CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. sgRNA Design and CRISPR/Cas9 Construct Generation
2.3. Protoplast Transfection and Genomic DNA Isolation of CRISPR/Cas9 Constructs
2.4. Agrobacterium-Infiltration in Tomato Fruit and Leaves
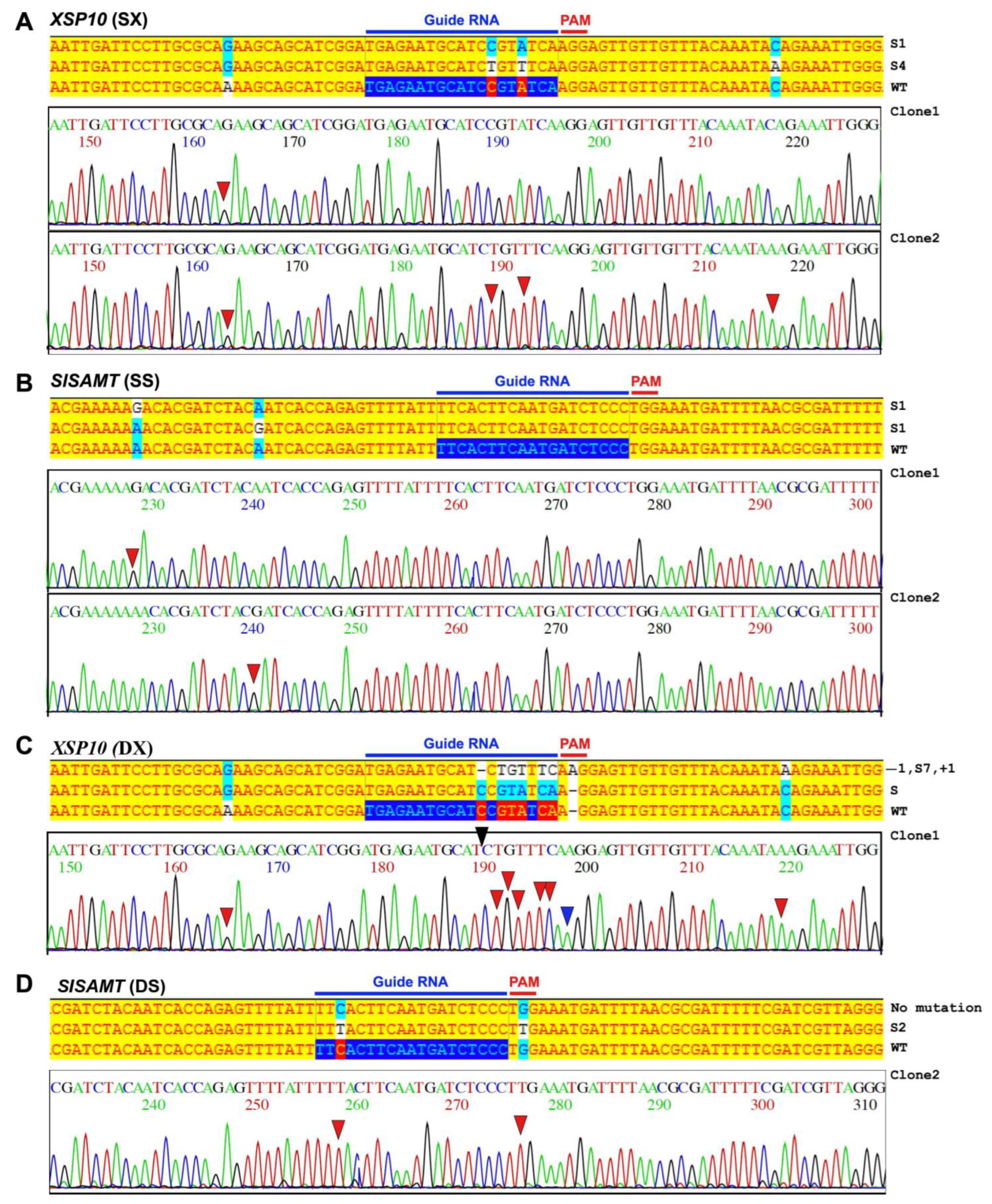
2.5. Molecular Analysis of CRISPR-Editing Events in Protoplast and Leaves
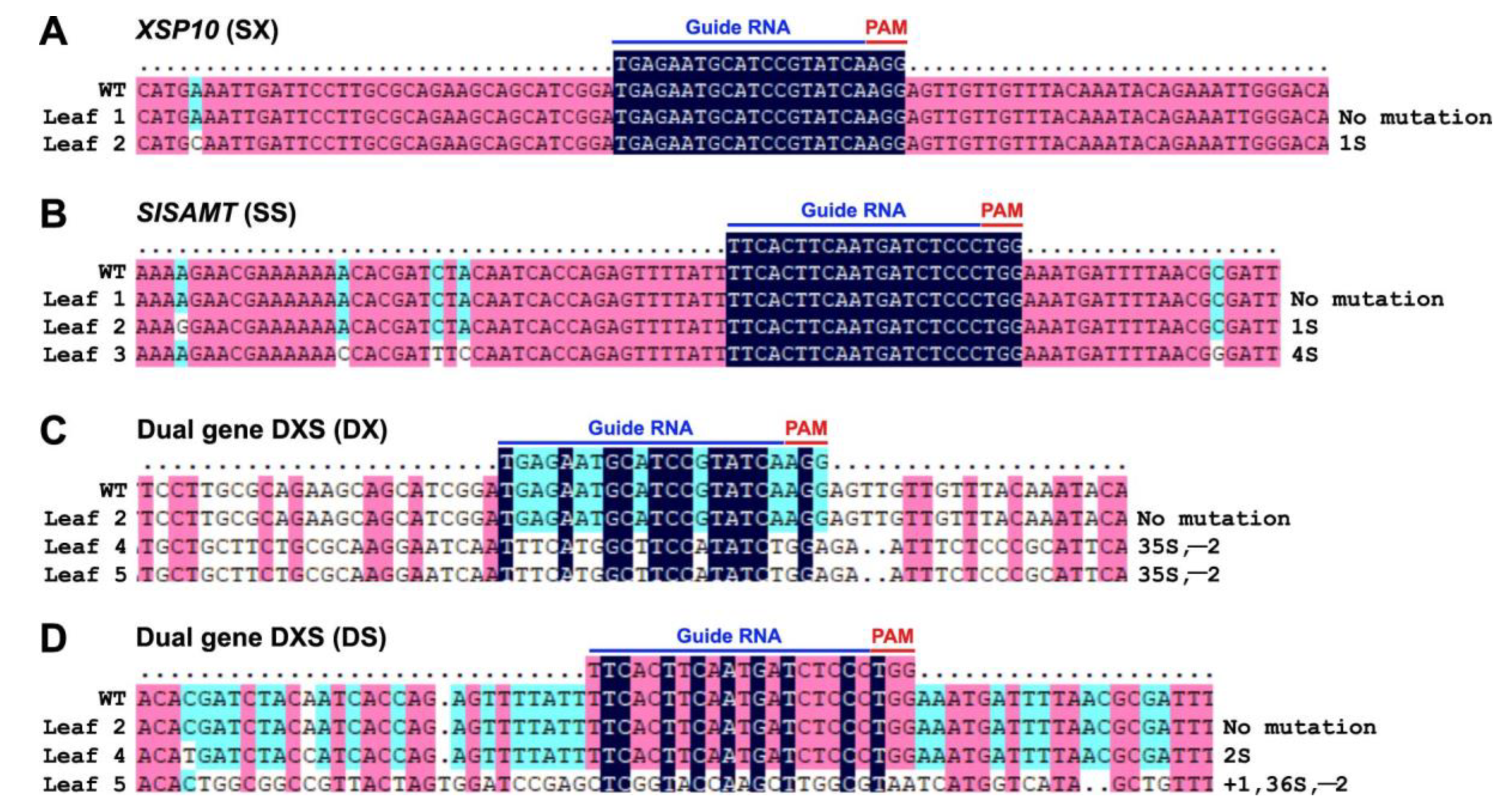
2.6. Stable Agrobacterium-Mediated Transformation of Tomato with the Binary CRISPR/Cas9 Constructs
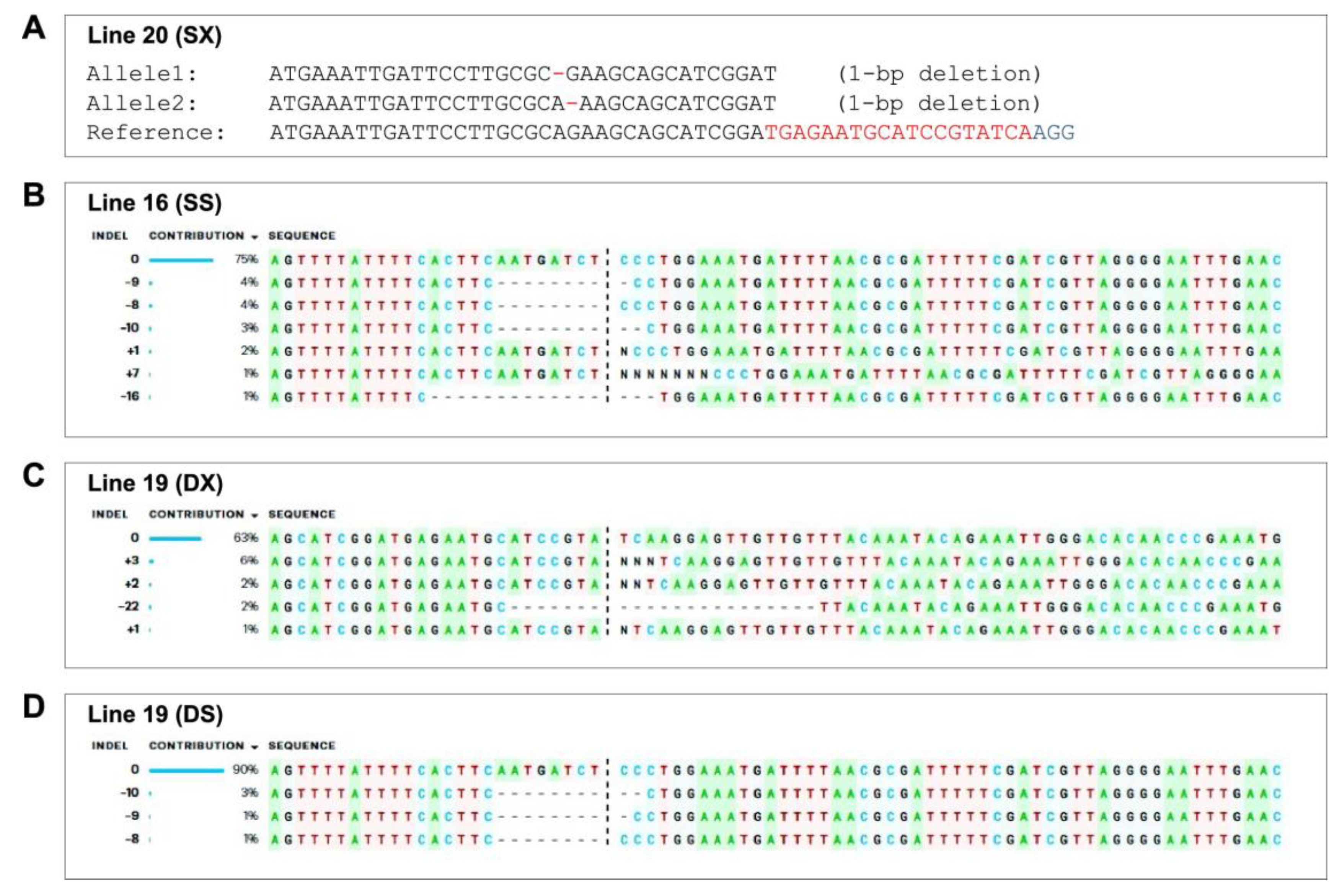
2.7. Genetic Analysis of CRISPR/Cas9 Editing Events and Off-Targets
2.8. Histochemical GUS Staining
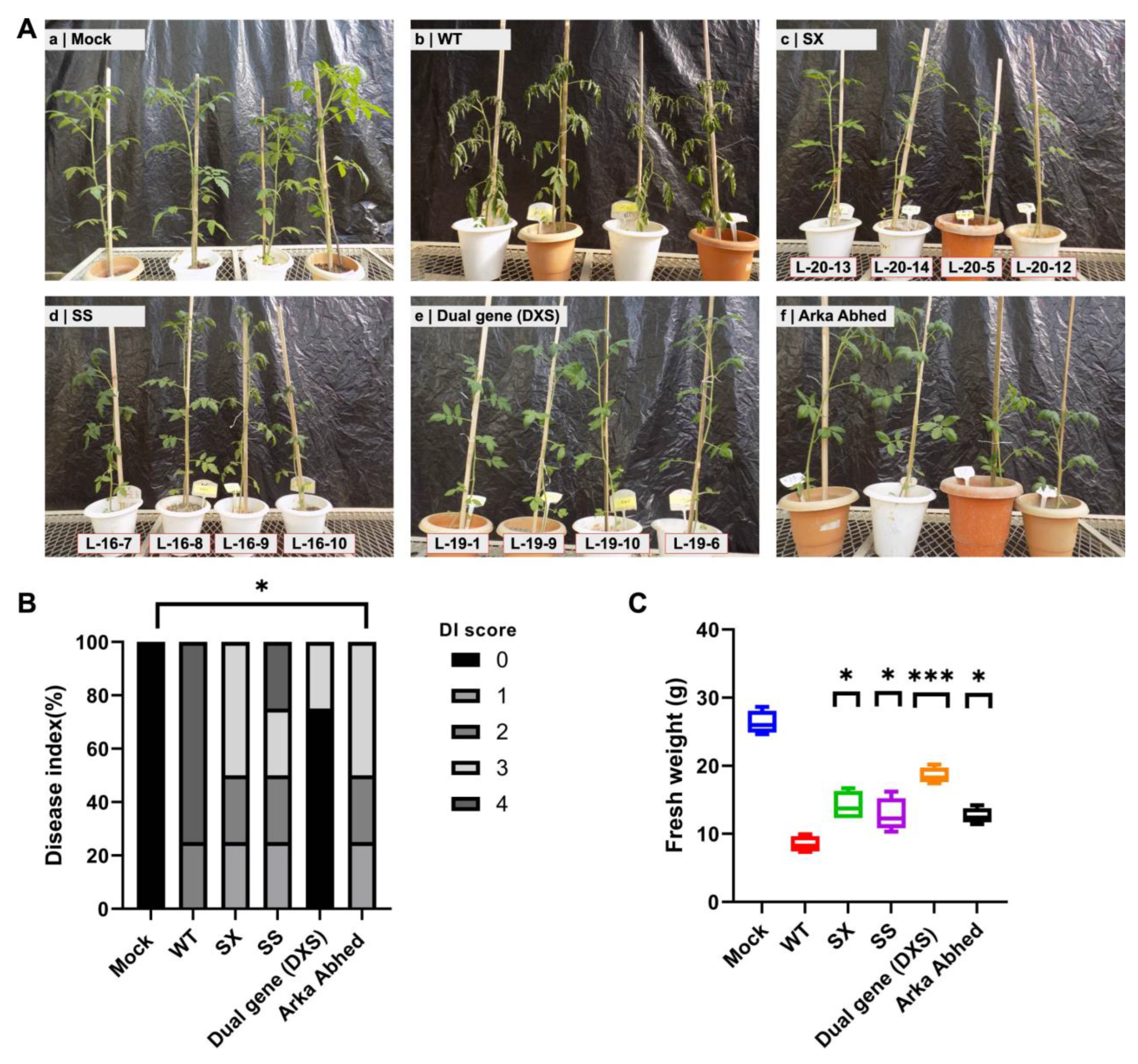
2.9. Pathogen (Fusarium) Leaf Detached and Wilt Assay
2.10. Microscopy and Fungal Outgrowth Assay
2.11. Histochemical Cell Death Staining and Quantification
2.12. Detection of Hydrogen Peroxide (H2O2)
2.13. Statistical Analysis
3. Results
3.1. Pre-Screening through Transient Analysis of the XSP10 and SlSAMT Single and Dual-Gene CRISPR/Cas9 Editing
3.2. Generation of CRISPR-Edited Lines of GE0 and GE1 in S. lycopersicum cv. AV
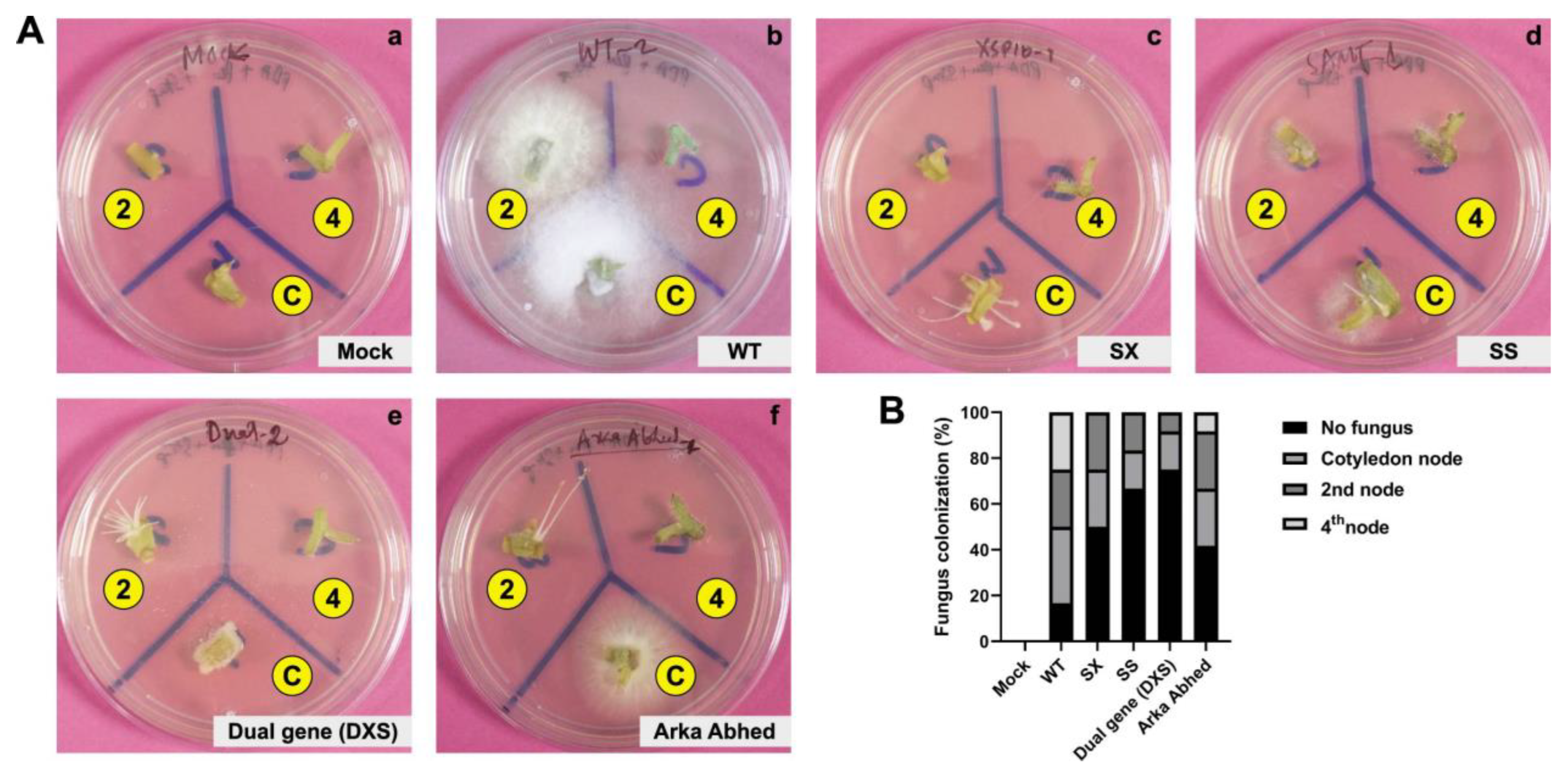
3.3. Phenotypic Evaluation of Single and Dual-Gene Editing of XSP10 and SlSAMT upon Fol Infection in S. lycopersicum cv. AV
3.4. Analysis of Potential Off-Targets of the XSP10 and SlSAMT Genes of S. lycopersicum L.
4. Discussion
4.1. Validation of CRISPR/Cas9 Editing at the Transient Level Showed Fol 1322 Tolerance in S. lycopersicum L.
4.2. CRISPR/Cas9 Induced Stable Gene Editing and Heritability in GE1 Generation
4.3. CRISPR-Edited Lines (CRELs) of XSP10 and SlSAMT Reduce the Susceptibility of Tomato to Fusarium Wilt
5. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- de Lamo, F.J.; Constantin, M.E.; Fresno, D.H.; Boeren, S.; Rep, M.; Takken, F.L.W. Xylem Sap Proteomics Reveals Distinct Differences Between R Gene- and Endophyte-Mediated Resistance Against Fusarium Wilt Disease in Tomato. Front. Microbiol. 2018, 9, 2977. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Sinha, N. Tomato (Solanum lycopersicum): A Model Fruit-Bearing Crop. Cold Spring Harb. Protoc. 2008, 2008, pdb.emo105. [Google Scholar] [CrossRef] [PubMed]
