Abstract
Impairments of the genes that encode enzymes that are involved in one-carbon metabolism because of the presence of gene polymorphisms can affect the methylation pattern. The altered methylation profiles of the genes involved in cardiogenesis may result in congenital heart defects (CHDs). The aim of this study was to investigate the association between the MTHFR rs1801133, MTHFR rs1801131, MTRR rs1801394, DNMT1 rs2228611, DNMT3A rs1550117, DNMT3B rs1569686, and DNMT3B rs2424913 gene polymorphisms and congenital heart defects in Down syndrome (DS) individuals. The study was conducted on 350 participants, including 134 DS individuals with CHDs (DSCHD+), 124 DS individuals without CHDs (DSCHD−), and 92 individuals with non-syndromic CHD. The genotyping was performed using the PCR–RFLP method. A statistically significant higher frequency of the DNMT3B rs2424913 TT in the DSCHD+ individuals was observed. The DNMT3B rs2424913 TT genotype, as well as the T allele, had significantly higher frequencies in the individuals with DS and atrial septal defects (ASDs) in comparison with the individuals with DS and other CHDs. Furthermore, our results indicate a statistically significant effect of the DNMT3B rs1569686 TT genotype in individuals with non-syndromic CHDs. The results of the study suggest that the DNMT3B rs2424913 TT genotypes may be a possible predisposing factor for CHDs in DS individuals, and especially those with ASDs.
1. Introduction
Congenital heart defects (CHDs) are the most common type of congenital malformations, with an incidence of 17.9/1000 [1], and they cause approximately 300,000 infant deaths per year [2]. CHDs are defined as a developmentally and clinically heterogeneous group of structural and functional congenital malformations of the heart and/or large blood vessels that result from the incomplete development of the heart during cardiac embryogenesis [3]. Chromosomal aberrations contribute to 10–12% of all CHD cases in live births [4]. In Down syndrome (DS), which is the most common chromosomal abnormality worldwide and is caused by a trisomy of human chromosome 21, CHDs are present in 40–50% of all cases [5]. The most common CHDs in individuals with DS are atrioventricular septal defects (AVSDs) (51%), ventricular septal defects (VSDs) (25%), atrial septal defects (ASDs) (9%), and tetralogy of Fallot (TF) (7%) [6]. The exact etiology that underlies CHDs is still unknown. A structurally normal heart is present in more than half of DS individuals; therefore, CHDs are not exclusively a consequence of trisomy 21 [7,8,9]. Two hypotheses have been proposed to explain DS-associated CHDs. According to the gene dosage hypothesis, CHDs could be the result of the increased expression of certain genes on chromosome 21 [10,11]. The gene mutation hypothesis states that various locus-specific mutations may be the underlying mechanism in the development of CHDs [7,11]. Today, we know that the development of CHDs requires the interaction of various genetic and epigenetic risk factors. Therefore, several risk factors have been proposed, such as locus-specific mutations, numerous single-nucleotide polymorphisms (SNPs), copy number variations (CNVs), microRNAs (miRNA), and various signaling and metabolic pathways [12,13]. One of the proposed risk factors is the carbon metabolic pathway, which includes the folate acid and homocysteine (Hcy) pathways. These metabolic pathways provide methyl groups for the cellular processes of nucleotide synthesis and methylation [14]. Methylenetetrahydrofolate reductase (MTHFR) is one of the key regulatory enzymes in the folate pathway. This enzyme catalyzes the conversion of methylenetetrahydrofolate to the main form of circulating folate, 5-methyltetrahydrofolate (5-metylTHF), which is required for the remethylation of Hcy to methionine. Methionine synthase reductase (MTRR) is another important enzyme for the conversion of the inactive form of methionine synthase to the active form, and it contributes to the transfer of the methyl group [15]. The methyl group is essential for DNA methylation and is the epigenetic mechanism involved in the regulation of many critical cellular processes during human development and throughout life, such as transcriptional regulation, genomic imprinting, chromosome maintenance, and genomic stability [16]. A number of studies suggest that aberrant DNA methylation may play an important role in many diseases [17]. The DNA methylation pattern is established and maintained by three DNA methyltransferases (DNMTs), with the addition of a methyl group to the 5′ position of the cytosine at a CpG site [18]. DNA methyltransferase 1 (DNMT1) authentically copies methylation information from parent strands to daughter strands during DNA replication. Because of this role, DNMT1 is referred to as a “maintenance” methyltransferase. Two other DNA methyltransferases, DNMT3A and DNMT3B, have de novo DNMT catalytic activities to establish methylation patterns during early embryogenesis [19]. An overview of the folate metabolic pathway is shown in the schematic in Figure 1 [20].
