Maps of Constitutive-Heterochromatin Distribution for Four Martes Species (Mustelidae, Carnivora, Mammalia) Show the Formative Role of Macrosatellite Repeats in Interspecific Variation of Chromosome Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampled Species and an Ethics Statement
2.2. Cell Culture, Chromosome Preparation, and Differential Staining
2.3. Preparation and Characterization of Chromosome-Specific Painting Probes and Detection of Nucleolus Organizer Regions (NORs) and Telomeric Repeats
2.4. Identification of Tandemly Arranged Repetitive DNA
2.5. FISH, Image Acquisition, and Data Processing
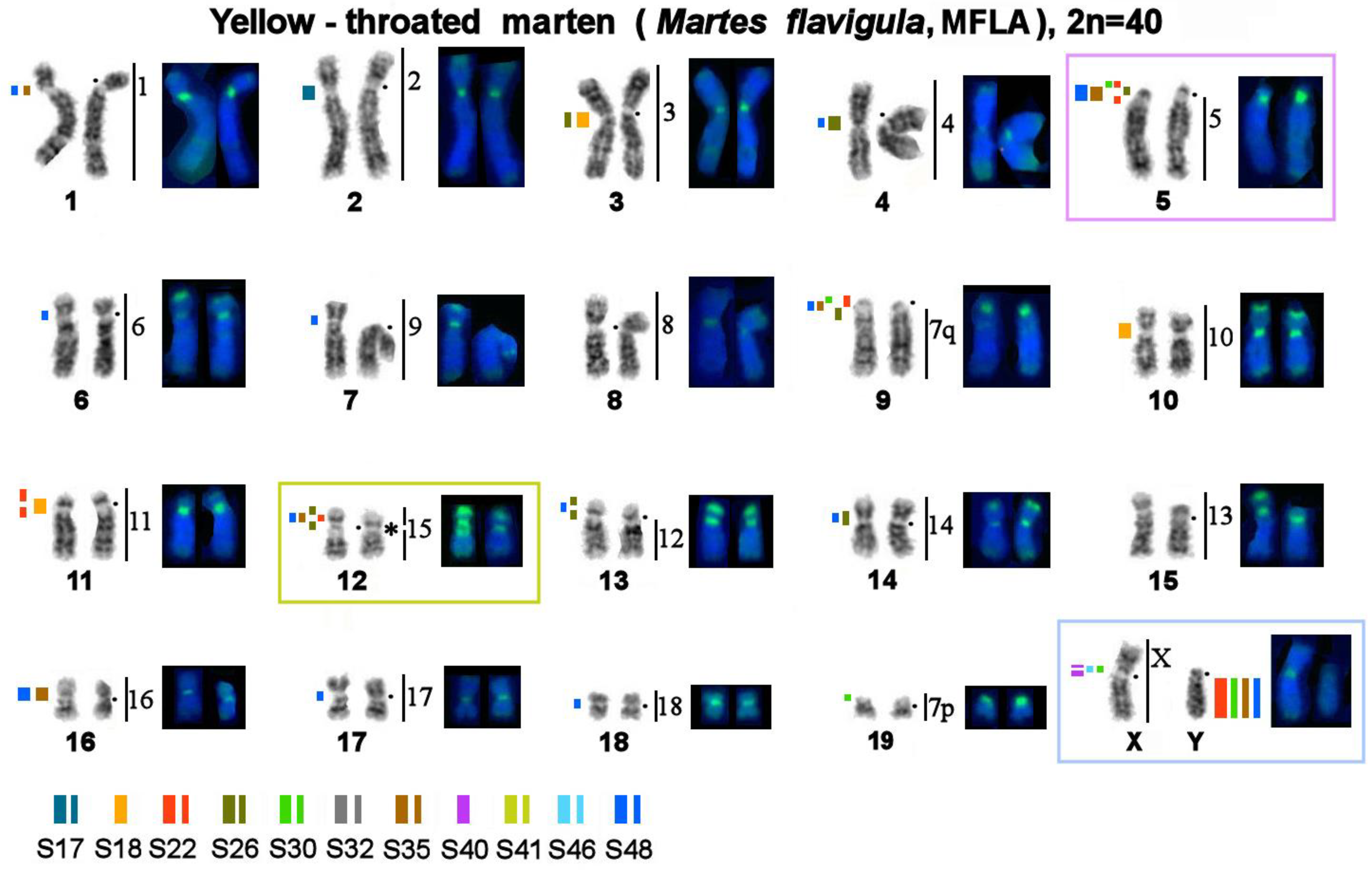
3. Results
3.1. Localization of MSRs in the Stone Marten Genome
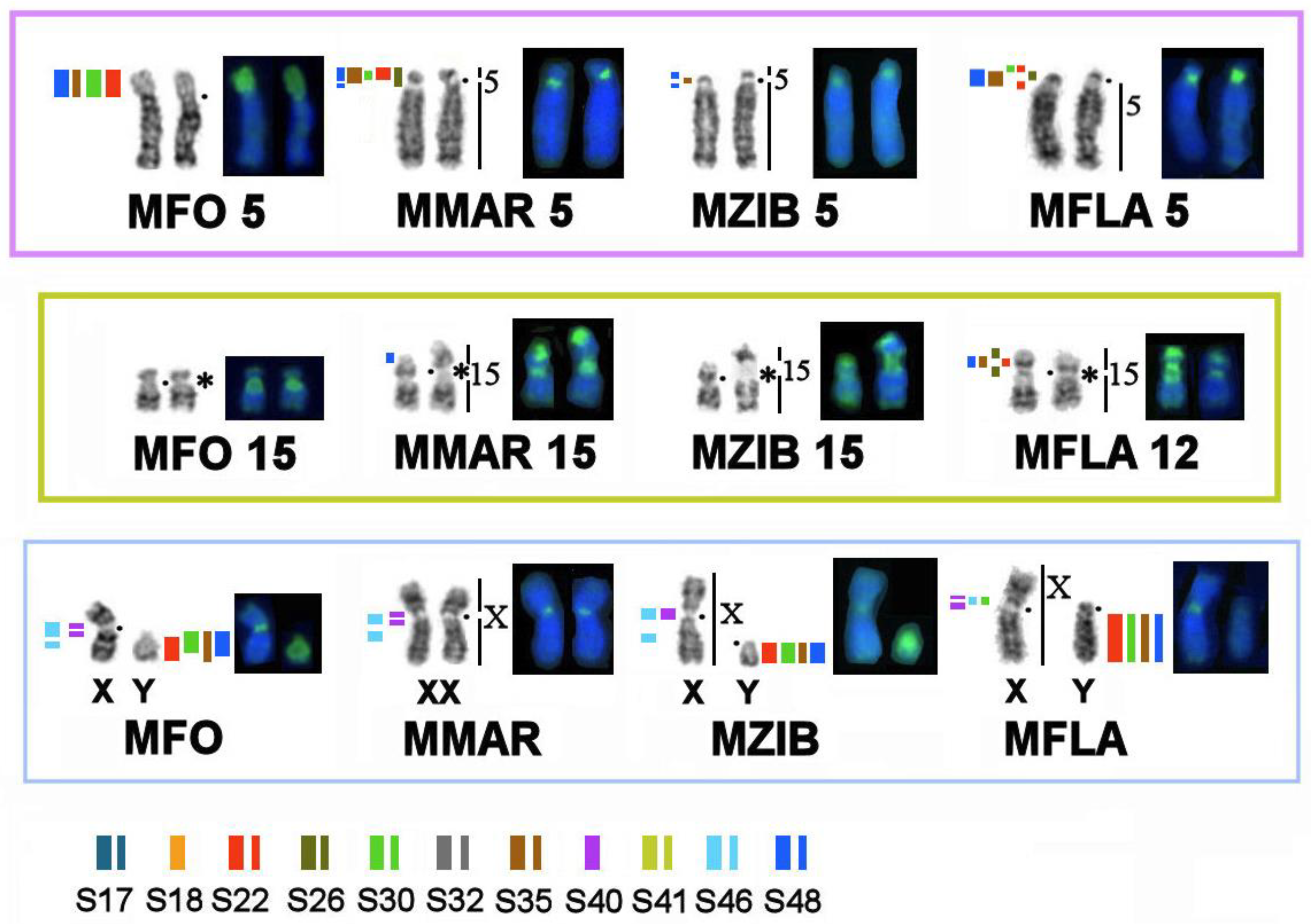
3.2. Homologous Elements in Karyotypes of the Stone Marten, Sable, Pine Marten, and Yellow-Throated Marten; Localization of Telomeric Sequences and Ribosomal Genes in Martes Species
3.3. Localization of MSRs in Genomes of the Pine Marten, Sable, and Yellow-Throated Marten
3.4. A Comparison of the Heterochromatin Segments Revealed by CDAG-Banding and by FISH with 11 MSRs’ Probes in the Four Martes Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Repetitive Elements in Humans. Int. J. Mol. Sci. 2021, 22, 2072. [Google Scholar] [CrossRef] [PubMed]
- Giacalone, J.; Friedes, J.; Francke, U. A novel GC–rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1992, 1, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Schaap, M.; Lemmers, R.J.L.F.; Maassen, R.; van der Vliet, P.J.; Hoogerheide, L.F.; van Dijk, H.K.; Baştürk, N.; de Knijff, P.; van der Maarel, S.M. Genome-Wide Analysis of Macrosatellite Repeat Copy Number Variation in Worldwide Populations: Evidence for Differences and Commonalities in Size Distributions and Size Restrictions. BMC Genom. 2013, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Dumbovic, G.; Forcales, S.-V.; Perucho, M. Emerging roles of macrosatellite repeats in genome organization and disease development. Epigenetics 2017, 12, 515–526. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Brahmachary, M.; Guilmatre, A.; Quilez, J.; Hasson, D.; Borel, C.; Warburton, P.; Sharp, A.J. Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats. PLoS Genet. 2014, 10, e1004418. [Google Scholar] [CrossRef]