- Nirmaladevi, D.; Venkataramana, M.; Srivastava, R.K.; Uppalapati, S.R.; Gupta, V.K.; Yli-Mattila, T.; Tsui, K.M.C.; Srinivas, C.; Niranjana, S.R.; Chandra, N.S. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Sci. Rep. 2016, 6, 21367. [Google Scholar] [CrossRef] [PubMed]
- van Schie, C.C.; Takken, F.L. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; De Boer, A.D.; Houterman, P.M.; De Koster, C.G.; Cornelissen, B.J.C. A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Lett. 2003, 534, 82–86. [Google Scholar] [CrossRef]
- Krasikov, V.; Dekker, H.L.; Rep, M.; Takken, F.L. The tomato xylem sap protein XSP10 is required for full susceptibility to Fusarium wilt disease. J. Exp. Bot. 2011, 62, 963–973. [Google Scholar] [CrossRef]
- Blein, J.-P.; Coutos-Thévenot, P.; Marion, D.; Ponchet, M. From elicitins to lipid-transfer proteins: A new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 2002, 7, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chao, M.; Wang, S.; Bu, J.; Tang, J.; Li, F.; Wang, Q.; Zhang, B. Proteome quantification of cotton xylem sap suggests the mechanisms of potassium-deficiency-induced changes in plant resistance to environmental stresses. Sci. Rep. 2016, 6, 21060. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Ross, J.R.; Koscheski, P.; Yang, Y.; Pichersky, E.; Noel, J.P. Structural Basis for Substrate Recognition in the Salicylic Acid Carboxyl Methyltransferase Family. Plant Cell 2003, 15, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Ament, K.; Krasikov, V.; Allmann, S.; Rep, M.; Takken, F.L.; Schuurink, R.C. Methyl salicylate production in tomato affects biotic interactions. Plant J. 2010, 62, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-P.; von Dahl, C.C.; Park, S.-W.; Klessig, D.F. Interconnection between Methyl Salicylate and Lipid-Based Long-Distance Signaling during the Development of Systemic Acquired Resistance in Arabidopsis and Tobacco. Plant Physiol. 2011, 155, 1762–1768. [Google Scholar] [CrossRef][Green Version]