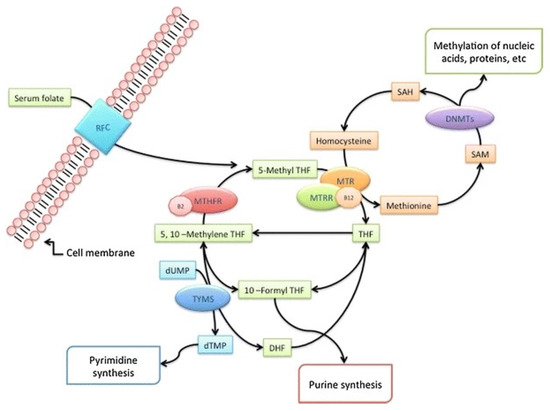
Figure 1.
Overview of the folate metabolic pathway (adapted from Coppedè 2015 [20] RightsLink No. 5490861127574). Dietary folates provide one-carbon units for DNA synthesis and methylation reactions. Enzymes: RFC1: reduced folate carrier; DNMT: DNA methyltransferases; MTR: methionine synthase; MTRR: methionine synthase reductase; MTHFR: methylenetetrahydrofolate reductase; TYMS: thymidylate synthase. Cofactors: B2: vitamin B2; B12: vitamin B12. Metabolites: SAH: S-adenosylhomocysteine; SAM: S-adenosylmethionine; 5,10-MTHF: 5,10 methylenetetrahydrofolate; THF: tetrahydrofolate; DHF: dihydrofolate; 10-formyl-THF: 10-formyl-tetrahydrofolate; 5-MTHF: 5-methyltetrahydrofolate; dUMP: deoxyuridine monophosphate; dTMP: deoxythymidine monophosphate.
The development of the heart is a complex process. DNA methylation plays a crucial role in the expression of cardiac genes during development [21]. Cardiac DNA methylation varies during life, and it is different in the embryonic stages, between neonates and adults, and between healthy individuals and patients [22]. Genetic variation caused by SNPs is the most common form of change in the gene structure, and it may influence gene expression. Literature evidence suggests that polymorphisms of the MTHFR, MTRR, and DNMTs genes could be a risk factor for CHDs; however, the results are inconsistent [23,24,25]. Specifically, the MTHFR 677C>T (rs1801133) polymorphism is a common genetic variation that affects the function of the MTHFR enzyme. The MTHFR 677TT genotype is associated with a decrease in MTHFR enzyme activity of about 70% compared with the wild-type CC genotype. The CT genotype is associated with a reduction in enzyme activity of about 40%. As a result, individuals with the TT or CT genotype may have lower levels of 5-methyl-THF and higher levels of homocysteine in their blood. Likewise, the MTHFR 1298A>C (rs1801131) polymorphism is another common variation in the MTHFR gene that affects enzyme activity. The MTHFR 1298CC genotype is associated with a reduction in MTHFR enzyme activity of about 30% compared with the wild-type AA genotype. The AC genotype is associated with a smaller reduction in enzyme activity of about 15% [26]. The most common polymorphism of the MTRR gene is MTRR 66A>G (rs1801394). Studies have shown that the MTRR 66A>G polymorphism leads to decreased MTRR enzyme activity and reduced efficiency in the remethylation of homocysteine to methionine [27]. The most studied DNMT1 SNPs are 97T>C (rs16999593), +32204A>G (rs2228611), and 327A>G (rs2228612), which exist in the coding regions and, consequently, may influence the DNMT1 expression [23,28,29]. Two common polymorphisms have been identified in the promoter region of the DNMT3B gene: −149C>T (rs2424913) and −579G>T (rs1569686). The DNMT3B −149C>T polymorphism is known to have an impact on DNMT3B gene expression [30]. The DNMT3A −448A>G (rs1550117) polymorphism is also present in the promoter region. The altered expressions of MTHFR, MTRR, and DNMTs can result in changes in DNA methylation patterns, which can have downstream effects on gene expressions relevant to heart development and function.
Because the MTHFR, MTRR, and DNMT genes are critical for maintaining DNA methylation patterns, we hypothesized that the polymorphisms in these genes might be the risk factors for CHDs in individuals with trisomy 21. Therefore, we selected the SNPs considering their influence on methylation during development and later in life, and we conducted a study to investigate the association between the MTHFR rs1801133, MTHFR rs1801131, MTRR rs1801394, DNMT1 rs2228611, DNMT3A rs1550117, DNMT3B rs1569686, and DNMT3B rs2424913 gene polymorphisms and CHDs in DS individuals and controls.
