Abstract
The broodiness traits of domestic geese are a bottleneck that prevents the rapid development of the goose industry. To reduce the broodiness of the Zhedong goose and thus improve it, this study hybridized it with the Zi goose, which has almost no broody behavior. Genome resequencing was performed for the purebred Zhedong goose, as well as the F2 and F3 hybrids. The results showed that the F1 hybrids displayed significant heterosis in growth traits, and their body weight was significantly greater than those of the other groups. The F2 hybrids showed significant heterosis in egg-laying traits, and the number of eggs laid was significantly greater than those of the other groups. A total of 7,979,421 single-nucleotide polymorphisms (SNPs) were obtained, and three SNPs were screened. Molecular docking results showed that SNP11 located in the gene NUDT9 altered the structure and affinity of the binding pocket. The results suggested that SNP11 is an SNP related to goose broodiness. In the future, we will use the cage breeding method to sample the same half-sib families to accurately identify SNP markers of growth and reproductive traits.
1. Introduction
China’s domestic geese output ranks first in the world, suggesting its great potential for the development of the goose industry. However, the broodiness of domesticated geese has always been a key problem limiting the development of goose industrialization [1]. Broody behavior varies widely among different varieties of geese [2]. The broodiness of the Zhedong goose is conspicuous and seriously affects the number of eggs laid [1]. The Zi goose, which lays a large number of eggs, shows hardly any broodiness. To reduce the broodiness of the Zhedong goose, in this study we used hybridization with the Zi goose to decrease the broody behavior of the Zhedong goose. Many methods have been explored to reduce the broodiness of geese, including hormone intervention [3,4], light adjustment [5,6,7], genetic improvement [8,9], and nutrition regulation [10]. Various numbers of SNPs have been reported in species of domestic geese [11,12,13], and these can possibly be used in livestock breeding to obtain superior genotypes.
One SNP of the growth differentiation factor 9 (GDF9) gene is homozygous AA in wild type ewe populations, and the mutant heterozygous Aa can increase the litter size. Interestingly, the homozygous SNP mutation aa type leads to infertility [14]. Modern breeding technology can use SNP chips to rapidly and efficiently determine the SNPs of individual animals. Animal SNP breeding chips have been developed and applied in pigs [15,16], cows [17], sheep [18], goats [19], and layers [20]. SNPs related to goose phenotypic traits have also been reported [11,12]. Currently, there are no reports concerning goose SNP chips. This study was begun in 2016 and was originally intended for commercial application. During the research, we found that the body weight and number of eggs laid by the F3 hybrids were decreased. We were concerned about GDF9, and thus we implemented whole genome resequencing to identify the SNP markers of growth and reproduction traits in the Zhedong and Zi crossbred geese. The purposes of this study were to verify whether there were SNPs that affected goose reproductive traits and to provide guidance for the commercial breeding of hybrid geese. These results can provide a reference for goose breeding research.
2. Materials and Methods
2.1. Ethics Statement
The study was approved by the Committee for Animal Welfare of the Institute of Animal Husbandry of Heilongjiang Academy of Agricultural Sciences, China (No. NKY-20140506), Ministry of Science and Technology.
2.2. Animals
The experimental geese were sourced from two farms; one was located at the Qiqihar Animal Husbandry and Veterinary Research Institute (47.35° N, 123.92° E), and the other was at the Shuangyashan Friendship Farm (46.63° N, 131.16° E). The two localities are very close; the monthly average local temperature ranges from −1 °C to 10 °C, and the minimum average temperature at night in January is −14 °C. The highest daytime average temperature in June is 37 °C. A total of 1200 Zhedong geese and 600 offspring were used as the experimental base population. In 2016 and 2019, 600 Zhedong geese were introduced from Xiangshan City, Zhejiang Province, to Heilongjiang Province. After at least one year of local domestication, they were used for hybridization experiments. The local domestication process gradually changed from the feed formula in Zhejiang Province to the feed formula used in this study. All geese were raised under natural light and temperature conditions. The geese engaged in free activities in the grounds during the day and entered their house at night. They had free access to feed and water daily. The male-to-female ratio was 1:4–5. The method of hybridization is shown in Figure 1. The nutritional standard of goose feed is listed in Table 1. Goslings hatched in May and June and started laying eggs in March of the following year.