- Plohl, M.; MešTrović, N.; Mravinac, B. Satellite DNA Evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef]
- Hartley, G.; O’Neill, R.J. Centromere Repeats: Hidden Gems of the Genome. Genes 2019, 10, 223. [Google Scholar] [CrossRef]
- Lu, W.; Zhang, Y.; Liu, D.; Songyang, Z.; Wan, M. Telomeres—Structure, Function, and Regulation. Exp. Cell Res. 2013, 319, 133–141. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar Organizer Regions: Genomic “Dark Matter” Requiring Illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Eickbush, D.G. Finely Orchestrated Movements: Evolution of the Ribosomal RNA. Genes Genet. 2007, 175, 477–485. [Google Scholar] [CrossRef]
- Clapp, J.; Mitchell, L.M.; Bolland, D.J.; Fantes, J.; Corcoran, A.E.; Scotting, P.J.; Armour, J.A.L.; Hewitt, J.E. Evolutionary Conservation of a Coding Function for D4Z4, the Tandem DNA Repeat Mutated in Facioscapulohumeral Muscular Dystrophy. Am. J. Hum. Genet. 2007, 81, 264–279. [Google Scholar] [CrossRef]
- Chadwick, B.P. Macrosatellite epigenetics: The two faces of DXZ4 and D4Z4. Chromosoma 2009, 118, 675–681. [Google Scholar] [CrossRef]
- Darrow, E.M.; Huntley, M.H.; Dudchenko, O.; Stamenova, E.K.; Durand, N.C.; Sun, Z.; Huang, S.-C.; Sanborn, A.L.; Machol, I.; Shamim, M.; et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. USA 2016, 113, E4504–E4512. [Google Scholar] [CrossRef]
- Adega, F.; Guedes-Pinto, H.; Chaves, R. Satellite DNA in the Karyotype Evolution of Domestic Animals—Clinical Considerations. Cytogenet. Genome Res. 2009, 126, 12–20. [Google Scholar] [CrossRef]
- Vieira-Da-Silva, A.; Louzada, S.; Adega, F.; Chaves, R. A High-Resolution Comparative Chromosome Map of Cricetus cricetus and Peromyscus eremicus Reveals the Involvement of Constitutive Heterochromatin in Breakpoint Regions. Cytogenet. Genome Res. 2015, 145, 59–67. [Google Scholar] [CrossRef]