2. Materials and Methods
2.1. Patients
This study was conducted on 350 participants, including 134 DS individuals with CHDs (DSCHD+), 124 DS individuals without CHDs (DSCHD−), and 92 individuals with non-syndromic CHDs (Table 1). The study population was of the same race (Caucasian). Participants with DSCHD+ and DSCHD− were selected from the Clinical Hospital Center Rijeka in collaboration with DS associations in Croatia. Participants with non-syndromic CHDs were selected from the Clinical Hospital Center Rijeka, the Pula General Hospital, and Istrian health centers. Trisomy 21 was confirmed by karyotyping, and CHDs were confirmed by ultrasound. Written informed consent was obtained from all participants (for minors, the consent was obtained from their legal guardians). The study protocol was approved by the Institutional Review Board of the Ethics Committee for Biomedical Research of the Faculty of Medicine (ethical approval number: 003-08/19-01/114; 2170-24-09-8-19-2), the University of Rijeka, the Clinical Hospital Center Rijeka, the Pula General Hospital, and Istrian health centers. The study was conducted in accordance with the Declaration of Helsinki.

Table 1.
Characteristics of the study populations.
2.2. DNA Isolation and Genotyping
Genomic DNA was isolated from peripheral blood leukocytes and buccal epithelial cells according to a standard procedure and using a commercially available kit (Qiagen FlexiGene® DNA Kit (250), Qiagen GmbH, Hilden, Germany and QIAamp® DNA Mini Kit (50), Qiagen GmbH, Hilden, Germany), and it was stored at −20 °C. Buccal epithelial cells were collected from participants who did not consent to blood sampling. The concentration of DNA was above 50 ng/μL. The A260/A280 ranged from 1.8 to 2.0. MTHFR 677C>T (rs1801133) and MTHFR 1298A>C (rs1801131), MTRR 66A>G (rs1801394), DNMT1 +32204A/G (rs2228611), DNMT3A −448A/G (rs1550117), DNMT3B −579G/T (rs1569686), and DNMT3B −149C/T (rs2424913) were analyzed using polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) (Supplementary Table S1). To assess the reproducibility of the results, the samples were run in duplicate, and at the end, 10% of all samples were randomly repeated. DNA samples of the wild-type and mutant homozygous genotypes were used as quality control samples in each enzyme digestion reaction.
2.3. Statistical Analysis
Statistical analysis was performed with Statistica for Windows, version 13.3 (StatSoft, Inc., Tulsa, OK, USA), and with MedCalc for Windows, version 14.12.0. The normality of the distribution was tested with the Kolmogorov–Smirnov test. A variable that was not normally distributed was expressed by the median and range. Nominal indicators were represented by frequency distributions by groups and proportion. A Pearson chi-square test and Fischer exact test were used to determine the differences in genotype and allele frequencies and the deviation from the Hardy–Weinberg equilibrium between groups. Odds ratios (ORs) with 95% confidence intervals (95% CIs) were estimated according to dominant, recessive, and codominant models for the analysis of the genetic association of genotypes and alleles. Statistical significance was considered at p ≤ 0.05.
3. Results
Table 1 shows the demographic characteristics of the study populations. All the genotype frequencies were consistent with Hardy–Weinberg expectations. Statistically significant higher frequencies of DNMT3B rs2424913 CT (χ2 = 5.37; p = 0.021) in the DSCHD− and TT (χ2 = 5.63; p = 0.019) in the DSCHD+ were observed (Table 2). No statistically significant associations were found for the other analyzed gene polymorphisms (Table 2). A significant risk for CHDs according to the dominant (CC + CT vs. TT) and codominant genetic (TT vs. CT) models was observed for DNMT3B rs2424913 in the DSCHD+ and DSCHD− groups (OR = 2.24; 95% CI = 1.13–4.42; p = 0.019; OR = 0.38; 95% CI = 0.18–0.77; p = 0.008, respectively) (Table 3). Statistically significant effects of the DNMT3B rs1569686 TT genotype (χ2 = 4.37; p = 0.039) (Table 2), as well as of the dominant (GG + TG vs. TT) (OR = 2.08; 95% CI = 1.03–4.20; p = 0.039) and codominant (TT vs. TG) genetic models (OR = 0.46; 95% CI = 0.22–0.98; p = 0.046) in the individuals with non-syndromic CHDs was observed (Table 3). The DNMT3B rs2424913 TT genotype and T allele had significantly higher frequencies in the DS individuals with ASDs compared with the DS individuals with other CHDs (χ2 = 4.97; p = 0.028; χ2 = 5.69; p = 0.018, respectively) (Table 4). The most frequent congenital heart defects in the DS and non-syndromic individuals were ASDs, followed by VSDs and AVSDs, with no difference in the sex distribution.