- Koo, Y.J.; Kim, M.A.; Kim, E.H.; Song, J.T.; Jung, C.; Moon, J.-K.; Kim, J.-H.; Seo, H.S.; Song, S.I.; Kim, J.-K.; et al. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol. Biol. 2007, 64, 1–15. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Van Esse, H.P.; Reuber, T.L.; Van Der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V.; Sorek, R.; Lawrence, C.; Wiedenheft, B.; Jinek, M.; Chylinski, K.; Fonfara, I.; et al. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef]
- Zaidi, S.S.-E.; Mukhtar, M.S.; Mansoor, S. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Z.; Li, J.-F. Targeted Gene Manipulation in Plants Using the CRISPR/Cas Technology. J. Genet. Genom. 2016, 43, 251–262. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, F.; Li, X.; Baller, J.A.; Qi, Y.; Starker, C.; Bogdanove, A.J.; Voytas, D.F. Transcription Activator-Like Effector Nucleases Enable Efficient Plant Genome Engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-S.; Hsu, C.-T.; Yang, L.-H.; Lee, L.-Y.; Fu, J.-Y.; Cheng, Q.-W.; Wu, F.-H.; Hsiao, H.C.-W.; Zhang, Y.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Debbarma, J.; Saikia, B.; Singha, D.L.; Maharana, J.; Velmuruagan, N.; Dekaboruah, H.; Arunkumar, K.P.; Chikkaputtaiah, C. XSP10 and SlSAMT, Fusarium wilt disease responsive genes of tomato (Solanum lycopersicum L.) express tissue specifically and interact with each other at cytoplasm in vivo. Physiol. Mol. Biol. Plants 2021, 27, 1559–1575. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Wang, J.-W.; Wang, A.; Li, K.; Wang, B.; Jin, S.; Reiser, M.; Lockey, R.F. CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning. Biotechniques 2015, 58, 161–170. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Natarajan, V.P.; Zhang, X.; Morono, Y.; Inagaki, F.; Wang, F. A Modified SDS-Based DNA Extraction Method for High Quality Environmental DNA from Seafloor Environments. Front. Microbiol. 2016, 7, 986. [Google Scholar] [CrossRef]