Figure 1.
Schematic diagram of Zhedong and Zi crossbred goose experiment. Purebred Zhedong geese were introduced in Xiangshan County in 2016 and 2019. The Zi goose is a local variety in Heilongjiang. The F1, F2 and F3 hybrids were obtained through hybridization, and some experimental populations were selected to measure body weight and laying rate as well as for whole genome resequencing analysis. If the primary generation Zhedong goose was homozygous for site AA, then gene type AB and AA could appear in the F2 and F3 generations. However, BB type was not possible.

Table 1.
Goose Feed Nutrition Standard.
2.3. Body Weight and Egg-Laying Phenotypic Measurements
The egg-laying data included a random selection of experimental mother geese from a large population. The ratio of males to females was 1:4–5, matching the male geese to form a test group. The number of eggs laid was recorded daily. The first egg was laid and recorded on 21 February 2017; thereafter February 21 each year was assigned as week 0. The bodyweight data were randomly collected from newly hatched goslings that were then marked on the neck with a dye representing a number. For example, red, yellow, blue, and green from top to bottom represented 0001. The body weight of each goose was tracked, measured, and recorded every two weeks until 12 weeks. The body weight was recorded when the goose entered and exited the goose house in order to minimize stress.
2.4. Genome Resequencing
Blood samples were collected for DNA extraction during the non-egg-laying stage. Five geese were randomly selected in each group to avoid as much stress as possible. Vacuum tubes containing ethylenediaminetetraacetic acid were used to collect wing blood. The blood samples were sent to Harbin Botai Gene Company for testing, and the quantity of clean data of each sample was not less than 11 GB, ensuring a Q30 value ≥ 80% (Table 2).

Table 2.
Sequencing data statistics.
2.5. SNP Calling and GWAS
We used SnpEff to annotate the mutated sites and to determine the corresponding gene information, synonymous and non-synonymous mutations, and the impact on amino acids of the mutated sites. This study applied the classical genome-wide association study (GWAS) analysis method but failed. There were four methods: (1) ADMIXTURE (Version 1.3.0) was used to analyze the population structure; (2) The genetic relationships analysis and principal component analysis of SNPs were performed using TASSEL 5.0; (3) The PopLDdecay software(Version 3.41) was used to analyze linkage disequilibrium (LD) decay; (4) The SNP sequences were employed using Mega X to construct an evolutionary tree via the neighbor-joining (NJ) method. The 15 samples could not be clustered, and thus the analysis failed.
Finally, the filtered SNPs were further screened based on the background of the project. For the screening principle, the parents were homozygous, and the two groups of offspring samples were either the heterozygous or homozygous Zhedong goose genotype (not the homozygous Zi goose genotype). To be specific, if the primary generation was homozygous for Zhedong goose site AA, then AB and AA could appear in the F2 and F3 generations. However, BB was not possible. See Figure 1 for the annotation. These sites may be candidate SNP sites related to the phenotype of egg laying. This process used the Sort method in Microsoft Excel. GO and KEGG enrichment analyses were conducted for the SNP sites.
2.6. Three-Dimensional (3D) Structure Prediction and Molecular Docking
NCBI was searched for reports of candidate SNP genes in recent years. Candidate genes for NUP37 (XP_013036270.1) and NUDT9 (XP_013045959.1) were found. These two candidate genes include SNP3, SNP4 and SNP11. For the candidate genes, we searched for protein changes caused by SNP transformation. The 3D structures of the target proteins containing SNPs were produced via SWISS-MODEL (https://swissmodel.expasy.org). The NUDT9 protein and adenosine diphosphate ribose (ADPR) were combined. AutoDock Vina software was used for docking analysis, and the results were visualized using Discovery Studio.