Table 2.
Genotype and allele frequencies of DNA methyltransferases (DNMTs), methylenetetrahydrofolate reductase (MTHFR), and 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR) gene polymorphisms in study groups.

Table 3.
Risk for congenital heart defects according to different genetic models in study groups.

Table 4.
Genotype and allele frequencies of DNMT3B rs2424913 gene polymorphism in Down syndrome individuals with atrial septal defects (ASDs) and other congenital heart defects (CHDs).
4. Discussion
To the best of our knowledge, this is the first study to assess the association between MTHFR, MTRR, and DNMT gene polymorphisms and CHDs in individuals with DS and individuals with non-syndromic CHDs. The results of our study indicate that the DNMT3B rs2424913 TT genotype and dominant and codominant genetic models of the same polymorphism may be potential predisposing factors for CHDs in individuals with DS, and particularly in individuals with ASDs. Furthermore, our results suggest that the DNMT3B rs1569686 TT genotype increased the CHD risk in the individuals with non-syndromic CHDs compared with the DS group with CHDs, which was also found under the dominant and codominant genetic models of the same polymorphism.
In the presence of the DNMT3B rs2424913 polymorphism, the expression of the DNMT3B gene is changed. The minor T allele increases the promoter activity by 30% [31,32] and affects the miRNA binding site [33]. Considering this, we propose that the increased DNMT gene expression due to the presence of the T allele of the rs2424913 SNP alters the DNA methylation of the relevant genes in cardiogenesis and potentially leads to CHDs. Moreover, this gene may be an additional risk factor for the altered methylation profiles of the genes on chromosome 21 (DSCAM, COL6A1, COL6A2, KCNJ6, RCAN1) related to the endocardial cushion type in DSCHD+, as suggested in a recent review [12,13]. In our study, we also found a significant risk of the DNMT3B rs1569686 TT genotype and dominant and codominant genetic models of the same polymorphism for CHDs in individuals with non-syndromic CHDs. The functional role of this polymorphism has not yet been clearly defined. Altered gene splicing, linkage disequilibrium with other polymorphisms, and the deactivation of illegitimate transcription factor binding are possible mechanisms by which rs1569686 polymorphisms affect gene expression [31,33,34].
Moreover, animal experiments have shown that one of the most active enzymes is Dnmt3b, which is strongly expressed in the early embryonic stages in DNA methylation reprogramming during cardiomyocyte differentiation [35,36,37]. The knockdown of Dnmt3b in mice leads to death due to ventricular defects [37]. Moreover, Chamberlain et al. found that Dnmt3b regulates the DNA methylation of the cardiac essential genes and, in combination with Dnmt3a, causes the repression of fetal cardiac genes [38]. Furthermore, a study by Zhang et al. has shown that approximately 60% of the RE1 silencing transcription factor target genes (REST target genes) in cardiac embryogenesis are at least partially controlled by Dnmt3b [39]. Interestingly, according to the methylation quantitative trait loci (mQTL) database, DNMT3B rs2424913 and rs1569686 are also the mQTLs that are active throughout the lifespan, and they are located in close proximity to multiple DNA methylation sites [40]. Although we did not find a significant association between DNMT3A rs1550117 and CHDs in syndromic or non-syndromic individuals, further studies are needed to clarify its role. Indeed, other studies have demonstrated correlations between DNMT3A and the tetralogy of Fallot [30] and cardiac fibrosis [41]. Moreover, previous Dnmt3a knockdown studies in animals revealed that Dnmt3a plays an important role in heart development by regulating embryonic cardiomyocyte gene expression, morphology, and function [36]. In contrast to the de novo catalytic activity of DNMT3A and DNMT3B, DNMT1 is responsible for maintaining methylation patterns [19]. Several previous studies have shown that DNMT1 polymorphisms (rs16999593, rs2228612) are associated with the risk of developing CHDs [23], and that the decreased expression of DNMT1 may play an important role in the pathogenesis of tetralogy of Fallot [30]. Researchers have also suggested that DNMT1 rs16999593 reduces the risk of the transposition of the great arteries [29]. Studies in animal models indicate that Dnmt1 is involved in embryonic cardiomyocytes by regulating DNA methylation, gene expression, gene splicing, and the cell’s function [42].