- Weise, A.; Kosyakova, N.; Voigt, M.; Aust, N.; Mrasek, K.; Löhmer, S.; Rubtsov, N.; Karamysheva, T.V.; Trifonov, V.A.; Hardekopf, D.; et al. Comprehensive Analyses of White-Handed Gibbon Chromosomes Enables Access to 92 Evolutionary Conserved Breakpoints Compared to the Human Genome. Cytogenet. Genome Res. 2015, 145, 42–49. [Google Scholar] [CrossRef]
- Chaves, R.; Louzada, S.; Meles, S.; Wienberg, J.; Adega, F. Praomys tullbergi (Muridae, Rodentia) genome architecture decoded by comparative chromosome painting with Mus and Rattus. Chromosome Res. 2012, 20, 673–683. [Google Scholar] [CrossRef]
- Damas, J.; Corbo, M.; Kim, J.; Turner-Maier, J.; Farré, M.; Larkin, D.M.; Ryder, O.A.; Steiner, C.; Houck, M.L.; Hall, S.; et al. Evolution of the ancestral mammalian karyotype and syntenic regions. Proc. Natl. Acad. Sci. USA 2022, 119, e2209139119. [Google Scholar] [CrossRef]
- Smalec, B.M.; Heider, T.N.; Flynn, B.L.; O’Neill, R.J. A centromere satellite concomitant with extensive karyotypic diversity across the Peromyscus genus defies predictions of molecular drive. Chromosome Res. 2019, 27, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, C.; Ko, B.J.; Yoo, D.A.; Won, S.; Phillippy, A.M.; Fedrigo, O.; Zhang, G.; Howe, K.; Wood, J.; et al. False gene and chromosome losses in genome assemblies caused by GC content variation and repeats. Genome Biol. 2022, 23, 204. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Novák, P.; Robledillo, L.Á.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Vondrak, T.; Ávila Robledillo, L.; Novák, P.; Koblížková, A.; Neumann, P.; Macas, J. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef]
- Heard, E.; Johnson, A.D.; Korbel, J.O.; Lee, C.; Snyder, M.P.; Sturgill, D. The X Chromosome from Telomere to Telomere: Key Achievements and Future Opportunities. Fac. Rev. 2021, 10, 63. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Bradshaw, W.N.; Hsu, T.C. Chromosomes of Peromyscus (Rodentia, Cricetidae): 3. Polymorphism in Peromyscus maniculatus. Cytogenetics 1972, 11, 436–451. [Google Scholar] [CrossRef]