3. Results
3.1. Analysis of Goose Body Weight and Egg-Laying Trait
The egg-laying data are presented in Figure 2A. From the figure, it can be seen that Zi geese began laying eggs in the middle of March and finished laying in early July. The average weekly egg-laying rate was 34.51%. From 2017 to 2020, the egg-laying rates were 17.35%, 9.07%, 14.13% and 18.65%, respectively. For the Zhedong and Zi crossed geese, the egg-laying rates of the F1, F2 and F3 hybrids were 30.05%, 70.98% and 15.02%, respectively. The duration of egg-laying for the F2 hybrids was surprising, lasting from March 4 to May 23. The birth weight (week 0) of the F3 hybrids was high, but the offspring were not used to the starter feed, resulting in a slow increase in the two-week-old body weight. When the feed was adjusted at 11 weeks, the F3 hybrids grew more rapidly. There was no significant difference between body weight and F1 hybrids at 12 weeks. The Zhedong geese in 2020 had the highest weight during the first five weeks, but this group was gradually surpassed by the F1 hybrids after changing the feed during the fifth week. At 12 weeks, the body weights of the F1 and F2 hybrids were significantly higher than those of the other experimental groups.


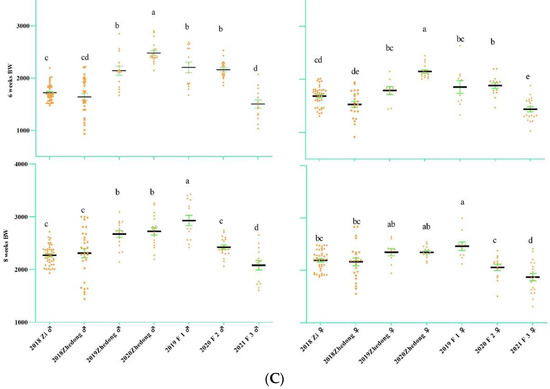

Figure 2.
Changes in the goose egg-laying trait and body weight in different generations. (A) Heat map of goose egg-laying data of different generations. The different colors represent different laying rates, and the abscissa is initially dated 21 February. The end time is 3 July; the total number of days is 133. On 29 February 2020, the experimental group did not start laying eggs. (B) Growth data of different generations of geese (0–4 weeks) (C) Growth data of different generations of geese (6–8 weeks) (D) Growth data of different generations of geese (10–12 weeks). The body weight of the F3 hybrids shows significant differences at 2 weeks and 12 weeks. The different letters indicate significant differences. The green bar represents SE. Body-weight unit: g.
3.2. Genome Resequencing and Whole Genome Resequencing
The sequences obtained in this project were compared with the reference genome, and 7,979,421 SNPs were obtained. The SNPs obtained were filtered using VCF tools, and finally 68,376 SNPs were obtained. To further narrow the scope, it was necessary to link the SNP data with the grouped experimental data. (1) See Supplementary Figure S1 for the results of the population structure analysis. (2) Genetic relationship and principal component analysis results are presented in Supplementary Figure S2A,B. (3) See Supplementary Figure S3 for the analysis results of the linkage disequilibrium (LD) decay. The results were disappointing and could not be analyzed according to our grouping. (4) The results of the NJ evolutionary tree construction are shown in Supplementary Figure S4. None of the 15 samples could be clustered. The above results show that the classical GWAS method was unable to achieve the expected result of the project.
3.3. Screening and Analysis of Candidate SNPs
Based on the background of this project, it was assumed that the parents were homozygous, and the two groups of offspring samples were heterozygous or homozygous for Zhedong goose sites. The offspring were grouped according to original purebred Zhedong geese, Zhedong and Zi crossbred geese, and F2, and F3 hybrid groups. A total of 3,370 candidate SNP loci were obtained. The GO enrichment analysis results are shown in Figure 3A, which lists the 25 GO terms with the highest p-values. These included DNA integration (GO:0015074, p = 4.21 × 10−19), the DNA metabolic process (GO: 0006259, p = 7.24 × 10−12), and the nitrogen compound metabolic process (GO: 0006807, p = 3.02 × 10−6) terms. The enrichment analysis results of level two GO terms are shown in Figure 3B. The KEGG enrichment analysis results are shown in Figure 3C. The distribution statistics of the candidate SNPs on chromosome positions are shown in Figure 3D. The 144 SNPs in the exons were further studied. Figure 3E shows the classification of the candidate SNPs distributed in the exons, and 39 SNPs caused missense mutations. These are shown in Table 3.