Although relationships between DNMT1 rs2228611 and CHDs were not found in our study, DNMT1 is still a good candidate for future investigation, considering that it plays an important role during cardiomyocyte development.
For the MTHFR rs1801131 and rs1801133 and MTRR rs1801394 polymorphisms, we did not find an association with the CHD risk in the case and control groups. Conflicting results have been shown in previous studies in which the associations of these polymorphisms and CHDs in the general population were investigated, as well as in the population with DS [24,25,27,43,44,45,46,47,48,49]. However, given the known associations of these polymorphisms with decreased enzyme activity, which may consequently lead to an increase in homocysteine levels and thus affect the methylation pattern [22], they still represent good candidates for the development of CHDs and should not be omitted in future studies. The potential limitations of our study include the small sample size and, especially, the subgroup analysis of specific defects. Furthermore, additional research should be focused on the possible influence of gene–gene and gene–environment interactions, considering the multiple etiologies of CHDs.
5. Conclusions
We examined the association between the MTHFR, MTRR, and DNMTs gene polymorphisms and CHDs in syndromic and non-syndromic individuals. We found that the DNMT3B rs2424913 TT genotypes and the genetic model CC + CT vs. TT could be predisposing factors for CHDs in individuals with Down syndrome and particularly those with ASDs. In addition, our results indicate that the DNMT3B rs1569686 TT genotypes increased the risk of CHDs in individuals with non-syndromic CHDs. Accordingly, these findings are noteworthy because they reveal new aspects of CHD etiology. Due to the small sample size of the research, further studies with larger cohorts focused on specific types of CHDs are needed to define the functional roles of the MTHFR, MTRR, and DNMTs gene polymorphisms in CHD etiology.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14030576/s1, Table S1: Primers and PCR-RFLP conditions for the genotyping of DNMT, MTHFR and MTRR genes polymorphisms with their mQTL timepoint [50,51,52,53,54,55].
Author Contributions
Conceptualization, D.M., A.B. and J.V.; methodology, D.M., A.B. and J.V.; validation, D.M., A.B., I.B.B., M.Š., I.B.Č., N.Č. and J.V.; formal analysis, D.M., A.B. and J.V.; investigation, D.M., A.B. and J.V.; resources, J.V. and M.Š.; data curation, D.M., A.B. and J.V.; writing—original draft preparation, D.M., A.B. and J.V.; writing—review and editing, D.M., A.B. and J.V.; visualization, D.M., A.B. and J.V.; supervision, I.B.B., M.Š., I.B.Č. and N.Č.; project administration, J.V.; funding acquisition, J.V. and M.Š. All authors have read and agreed to the published version of the manuscript.
Funding
The research reported in this publication was supported by research grant number (918.10.0230/uniri.-biomed 18-120) (funding: University of Rijeka, Croatia) and the institutional research fund of Juraj Dobrila University of Pula, Croatia.
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board of the Ethics Committee for Biomedical Research of the Faculty of Medicine, the University of Rijeka, the Clinical Hospital Center Rijeka, and it was performed in accordance with the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the participants to publish this paper.Ethical approval number: 003-08/19-01/114; 2170-24-09-8-19-2.
Data Availability Statement
All data from this research are available from the corresponding author upon reasonable request.
