.51 | 1 | 2.28 | 0.74 | 1 |
| AO | 4.09 | 1.27 | 6 | 1.65 | 0.50 | 2 | 2.79 | 0.85 | 2 |
| SNAP | 4.16 | 1.32 | 7 | 2.19 | 0.68 | 8 | 3.35 | 1.05 | 3 |
| CYP95 | 3.51 | 1.07 | 2 | 2.39 | 0.68 | 10 | 3.39 | 1.00 | 4 |
| ACT8 | 3.52 | 1.10 | 3 | 3.87 | 1.16 | 14 | 3.54 | 1.09 | 5 |
| CYP23 | 3.86 | 1.15 | 5 | 2.90 | 0.82 | 12 | 3.58 | 1.04 | 6 |
| TIP41 | 4.81 | 1.48 | 9 | 2.06 | 0.61 | 6 | 3.59 | 1.08 | 7 |
| PP2A | 5.40 | 1.62 | 11 | 1.88 | 0.55 | 3 | 3.73 | 1.10 | 8 |
| CYP22 | 4.88 | 1.55 | 10 | 2.12 | 0.62 | 7 | 4.35 | 1.33 | 9 |
| EF-1α | 5.78 | 1.47 | 13 | 3.05 | 0.74 | 13 | 4.45 | 1.10 | 10 |
| TFIIS | 4.44 | 1.25 | 8 | 2.24 | 0.58 | 9 | 4.48 | 1.22 | 11 |
| RPL | 3.58 | 1.10 | 4 | 2.78 | 0.77 | 11 | 5.15 | 1.51 | 12 |
| OBP | 6.57 | 1.86 | 15 | 2.05 | 0.53 | 5 | 5.24 | 1.43 | 13 |
| ACT7 | 5.51 | 1.51 | 12 | 4.39 | 1.08 | 15 | 6.46 | 1.68 | 14 |
| TUB | 6.02 | 1.86 | 14 | 2.01 | 0.53 | 4 | 9.35 | 2.68 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Qiao, F.; Geng, G.; Lu, Y. Evaluation of Candidate Reference Genes for Gene Expression Analysis in Wild Lamiophlomis rotata. Genes 2023, 14, 573. https://doi.org/10.3390/genes14030573
Wang L, Qiao F, Geng G, Lu Y. Evaluation of Candidate Reference Genes for Gene Expression Analysis in Wild Lamiophlomis rotata. Genes. 2023; 14(3):573. https://doi.org/10.3390/genes14030573
Chicago/Turabian StyleWang, Luhao, Feng Qiao, Guigong Geng, and Yueheng Lu. 2023. "Evaluation of Candidate Reference Genes for Gene Expression Analysis in Wild Lamiophlomis rotata" Genes 14, no. 3: 573. https://doi.org/10.3390/genes14030573
APA StyleWang, L., Qiao, F., Geng, G., & Lu, Y. (2023). Evaluation of Candidate Reference Genes for Gene Expression Analysis in Wild Lamiophlomis rotata. Genes, 14(3), 573. https://doi.org/10.3390/genes14030573