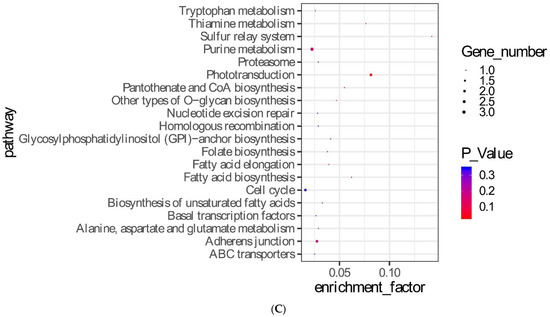

Figure 3.
Screening and analysis of candidate SNPs. (A) Gene Ontology (GO) analysis for 3.370 candidate SNPs. (B) Level two GO analysis of candidate SNPs. (C) KEGG enrichment of candidate SNPs. (D) Distribution statistics of candidate SNPs on chromosome positions. (E) Classified statistics of candidate SNPs distributed in the exon.

Table 3.
A total of 39 candidate SNPs were screened in this study.
3.4. Candidate Genes’ 3D Structure Prediction and Molecular Docking
The results of SNP3 and SNP4 affecting the NUP37 protein are shown in Figure 4. The SNPs did not cause variation in protein structure, but they did change a single amino acid residue. SNP3 and SNP4 had very limited impact on the NUP37 protein. SNP11 also changed the local amino acid of NUDT9 located at the 218th amino acid of the binding site. Figure 5 shows that the amino acids were combined by 218 Thr (affinity, −5.1 kcal/mol), and the ADPR was 219 Gln, 217 Arg, and 212 Lys. The amino acids were combined by 218 Ala (affinity, −4.8 kcal/mol), and the ADPR was 210 Glu, 217 Arg, 170 Glu, and 214 Arg. SNP11 will affect the biological activity of NUDT9.
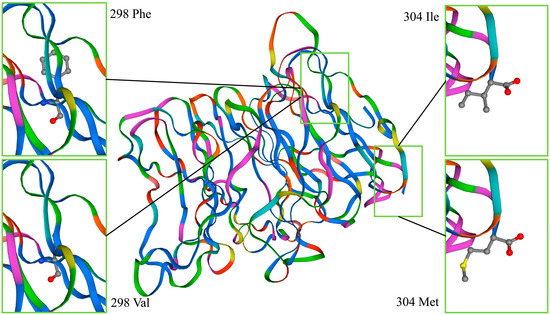
Figure 4.
SNP3 and SNP4 affect the NUP37 protein. The structure of the NUP37 protein was unchanged due to SNP3 and SNP4. The 304th amino acid of SNP3 in the upper right corner is Ile, and in the lower right corner is Met. The 298th amino acid of SNP4 in the upper left corner is Phe, and in the lower left corner is Val.

Figure 5.
SNP11 affects NUDT9 molecular docking. SNP11 does not affect the structure of the NUDT9 protein. To display the docking of amino acids, different angles were rotated. The left side shows 218 Thr combined with ADPR. The right side shows 218 Ala combined with ADPR. The combination box has changed.
4. Discussion
The broodiness trait of domestic geese is a bottleneck preventing the rapid development of the goose industry [1]. The advantage of the Zhedong goose is its large body weight and superior meat quality [1]. The advantage of the Zi goose is that it has desirable egg-laying traits. Our research results show that the F1 hybrids displayed clear heterosis in growth traits, and body weight was significantly greater than those in other groups. The F2 hybrids showed significant heterosis in the egg-laying trait, and the number of eggs laid was significantly greater than those of the other groups. Our hypothesis is the SNP11 may affect the goose egg-laying trait, and the verification experiment will continue in the future.
Based on the breeding season, geese of different varieties can be divided into three types: type one and type two are long sunshine geese that inhabit the high-latitude or middle-latitude (30–40° N) temperate zone. Type three refers to the short sunshine geese distributed in subtropical regions [3]. The Zhedong goose is a short sunshine variety, and the Zi goose is a long sunshine variety. Light regulates the secretion of goose melatonin and further regulates egg-laying traits [5]. A transcriptome analysis indicated that broodiness behavior was consistent with gene expression in the pineal gland [21]. This indirectly indicated that light could change the egg-laying timing of a goose. Our results also show that the egg-laying traits of Zhedong geese changed under different light and temperature conditions when the geese migrated from Zhejiang Province to Heilongjiang Province.