- Pathak, S.; Hsu, T.C.; Arrighi, F.E. Chromosomes of Peromyscus (Rodentia, Cricetidae): IV. The Role of Heterochromatin in Karyotypic Evolution. Cytogenet. Cell Genet. 1973, 12, 315–326. [Google Scholar] [CrossRef]
- Graphodatsky, A.S.; Ternovsky, D.V.; Isaenko, A.A.; Radhzabli, S.I. Constitutive heterochromatin and DNA content in some mustelids (Mustelidae, Carnivora). Genetika 1972, 13, 2123–2128. [Google Scholar]
- Graphodatsky, A.S.; Radjabli, S.I. Comparative cytogenetics of three canids species (Carnivora, Canidae): II. Distribution of C-heterochromatin. Genetika 1981, 17, 1504–1507. [Google Scholar]
- Wilson, D.E.; Reeder, D.M. Mammal Species of the World: A Taxonomic and Geographic Reference; JHU Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Wurster-Hill, D.H.; Centerwall, W.R. The interrelationships of chromosome banding patterns in canids, mustelids, hyena, and felids. Cytogenet. Genome Res. 1982, 34, 178–192. [Google Scholar] [CrossRef]
- Obara, Y. G-band homology and C-band variation in the Japanese mustelids, Mustela erminea nippon and M. sibirica itatsi. Genetica 1985, 68, 59–64. [Google Scholar] [CrossRef]
- Hameister, H.; Klett, C.; Bruch, J.; Dixkens, C.; Vogel, W.; Christensen, K. Zoo-FISH analysis: The American mink (Mustela vison) closely resembles the cat karyotype. Chromosome Res. 1997, 5, 5–11. [Google Scholar] [CrossRef]
- Rettenberger, G.; Klett, C.; Zechner, U.; Bruch, J.; Just, W.; Vogel, W.; Hameister, H. ZOO-FISH analysis: Cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chromosome Res. 1995, 3, 479–486. [Google Scholar] [CrossRef]
- Cavagna, P.; Menotti, A.; Stanyon, R. Genomic homology of the domestic ferret with cats and humans. Mamm. Genome 2000, 11, 866–870. [Google Scholar] [CrossRef]
- Nie, W.; Wang, J.; O’Brien, P.C.; Fu, B.; Ying, T.; Ferguson-Smith, M.A.; Yang, F. The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding. Chromosome Res. 2002, 10, 209–222. [Google Scholar] [CrossRef]
- Graphodatsky, A.S.; Yang, F.; Perelman, P.L.; O’Brien, P.C.M.; Serdukova, N.A.; Milne, B.S.; Biltueva, L.S.; Fu, B.; Vorobieva, N.V.; Kawada, S.-I.; et al. Comparative molecular cytogenetic studies in the order Carnivora: Mapping chromosomal rearrangements onto the phylogenetic tree. Cytogenet. Genome Res. 2002, 96, 137–145. [Google Scholar] [CrossRef]