Acknowledgments
The authors thank all the participants in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wu, W.; He, J.; Shao, X. Incidence and mortality trend of congenital heart disease at the global, regional, and national level, 1990–2017. Medicine 2020, 99, e20593. [Google Scholar] [CrossRef]
- GBD 2017 Congenital Heart Disease Collaborators. Global, regional, and national burden of congenital heart disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Child Adolesc. Health 2020, 4, 185–200. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Zühlke, L.; Black, G.C.; Choy, M.K.; Li, N.; Keavney, B.D. Global birth prevalence of congenital heart defects 1970–2017: Updated systematic review and meta-analysis of 260 studies. Int. J. Epidemiol. 2019, 48, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Lalani, S.R.; Belmont, J.W. Genetic basis of congenital cardiovascular malformations. Eur. J. Med. Genet. 2014, 57, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Landis, B.J.; Cooper, D.S.; Hinton, R.B. CHD associated with syndromic diagnoses: Peri-Operative risk factors and early outcomes. Cardiol. Young 2016, 26, 30–52. [Google Scholar] [CrossRef] [PubMed]
- Pfitzer, C.; Helm, P.C.; Rosenthal, L.M.; Berger, F.; Bauer, U.M.M.; Schmitt, K.R. Dynamics in prevalence of Down syndrome in children with congenital heart disease. Eur. J. Pediatr. 2018, 177, 107–115. [Google Scholar] [CrossRef]
- Ackerman, C.; Locke, A.E.; Feingold, E.; Reshey, B.; Espana, K.; Thusberg, J.; Mooney, S.; Bean, L.J.; Dooley, K.J.; Cua, C.L.; et al. An excess of deleterious variants in VEGF-A pathway genes in Down-syndrome-associated atrioventricular septal defects. Am. J. Hum. Genet. 2012, 91, 646–659. [Google Scholar] [CrossRef]
- Benhaourech, S.; Drighil, A.; Hammiri, A.E. Congenital heart disease and Down syndrome: Various aspects of a confirmed association. Cardiovasc. J. Afr. 2016, 27, 287–290. [Google Scholar] [CrossRef]
- Zaidi, S.; Brueckner, M. Genetics and Genomics of Congenital Heart Disease. Circ. Res. 2017, 120, 923–940. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Lyle, R.; Dermitzakis, E.T.; Reymond, A.; Deutsch, S. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nat. Rev. Genet. 2004, 5, 725–738. [Google Scholar] [CrossRef]
- Letourneau, A.; Santoni, F.A.; Bonilla, X.; Sailani, M.R.; Gonzalez, D.; Kind, J.; Chevalier, C.; Thurman, R.; Sandstrom, R.S.; Hibaoui, Y.; et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature 2014, 508, 345–350. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Tian, J. Molecular mechanisms of congenital heart disease in down syndrome. Genes Dis. 2019, 6, 372–377. [Google Scholar] [CrossRef]
- Asim, A.; Agarwal, S. Congenital heart defects among Down’s syndrome cases: An updated review from basic research to an emerging diagnostics technology and genetic counselling. J. Genet. 2021, 100, 45. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F. The genetics of folate metabolism and maternal risk of birth of a child with Down syndrome and associated congenital heart defects. Front. Genet. 2015, 6, 223. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F.; Stoccoro, A.; Tannorella, P.; Gallo, R.; Nicolì, V.; Migliore, L. Association of Polymorphisms in Genes Involved in One-Carbon Metabolism with MTHFR Methylation Levels. Int. J. Mol. Sci. 2019, 20, 3754. [Google Scholar] [CrossRef] [PubMed]
- Villicaña, S.; Bell, J.T. Genetic impacts on DNA methylation: Research findings and future perspectives. Genome Biol. 2021, 22, 127. [Google Scholar] [CrossRef]
- Jin, Z.; Liu, Y. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Moore, L.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Wang, H.; Lou, D.; Wang, Z. Crosstalk of Genetic Variants, Allele-Specific DNA Methylation, and Environmental Factors for Complex Disease Risk. Front. Genet. 2019, 9, 695. [Google Scholar] [CrossRef]
- Coppedè, F. Risk factors for Down syndrome. Arch. Toxicol. 2016, 90, 2917–2929. [Google Scholar] [CrossRef]