The egg production of domestic geese is a quantitative trait regulated by multiple genes and is affected by environmental factors [22]. The egg-laying characteristic of the Zhedong goose comprises repeating the cycle of egg-laying and broodiness. Birds in nature also have similar behaviors [23]. Broodiness is essential for the breeding of wild birds [24], with the exception of brood parasitism by cuckoos [25]. The artificial domestication of geese is far from the effect in hens. After years of selection and elimination, hens have completely lost their broodiness ability [20].
The broodiness of the Zhedong goose is strong, and as such the goose is an ideal animal model for studying the mechanism of broody behavior [1,4,26]. In Zhejiang Province, the egg-laying period of the Zhedong goose lasts from October to April of the next year, during which three to four laying cycles (laying–broodiness–recovery) are experienced [27]. The broody behavior of a goose is regulated by the hypothalamic–pituitary–gonadal axis [28]. Many studies on goose broodiness have focused on the ovary [27,29,30,31]. The signal pathways involved in the ovarian regulation of broodiness include the autophagic pathway [29] and the GnRH pathway [32]. Our previous research showed that light affects the number of eggs laid by geese [5]. Mitochondrial dysfunction in follicles affects the broodiness of the Zhedong goose [1]. Injection of prolactin (PRL) fusion protein improved the number of eggs laid by Zhedong geese [3].
Before 2019, geese were generally raised on the ground [2]. By 2021, raising geese in cages had become popular [22]. This research started in 2016 with more than 200 geese in each group. It was impossible to select individual experimental geese based on full-sib or half-sib families. All the SNPs found included growth and reproduction traits as well as other traits. Finally, we chose the SNP screening method from the above methods. The main purpose was to gradually narrow the scope of the investigation. The first step in the screening criteria was to narrow the scope according to Mendelian genetics. The second step was to find SNPs located in the exons, and the third step was to conduct a comparative analysis according to other studies. Finally, three candidate SNPs were determined.
NUP37 is a biomarker in breast cancer [33]. NUP37 regulates the YAP/TEAD signaling pathway [34]. The Yes-associated protein (YAP) regulates muscle growth [35], cell division [36], and regeneration [37]. Our results showed that SNP3 and SNP4 changed an amino acid residue of NUP37, but the impact of this change needs to be further studied. This is because the change did not cause changes in the advanced structure of the NUP37 protein, and it also did not affect the structures of the binding pockets.
The newly discovered SNP11 in this study is located in NUDT9, a gene that can regulate the menstrual cycle [38]. This suggests that SNP11 is an SNP related to goose broodiness. NUDT9 plays a role in breast cancer [39]. The NUDT9 protein can dock with ADPR, and 218 amino acids directly participate in the docking [40]. We simulated the docking and found that the goose NUDT9 protein could dock with ADPR. Neither 218 Thr nor 218 Ala corresponding to SNP11 directly participated in the link, but the structures of the binding pockets were altered. The 217 and 219 amino acids participated in the docking, and the affinity was altered.
5. Conclusions
In terms of growth traits, the F1 hybrids of Zhedong and Zi crossbred geese showed significant heterosis, and the body weight was significantly greater than in other groups. The F2 hybrids showed significant heterosis in egg-laying traits, and the number of eggs laid was significantly greater than those of the other groups. SNP11, located in NUDT9, is an SNP related to goose breeding. Moving from Zhejiang Province to Heilongjiang Province caused the egg-laying performance of the Zhedong goose to change under different lighting and temperature conditions. In the future, we will use the cage breeding method to sample the same half-sib family to accurately identify the SNP markers of growth and reproduction traits.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14020487/s1, Figure S1. Cross Entropy Line Chart. The minimum value of cross entropy is generally selected for population result analysis. There is no obvious population structure in this analysis. Figure S2. Genetic relationship and principal component analysis (PCA). (A) Kinship analysis heat map. The different colors represent kinship. There is no kinship between the samples. In the figure, C represents the original Zhedong goose, B represents F2, and O represents F3. Figure S3. Analyze the linkage disequilibrium (LD) decal. It is not possible to group using different generations based on the distance in the picture. Figure S4. NJ evolutionary tree. Grouping explicitly based on the distance in the picture is not possible.