- Breen, M.; Thomas, R.; Binns, M.M.; Carter, N.P.; Langford, C.F. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics 1999, 61, 145–155. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C.; Milne, B.S.; Graphodatsky, A.S.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.; Ferguson-Smith, M.A. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 1999, 62, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wang, J.; Su, W.; Wang, D.; Tanomtong, A.; Perelman, P.L.; Graphodatsky, A.S.; Yang, F. Chromosomal rearrangements and karyotype evolution in carnivores revealed by chromosome painting. Heredity 2012, 108, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Beklemisheva, V.R.; Perelman, P.L.; Lemskaya, N.A.; Proskuryakova, A.A.; Serdyukova, N.A.; Burkanov, V.N.; Gorshunov, M.B.; Ryder, O.; Thompson, M.; Lento, G.; et al. Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics. Genes 2020, 11, 1485. [Google Scholar] [CrossRef] [PubMed]
- Beklemisheva, V.R.; Perelman, P.L.; Lemskaya, N.A.; Kulemzina, A.I.; Proskuryakova, A.A.; Burkanov, V.N.; Graphodatsky, A.S. The Ancestral Carnivore Karyotype As Substantiated by Comparative Chromosome Painting of Three Pinnipeds, the Walrus, the Steller Sea Lion and the Baikal Seal (Pinnipedia, Carnivora). PLoS ONE 2016, 11, e0147647. [Google Scholar] [CrossRef]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 298, 971–972. [Google Scholar] [CrossRef]
- Lemskaya, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Biltueva, L.S.; Proskuryakova, A.A.; Hallenbeck, J.M.; Perelman, P.L.; Graphodatsky, A.S. A combined banding method that allows the reliable identification of chromosomes as well as differentiation of AT- and GC-rich heterochromatin. Chromosome Res. 2018, 26, 307–315. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Lisachov, A.; Andreyushkova, D.; Davletshina, G.; Prokopov, D.; Romanenko, S.; Galkina, S.; Saifitdinova, A.; Simonov, E.; Borodin, P.; Trifonov, V. Amplified Fragments of an Autosome-Borne Gene Constitute a Significant Component of the W Sex Chromosome of Eremias velox (Reptilia, Lacertidae). Genes 2021, 12, 779. [Google Scholar] [CrossRef]