- Chowdhury, S.; Cleves, M.A.; MacLeod, S.L.; James, S.J.; Zhao, W.; Hobbs, C.A. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wu, Q.; Huang, Y.; Wang, L.; Su, Z.; Ye, H. The role of DNA methylation in syndromic and non-syndromic congenital heart disease. Clin. Epigenet. 2021, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhou, S.; Wang, Y.; Wang, L.; Zhou, J.; Wang, H.; Li, C.; Chang, M. Association of DNMT1 Gene Polymorphisms with Congenital Heart Disease in Child Patients. Pediatr. Cardiol. 2015, 36, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Wang, W.; Jiang, X. The roles of MTRR and MTHFR gene polymorphisms in congenital heart diseases: A me-ta-analysis. Biosci. Rep. 2018, 38, BSR20181160. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhuang, Z.; Wen, Z.; Zang, X.; Mo, X. MTHFR A1298C polymorphisms reduce the risk of congenital heart defects: A meta-analysis from 16 case-control studies. Ital. J. Pediatr. 2017, 43, 108. [Google Scholar] [CrossRef] [PubMed]
- Castiglia, P.; Sanna, V.; Azara, A.; De Miglio, M.R.; Murgia, L.; Pira, G.; Sanges, F.; Fancellu, A.; Carru, C.; Bisail, M.; et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms in breast cancer: A Sardinian preliminary case-control study. Int. J. Med. Sci. 2019, 16, 1089–1095. [Google Scholar] [CrossRef]
- Cai, B.; Zhang, T.; Zhong, R.; Zou, L.; Zhu, B.; Chen, W.; Shen, N.; Ke, J.; Lou, J.; Wang, Z.; et al. Genetic variant in MTRR, but not MTR, is associated with risk of congenital heart disease: An integrated meta-analysis. PLoS ONE 2014, 9, e89609. [Google Scholar] [CrossRef]
- Sheng, W.; Qian, Y.; Wang, H.; Ma, X.; Zhang, P.; Chen, L.; Ma, D.; Huang, G. Association between mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2 and LINE-1 methylation status in infants with tetralogy of Fallot. Int. J. Mol. Med. 2013, 32, 694–702. [Google Scholar] [CrossRef]
- Lei, L.; Lin, H.; Zhong, S.; Zhang, Z.; Chen, J.; Yu, X.; Liu, X.; Zhang, C.; Nie, Z.; Zhuang, J. DNA methyltransferase 1 rs16999593 genetic polymorphism decreases risk in patients with transposition of great arteries. Gene 2017, 615, 50–56. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, H.; Tang, Y.; Liu, P.; Wang, J. DNMT3B polymorphisms and cancer risk: A meta analysis of 24 case-control studies. Mol. Biol. Rep. 2012, 39, 4429–4437. [Google Scholar] [CrossRef]
- Shen, H.; Wang, L.; Spitz, M.R.; Hong, W.K.; Mao, L.; Wei, Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002, 62, 4992–4995. [Google Scholar]
- Xiao, Y.; Word, B.; Hammons, G.; Lyn-Cook, B. Transcriptional activity of DNMT3B in pancreatic cancer cells: Effects of −149 (C→T) promoter polymorphism. Biochem. Biophys. Res. Commun. 2011, 415, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Saradalekshmi, K.R.; Neetha, N.V.; Sathyan, S.; Nair, I.V.; Nair, C.M.; Banerjee, M. DNA methyl transferase (DNMT) gene polymorphisms could be a primary event in epigenetic susceptibility to schizophrenia. PLoS ONE 2014, 9, e98182. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F.; Boco, P.; Tannorella, P.; Romano, C.; Antonucci, I.; Stuppia, L.; Romano, C.; Migliore, L. DNMT3B promoter polymorphisms and maternal risk of birth of a child with Down syndrome. Hum. Reprod. 2013, 28, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, A.A.; Lin, M.; Lister, R.L.; Maslov, A.A.; Wang, Y.; Suzuki, M.; Wu, B.; Greally, J.M.; Zheng, D.; Zhou, B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 2014, 3, e000976. [Google Scholar] [CrossRef]
- Fang, X.; Poulsen, R.R.; Wang-Hu, J.; Shi, O.; Calvo, N.S.; Simmons, C.S.; Rivkees, S.A.; Wendler, C.C. Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. FASEB J. 2016, 30, 3238–3255. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Gilsbach, R.; Preissl, S.; Grüning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Würch, A.; Bönisch, U.; Günther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, B.; Wang, P.; Wang, Y.; Lu, P.; Nechiporuk, T.; Floss, T.; Greally, J.M.; Zheng, D.; Zhou, B. Non-CpG methylation by DNMT3B facilitates REST binding and gene silencing in developing mouse hearts. Nucleic Acids Res. 2017, 45, 3102–3115. [Google Scholar] [CrossRef]