Author Contributions
Conceptualization, G.L. and Z.G.; formal analysis, G.L., Z.G., Z.C. and Z.M.; data curation, G.L., X.Z., J.S., S.Y. and M.L.; writing—original draft preparation, G.L. and Z.G.; writing—review and editing, Z.G.; supervision, Z.G. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the China Agricultural Research System (CARS-42-24).
Institutional Review Board Statement
The study was approved by the Committee for Animal Welfare of the Institute of Animal Husbandry of Heilongjiang Academy of Agricultural Sciences, China (No. NKY-20140506), Ministry of Science and Technology.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yu, J.; Guo, C. Mitochondrial dysfunction in follicles is associated with broodiness in Zhedong white goose. Anim. Reprod. Sci. 2022, 243, 107032. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, Y.Z.; Gu, T.T.; Cao, Z.F.; Zhao, W.M.; Qin, H.R.; Xu, Q.; Chen, G.H. Comparison of the broody behavior characteristics of different breeds of geese. Poult. Sci. 2019, 98, 5226–5233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Z.Y.; An, C.; Weng, K.Q.; Cao, Z.F.; Xu, Q.; Chen, G.H. Effect of active immunization with recombinant-derived goose INH-alpha, AMH, and PRL fusion protein on broodiness onset and egg production in geese (Anser cygnoides). Poult. Sci. 2021, 100, 101452. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, L.; Zhang, Y.; Yao, Y.; Zhao, W.; Xu, Q.; Chen, G. Characterization of ovarian morphology and reproductive hormones in Zhedong white geese (Anser cygnoides domesticus) during the reproductive cycle. J. Anim. Physiol. Anim. Nutr. 2021, 105, 938–945. [Google Scholar] [CrossRef]
- Liu, G.J.; Chen, Z.F.; Zhao, X.H.; Li, M.Y.; Guo, Z.H. Meta-analysis: Supplementary artificial light and goose reproduction. Anim. Reprod. Sci. 2020, 214, 106278. [Google Scholar] [CrossRef]
- Zhu, H.X.; Liu, X.Q.; Hu, M.D.; Lei, M.M.; Chen, Z.; Ying, S.J.; Yu, J.N.; Dai, Z.C.; Shi, Z.D. Endocrine and molecular regulation mechanisms of the reproductive system of Hungarian White geese investigated under two artificial photoperiodic programs. Theriogenology 2019, 123, 167–176. [Google Scholar] [CrossRef]
- Zhu, H.X.; Hu, M.D.; Guo, B.B.; Qu, X.L.; Lei, M.M.; Chen, R.; Chen, Z.; Shi, Z.D. Effect and molecular regulatory mechanism of monochromatic light colors on the egg-laying performance of Yangzhou geese. Anim. Reprod. Sci. 2019, 204, 131–139. [Google Scholar] [CrossRef]
- Ouyang, Q.; Hu, S.; Wang, G.; Hu, J.; Zhang, J.; Li, L.; Hu, B.; He, H.; Liu, H.; Xia, L.; et al. Comparative Transcriptome Analysis Suggests Key Roles for 5-Hydroxytryptamlne Receptors in Control of Goose Egg Production. Genes 2020, 11, 455. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, J.; Zhang, X.; Xu, Z.; Lin, Z.; Li, H.; Lin, W.; Xie, Q. Genome-Wide Association Analysis Reveals Key Genes Responsible for Egg Production of Lion Head Goose. Front. Genet. 2019, 10, 1391. [Google Scholar] [CrossRef]
- Ye, M.; Sun, L.; Yang, R.; Wang, Z.; Qi, K. The optimization of fermentation conditions for producing cellulase of Bacillus amyloliquefaciens and its application to goose feed. R. Soc. Open Sci. 2017, 4, 171012. [Google Scholar] [CrossRef]
- Melak, S.; Wang, Q.; Tian, Y.; Wei, W.; Zhang, L.; Elbeltagy, A.; Chen, J. Identification and Validation of Marketing Weight-Related SNP Markers Using SLAF Sequencing in Male Yangzhou Geese. Genes 2021, 12, 1203. [Google Scholar] [CrossRef] [PubMed]