- Nagel, R.; Elliott, A.; Masel, A.; Birch, R.G.; Manners, J.M. Electroporation of binary Ti plasmid vector into Agrobacterium tumefaciens and Agrobacterium rhizogenes. FEMS Microbiol. Lett. 1990, 67, 325–328. [Google Scholar] [CrossRef]
- Hoshikawa, K.; Fujita, S.; Renhu, N.; Ezura, K.; Yamamoto, T.; Nonaka, S.; Ezura, H.; Miura, K. Efficient transient protein expression in tomato cultivars and wild species using agroinfiltration-mediated high expression system. Plant Cell Rep. 2019, 38, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Fister, A.S.; Shi, Z.; Zhang, Y.; Helliwell, E.E.; Maximova, S.N.; Guiltinan, M.J. Protocol: Transient expression system for functional genomics in the tropical tree Theobroma cacao L. Plant Methods 2016, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Manamohan, M.; Prakash, M.; Chandra, G.S.; Asokan, R.; Nagesha, S. An improved protocol for rapid and efficient Agrobacterium mediated transformation of tomato (Solanum lycopersicum L.). J. Appl. Hortic. 2011, 13, 3–7. [Google Scholar] [CrossRef]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.-G. DSDecode: A Web-Based Tool for Decoding of Sequencing Chromatograms for Genotyping of Targeted Mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chen, L.; Zhu, Q.; Chen, Y.; Liu, Y.-G. Rapid Decoding of Sequence-Specific Nuclease-Induced Heterozygous and Biallelic Mutations by Direct Sequencing of PCR Products. Mol. Plant 2015, 8, 1285–1287. [Google Scholar] [CrossRef]
- Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Pino, L.E.; Lombardi-Crestana, S.; Azevedo, M.S.; Scotton, D.C.; Borgo, L.; Quecini, V.; Figueira, A.; Peres, L.E. The Rg1 allele as a valuable tool for genetic transformation of the tomato ‘Micro-Tom’ model system. Plant Methods 2010, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Ezura, K.; Lee, J.; Ariizumi, T.; Ezura, H. Genetic engineering of parthenocarpic tomato plants using transient SlIAA9 knockdown by novel tissue-specific promoters. Sci. Rep. 2019, 9, 18871. [Google Scholar] [CrossRef]