- Gaunt, T.R.; Shihab, H.A.; Hemani, G.; Min, J.L.; Woodward, G.; Lyttleton, O.; Zheng, J.; Duggirala, A.; McArdle, W.L.; Ho, K.; et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016, 17, 61. [Google Scholar] [CrossRef]
- Tao, H.; Yang, J.J.; Chen, Z.W.; Xu, S.S.; Zhou, X.; Zhan, H.; Shi, K.H. DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology 2014, 323, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Poulsen, R.; Zhao, L.; Wang, J.; Rivkees, S.A.; Wendler, C.C. Knockdown of DNA methyltransferase 1 reduces DNA methylation and alters expression patterns of cardiac genes in embryonic cardiomyocytes. FEBS Open Bio. 2021, 11, 2364–2382. [Google Scholar] [CrossRef] [PubMed]
- Junker, R.; Kotthoff, S.; Vielhaber, H.; Halimeh, S.; Kosch, A.; Koch, H.G.; Kassenböhmer, R.; Heineking, B.; Nowak-Göttl, U. Infant methylenetetrahydrofolate reductase 677TT genotype is a risk factor for congenital heart disease. Cardiovasc. Res. 2001, 51, 251–254. [Google Scholar] [CrossRef] [PubMed]
- van Beynum, I.M.; den Heijer, M.; Blom, H.J.; Kapusta, L. The MTHFR 677C->T polymorphism and the risk of congenital heart defects: A literature review and meta-analysis. QJM 2007, 100, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.N.; Wang, H.D.; Tie, L.Z.; Li, T.; Xiao, H.; Long, J.G.; Liao, S.X. Parental Genetic Variants, MTHFR 677C>T and MTRR 66A>G, Associated Differently with Fetal Congenital Heart Defect. Biomed. Res. Int. 2017, 2017, 3043476. [Google Scholar] [CrossRef]
- Asim, A.; Agarwal, S.; Panigrahi, I.; Saiyed, N.; Bakshi, S. MTHFR promoter hypermethylation may lead to congenital heart defects in Down syndrome. Intractable Rare Dis. Res. 2017, 6, 295–298. [Google Scholar] [CrossRef]
- Božović, I.B.; Vraneković, J.; Cizmarević, N.S.; Mahulja-Stamenković, V.; Prpić, I.; Brajenović-Milić, B. MTHFR C677T and A1298C polymorphisms as a risk factor for congenital heart defects in Down syndrome. Pediatr. Int. 2011, 53, 546–550. [Google Scholar] [CrossRef]
- Vraneković, J.; Slivšek, G.; Majstorović, D. Methyltetrahydrofolate-homocysteine methyltransferase reductase gene and congenital heart defects in Down syndrome. Genet. Appl. 2020, 41, 12–17. [Google Scholar] [CrossRef]
- Ganguly, A.; Halder, P.; Pal, U.; Sarkar, S.; Datta, S.; Pati, S.; Ghosh, S. Risk of Atrioventricular Septal Defects in Down syndrome: Association of MTHFR C677T and RFC1 A80G polymorphisms in Indian Bengali cohort. J. Hum. Genet. Genom. 2021, 5, 1–12. [Google Scholar] [CrossRef]
- Khatami, F.; Noorinayer, B.; Ghiasi, S.; Mohebi, R.; Hashemi, M.; Zali, M.R. Lack of Effects of Single Nucleotide Polymorphisms of the DNA Methyltransferase 1 Gene on Gastric Cancer in Iranian Patients: A Case Control Study. Asian Pac. J. Cancer Prev. 2009, 10, 1177–1182. [Google Scholar]
- Fan, H.; Liu, D.; Qiu, X.; Qiao, F.; Wu, Q.; Su, X.; Zhang, F.; Song, Y.; Zhao, Z.; Xie, W. A functional polymorphism in the DNA methyltransferase-3A promoter modifies the susceptibility in gastric cancer but not in esophageal carcinoma. BMC Med. 2010, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhang, F.; Hu, J.; Liu, D.; Zhao, Z. Promoter polymorphisms of DNMT3B and the risk of colorectal cancer in Chinese: A case-control study. J. Exp. Clin. Cancer Res. CR 2008, 27, 24. [Google Scholar] [CrossRef] [PubMed]
- Mostowska, A.; Sajdak, S.; Pawlik, P.; Lianeri, M.; Jagodzinski, P.P. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol. Biol. Rep. 2013, 40, 4893–4899. [Google Scholar] [CrossRef] [PubMed]
- Coppedè, F.; Marini, G.; Bargagna, S.; Stuppia, L.; Minichilli, F.; Fontana, I.; Colognato, R.; Astrea, G.; Palka, G.; Migliore, L. Folate gene polymorphisms and the risk of Down syndrome pregnancies in young Italian women. Am. J. Med. Genet. Part A 2006, 140, 1083–1091. [Google Scholar] [CrossRef]
- Jacques, P.F.; Bostom, A.G.; Selhub, J.; Rich, S.; Ellison, R.C.; Eckfeldt, J.H.; Gravel, R.A.; Rozen, R.; National Heart, Lung and Blood Institute; National Institutes of Health. Effects of polymorphisms of methionine synthase and methionine synthase reductase on total plasma homocysteine in the NHLBI Family Heart Study. Atherosclerosis 2003, 66, 49–55. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).