- Abdel Moniem, H.; Yusuf, M.S.; Chen, G. Ecology and population structure of some indigenous geese breeds and the impact of four GH and Pit-1 SNPs on their body weights. Environ. Sci. Pollut. Res. Int. 2021, 28, 37603–37615. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Chen, P.; Zhou, C.; Zhao, X.; Zhang, K.; Wu, R.; Zhang, C.; Wang, Y.; Xie, Y.; Wang, Q. Genome-wide association study for reproduction-related traits in Chinese domestic goose. Br. Poult. Sci 2022, 63, 754–760. [Google Scholar] [CrossRef]
- Souza, C.J.; McNeilly, A.S.; Benavides, M.V.; Melo, E.O.; Moraes, J.C. Mutation in the protease cleavage site of GDF9 increases ovulation rate and litter size in heterozygous ewes and causes infertility in homozygous ewes. Anim. Genet. 2014, 45, 732–739. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.H.; Park, H.B.; Kim, J.M. Identification of key adipogenic transcription factors for the pork belly parameters via the association weight matrix. Meat Sci. 2023, 195, 109015. [Google Scholar] [CrossRef]
- Yuan, J.; Zhou, X.; Xu, G.; Xu, S.; Liu, B. Genetic diversity and population structure of Tongcheng pigs in China using whole-genome SNP chip. Front. Genet. 2022, 13, 910521. [Google Scholar] [CrossRef] [PubMed]
- Mesbah-Uddin, M.; Guldbrandtsen, B.; Capitan, A.; Lund, M.S.; Boichard, D.; Sahana, G. Genome-wide association study with imputed whole-genome sequence variants including large deletions for female fertility in 3 Nordic dairy cattle breeds. J. Dairy Sci. 2022, 105, 1298–1313. [Google Scholar] [CrossRef]
- Guo, Y.; Bai, F.; Wang, J.; Fu, S.; Zhang, Y.; Liu, X.; Zhang, Z.; Shao, J.; Li, R.; Wang, F.; et al. Design and characterization of a high-resolution multiple-SNP capture array by target sequencing for sheep. J. Anim. Sci. 2022, 101, skac383. [Google Scholar] [CrossRef]
- Moaeen-Ud-Din, M.; Danish Muner, R.; Khan, M.S. Genome wide association study identifies novel candidate genes for growth and body conformation traits in goats. Sci. Rep. 2022, 12, 9891. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Lubritz, D.; Arango, J.; Fulton, J.; Settar, P.; Rowland, K.; Cheng, H.; Wolc, A. Genome-wide association studies for egg quality traits in White Leghorn layers using low-pass sequencing and SNP chip data. J. Anim. Breed. Genet. 2022, 139, 380–397. [Google Scholar] [CrossRef]
- Yuan, X.; Lan, G.; Li, L.; He, H.; Wang, J.; Hu, S. Differential gene expression profiling of the goose pineal gland. Br. Poult. Sci. 2020, 61, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Gao, D.; Zhao, X.; Xu, S.; Zhang, K.; Wu, R.; Yin, C.; Li, J.; Xie, Y.; Hu, S.; et al. Genome-Wide Association Study-Based Identification of SNPs and Haplotypes Associated With Goose Reproductive Performance and Egg Quality. Front. Genet. 2021, 12, 602583. [Google Scholar] [CrossRef] [PubMed]
- Farrar, V.S.; Flores, L.; Viernes, R.C.; Ornelas Pereira, L.; Mushtari, S.; Calisi, R.M. Prolactin promotes parental responses and alters reproductive axis gene expression, but not courtship behaviors, in both sexes of a biparental bird. Horm. Behav. 2022, 144, 105217. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.R.; Martin, K.; Cockle, K.L. Weather and nest cavity characteristics influence fecundity in mountain chickadees. PeerJ 2022, 10, e14327. [Google Scholar] [CrossRef] [PubMed]
- Morelli, F.; Benedetti, Y.; Pape Moller, A. Diet specialization and brood parasitism in cuckoo species. Ecol. Evol. 2020, 10, 5097–5105. [Google Scholar] [CrossRef]