- Kostov, K.; Christova, P.; Slavov, S.; Batchvarova, R. Constitutive Expression of a Radish Defensin Gene Rs-AFP2 in Tomato Increases the Resisstance to Fungal Pathogens. Biotechnol. Biotechnol. Equip. 2009, 23, 1121–1125. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W.; Instrumentation, C. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Mes, J.J.; Weststeijn, E.A.; Herlaar, F.; Lambalk, J.J.M.; Wijbrandi, J.; Haring, M.A.; Cornelissen, B.J.C. Biological and Molecular Characterization of Fusarium oxysporum f. sp. lycopersici Divides Race 1 Isolates into Separate Virulence Groups. Phytopathology 1999, 89, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, H.C.; Constantin, M.E.; Houterman, P.M.; Takken, F.L.W.; Cornelissen, B.J.C.; Haring, M.A.; Burg, H.A.V.D.; Rep, M. Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol. 2019, 154, 55–65. [Google Scholar] [CrossRef]
- Prihatna, C.; Barbetti, M.J.; Barker, S.J. A Novel Tomato Fusarium Wilt Tolerance Gene. Front. Microbiol. 2018, 9, 1226. [Google Scholar] [CrossRef] [PubMed]
- Constantin, M.E.; de Lamo, F.J.; Vlieger, B.V.; Rep, M.; Takken, F.L.W. Endophyte-Mediated Resistance in Tomato to Fusarium oxysporum Is Independent of ET, JA, and SA. Front. Plant Sci. 2019, 10, 979. [Google Scholar] [CrossRef]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic Acid–Independent ENHANCED DISEASE SUSCEPTIBILITY1 Signaling in Arabidopsis Immunity and Cell Death Is Regulated by the Monooxygenase FMO1 and the Nudix Hydrolase NUDT7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Xu, Q. CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Straus, M.R.; Rietz, S.; Ver Loren Van Themaat, E.; Bartsch, M.; Parker, J.E. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 2010, 62, 628–640. [Google Scholar] [CrossRef]
- Liu, N.; Ma, X.; Zhou, S.; Wang, P.; Sun, Y.; Li, X.; Hou, Y. Molecular and Functional Characterization of a Polygalacturonase-Inhibiting Protein from Cynanchum komarovii That Confers Fungal Resistance in Arabidopsis. PLoS ONE 2016, 11, e0146959. [Google Scholar] [CrossRef] [PubMed]
- Meitei, A.L.; Bhattacharjee, M.; Dhar, S.; Chowdhury, N.; Sharma, R.; Acharjee, S.; Sarmah, B.K. Activity of defense related enzymes and gene expression in pigeon pea (Cajanus cajan) due to feeding of Helicoverpa armigera larvae. J. Plant Interact. 2018, 13, 231–238. [Google Scholar] [CrossRef]
- Li, J.; Aach, J.; Norville, J.E.; Mccormack, M.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and Homologous Recombination-Mediated Plant Genome Editing via Guide RNA/Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, G.; Ma, S.; Xie, X.; Wu, X.; Zhang, X.; Wu, Y.; Zhao, P.; Xia, Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef]
- Jiang, W.; Yang, B.; Weeks, D.P. Efficient CRISPR/Cas9-Mediated Gene Editing in Arabidopsis thaliana and Inheritance of Modified Genes in the T2 and T3 Generations. PLoS ONE 2014, 9, e99225. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Kapusi, E.; Corcuera-Gómez, M.; Melnik, S.; Stoger, E. Heritable Genomic Fragment Deletions and Small Indels in the Putative ENGase Gene Induced by CRISPR/Cas9 in Barley. Front. Plant Sci. 2017, 8, 540. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.-Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, G.; Zhou, J.; Ren, Q.; You, Q.; Tian, L.; Xin, X.; Zhong, Z.; Liu, B.; Zheng, X.; et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018, 19, 84. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Lin, Y.; Cradick, T.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A Software Package for Designing CRISPR sgRNA and Evaluating Potential Off-Target Cleavage Sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]