- Chen, F.; Li, J.; Zhang, H.; Xu, J.; Tao, Z.; Shen, J.; Shen, J.; Lu, L.; Li, C. Identification of differentially expressed known and novel miRNAs in broodiness of goose. Mol. Biol. Rep. 2014, 41, 2767–2777. [Google Scholar] [CrossRef]
- Yu, J.; Lou, Y.; Zhao, A. Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose. Sci. Rep. 2016, 6, 36877. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Li, L.; Han, C.; He, H.; Xu, H. Transcriptome analysis revealed the possible regulatory pathways initiating female geese broodiness within the hypothalamic-pituitary-gonadal axis. PLoS ONE 2018, 13, e0191213. [Google Scholar] [CrossRef]
- Lou, Y.; Yu, W.; Han, L.; Yang, S.; Wang, Y.; Ren, T.; Yu, J.; Zhao, A. ROS activates autophagy in follicular granulosa cells via mTOR pathway to regulate broodiness in goose. Anim. Reprod. Sci. 2017, 185, 97–103. [Google Scholar] [CrossRef]
- Qin, H.; Li, X.; Wang, J.; Sun, G.; Mu, X.; Ji, R. Ovarian transcriptome profile from pre-laying period to broody period of Xupu goose. Poult. Sci. 2021, 100, 101403. [Google Scholar] [CrossRef]
- Hou, L.; Ji, W.; Gu, T.; Weng, K.; Liu, D.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. MiR-34c-5p promotes granulosa cells apoptosis by targeting Bcl2 in broody goose (Anser cygnoides). Anim. Biotechnol. 2022, 33, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lou, Y.; He, K.; Yang, S.; Yu, W.; Han, L.; Zhao, A. Goose broodiness is involved in granulosa cell autophagy and homeostatic imbalance of follicular hormones. Poult. Sci. 2016, 95, 1156–1164. [Google Scholar] [CrossRef]
- Li, K.; Liu, T. Evaluation of Oncogene NUP37 as a Potential Novel Biomarker in Breast Cancer. Front. Oncol. 2021, 11, 669655. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, Y.; Feng, W.; Lei, L.; Du, Y.; Wu, J.; Wang, S. NUP37, a positive regulator of YAP/TEAD signaling, promotes the progression of hepatocellular carcinoma. Oncotarget 2017, 8, 98004–98013. [Google Scholar] [CrossRef] [PubMed]
- Esteves de Lima, J.; Bonnin, M.A.; Birchmeier, C.; Duprez, D. Muscle contraction is required to maintain the pool of muscle progenitors via YAP and NOTCH during fetal myogenesis. Elife 2016, 5, e15593. [Google Scholar] [CrossRef]
- Cao, X.; Pfaff, S.L.; Gage, F.H. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008, 22, 3320–3334. [Google Scholar] [CrossRef]
- Rudolf, M.A.; Andreeva, A.; Kozlowski, M.M.; Kim, C.E.; Moskowitz, B.A.; Anaya-Rocha, A.; Kelley, M.W.; Corwin, J.T. YAP Mediates Hair Cell Regeneration in Balance Organs of Chickens, But LATS Kinases Suppress Its Activity in Mice. J. Neurosci. 2020, 40, 3915–3932. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.; Chen, T.H.; Wen, P.Y.; Chou, C.H.; Ying, T.H.; Chang, S.P.; Ma, G.C.; Chen, M. Differential expression of NUDT9 at different phases of the menstrual cycle and in different components of normal and neoplastic human endometrium. Taiwan J. Obstet. Gynecol. 2009, 48, 96–107. [Google Scholar] [CrossRef]
- Wright, R.H.G.; Beato, M. Role of the NUDT Enzymes in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 2267. [Google Scholar] [CrossRef]
- Gattkowski, E.; Rutherford, T.J.; Mockl, F.; Bauche, A.; Sander, S.; Fliegert, R.; Tidow, H. Analysis of ligand binding and resulting conformational changes in pyrophosphatase. NUDT9. FEBS J. 2021, 288, 6769–6782. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).