- de Lamo, F.J.; Takken, F.L.W. Biocontrol by Fusarium oxysporum Using Endophyte-Mediated Resistance. Front. Plant Sci. 2020, 11, 37. [Google Scholar] [CrossRef]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.; Takken, F.L.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Lund, S.T.; Stall, R.E.; Klee, H.J. Ethylene Regulates the Susceptible Response to Pathogen Infection in Tomato. Plant Cell 1998, 10, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Ino, Y.; Kimura, Y.; Tago, A.; Shimizu, M.; Natsume, S.; Sano, Y.; Fujimoto, R.; Kaneko, K.; Shea, D.J.; et al. Changes in the Proteome of Xylem Sap in Brassica oleracea in Response to Fusarium oxysporum Stress. Front. Plant Sci. 2016, 7, 31. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A. Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 2008, 13, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mallick, N.; Mitra, A. Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol. Biochem. 2009, 47, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Zehra, A.; Meena, M.; Dubey, M.K.; Aamir, M.; Upadhyay, R.S. Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot. Stud. 2017, 58, 44. [Google Scholar] [CrossRef] [PubMed]
- Mock, H.-P.; Keetman, U.; Kruse, E.; Rank, B.; Grimm, B. Defense Responses to Tetrapyrrole-Induced Oxidative Stress in Transgenic Plants with Reduced Uroporphyrinogen Decarboxylase or Coproporphyrinogen Oxidase Activity1. Plant Physiol. 1998, 116, 107–116. [Google Scholar] [CrossRef]
- Chen, S.-C.; Ren, J.-J.; Zhao, H.-J.; Wang, X.-L.; Wang, T.-H.; Jin, S.-D.; Wang, Z.-H.; Li, C.-Y.; Liu, A.-R.; Lin, X.-M.; et al. Trichoderma harzianum Improves Defense Against Fusarium oxysporum by Regulating ROS and RNS Metabolism, Redox Balance, and Energy Flow in Cucumber Roots. Phytopathology 2019, 109, 972–982. [Google Scholar] [CrossRef] [PubMed]

| No. of Plants Examined | Cas9 Positive Efficiency (%) | Editing Efficiency (%) | |||
|---|---|---|---|---|---|
| Single Gene (SX) | Single Gene (SS) | Dual Gene (DXS) | Combined (SX, SS and DXS) | ||
| 73 | 56.16% (41/73) | 11.53% (3/26 *) | 19.23% (5/26 *) | 3.8% (1/26 *) | 34.6% (9/26 *) |
| GE0 Segregation | GE1 Segregation | ||||
|---|---|---|---|---|---|
| Target Gene | Lines | Cas9 | Mutation | Mutation | Cas9/T-DNA |
| XSP10 (SX) | 20 | + | Bi-allelic | 2 Bi/2 He | + |
| XSP10 (SX) | 34 | + | Chimera | NA | NA |
| XSP10 (SX) | 40 | + | Heterozygote | NA | NA |
| XSP10 (DX) | 19 | + | Chimera | 3 Chi/2 He | + |
| SlSAMT (SS) | 16 | + | Chimera | 1 Bi/1 He/6 Chi | + |
| SlSAMT (SS) | 7 | + | Chimera | NA | NA |
| SlSAMT (SS) | 23 | + | Chimera | NA | NA |
| SlSAMT (SS) | 33 | + | Chimera | NA | NA |
| SlSAMT (SS) | 52 | + | Chimera | NA | NA |
| SlSAMT (DS) | 19 | + | Chimera | 3 Chi/2 He | + |
| Target | Potential Off-Target Sequence | No. of Mismatch Bases | Total No. of Plants Tested | No. of Mutation |
|---|---|---|---|---|
| Solyc05g016300 | TTTGAATGCTTCCATATCAGGG | 4 | 5 | 0 |
| Solyc12g016150 | TGATATTGGACCCGTATCACGG | 4 | 5 | 0 |
| Solyc02g032110 | TTATATTGAATCCGTATCA CGG | 4 | 5 | 0 |
| Solyc01g057280 | ATCACTAAAATGTTCTCCCAGG | 4 | 5 | 0 |
| Solyc09g060120 | CTCATTTGAATGATCACCCAGG | 4 | 5 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Debbarma, J.; Saikia, B.; Singha, D.L.; Das, D.; Keot, A.K.; Maharana, J.; Velmurugan, N.; Arunkumar, K.P.; Reddy, P.S.; Chikkaputtaiah, C. CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.). Genes 2023, 14, 488. https://doi.org/10.3390/genes14020488
Debbarma J, Saikia B, Singha DL, Das D, Keot AK, Maharana J, Velmurugan N, Arunkumar KP, Reddy PS, Chikkaputtaiah C. CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.). Genes. 2023; 14(2):488. https://doi.org/10.3390/genes14020488
Chicago/Turabian StyleDebbarma, Johni, Banashree Saikia, Dhanawantari L. Singha, Debajit Das, Ajay Kumar Keot, Jitendra Maharana, Natarajan Velmurugan, Kallare P. Arunkumar, Palakolanu Sudhakar Reddy, and Channakeshavaiah Chikkaputtaiah. 2023. "CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.)" Genes 14, no. 2: 488. https://doi.org/10.3390/genes14020488
APA StyleDebbarma, J., Saikia, B., Singha, D. L., Das, D., Keot, A. K., Maharana, J., Velmurugan, N., Arunkumar, K. P., Reddy, P. S., & Chikkaputtaiah, C. (2023). CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.). Genes, 14(2), 488. https://doi.org/10.3390/genes14020488