- Romanenko, S.A.; Biltueva, L.S.; Serdyukova, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Gladkikh, O.L.; Lemskaya, N.A.; Interesova, E.A.; Korentovich, M.A.; Vorobieva, N.V.; et al. Segmental paleotetraploidy revealed in sterlet (Acipenser ruthenus) genome by chromosome painting. Mol. Cytogenet. 2015, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Law, C.J.; Slater, G.J.; Mehta, R.S. Lineage Diversity and Size Disparity in Musteloidea: Testing Patterns of Adaptive Radiation Using Molecular and Fossil-Based Methods. Syst. Biol. 2018, 67, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.; Perelman, P.L.; O’Brien, S.J. Atlas of Mammalian Chromosomes; Wiley Blackwell: Hoboken, NJ, USA, 2020. [Google Scholar]
- Graphodatsky, A.S.; Sharshov, A.A.; Ternovsky, D.V.; Ternovskaya, Y.G. Comparative cytogenetics of Mustelidae (Carnivora). Zool. Zhurnal 1989, 68, 96–106. [Google Scholar]
- Lushnikova, T.P.; Grafodatskiĭ, A.S.; Ivanov, S.V.; Romashchenko, A.G.; Ternovskiĭ, L.V.; Ternovskaia, I.G.; Radzhabli, S.I. EcoRI- and BamHI-families of repeated sequences in mustelids. Genetika 1989, 25, 1449–1461. [Google Scholar]
- Kim, J.; Farré, M.; Auvil, L.; Capitanu, B.; Larkin, D.M.; Ma, J.; Lewin, H.A. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E5379–E5388. [Google Scholar] [CrossRef]
- Hsu, T.C.; Rearden, H.H. Further karyological studies on Felidae. Chromosoma 1965, 16, 365–371. [Google Scholar] [CrossRef]
- Li, B.; Wolsan, M.; Wu, D.; Zhang, W.; Xu, Y.; Zeng, Z. Mitochondrial genomes reveal the pattern and timing of marten (Martes), wolverine (Gulo), and fisher (Pekania) diversification. Mol. Phylogenet. Evol. 2014, 80, 156–164. [Google Scholar] [CrossRef]
- Mestrovic, N.; Plohl, M.; Mravinac, B.; Ugarkovic, D. Evolution of satellite DNAs from the genus Palorus--experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef]
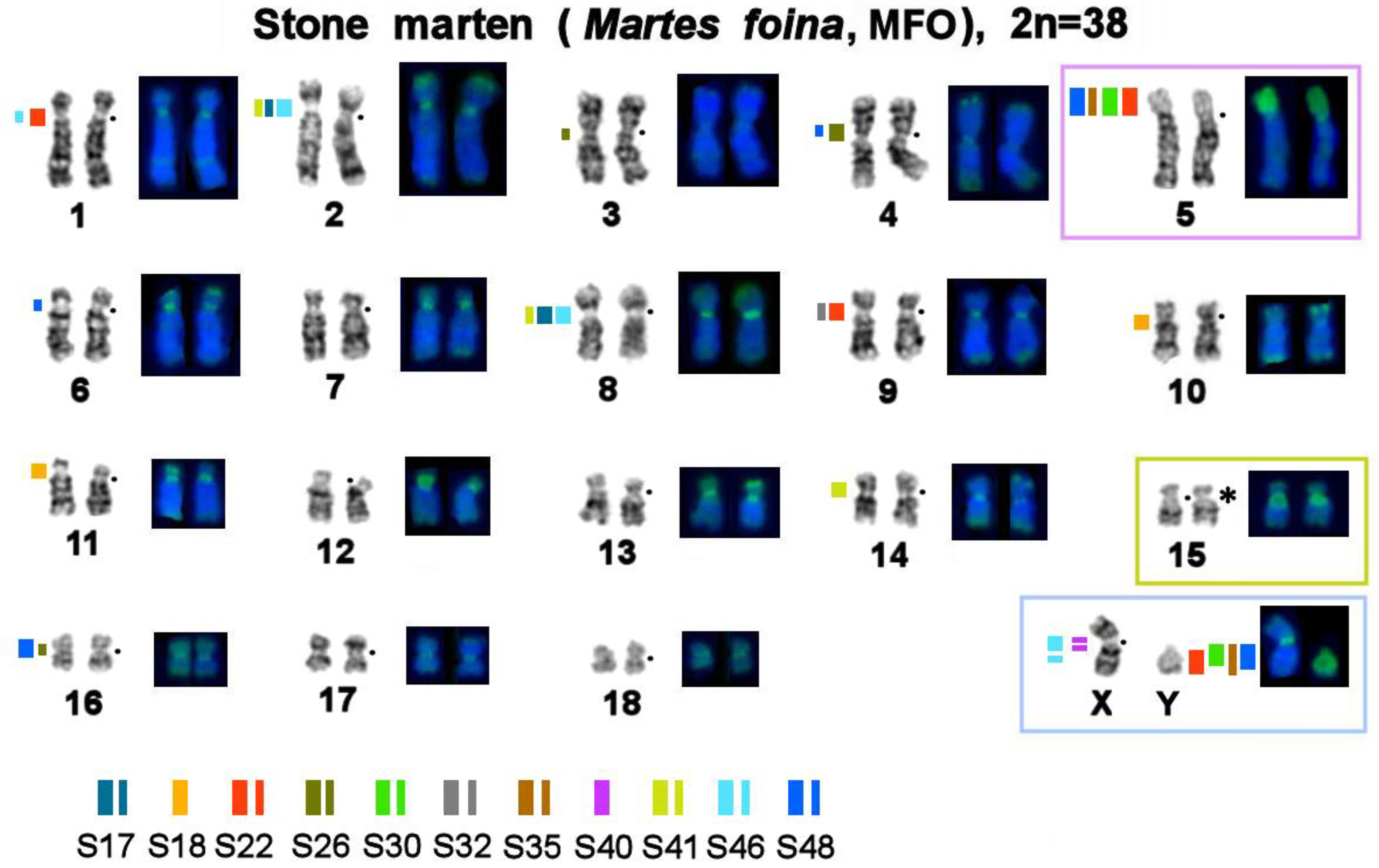

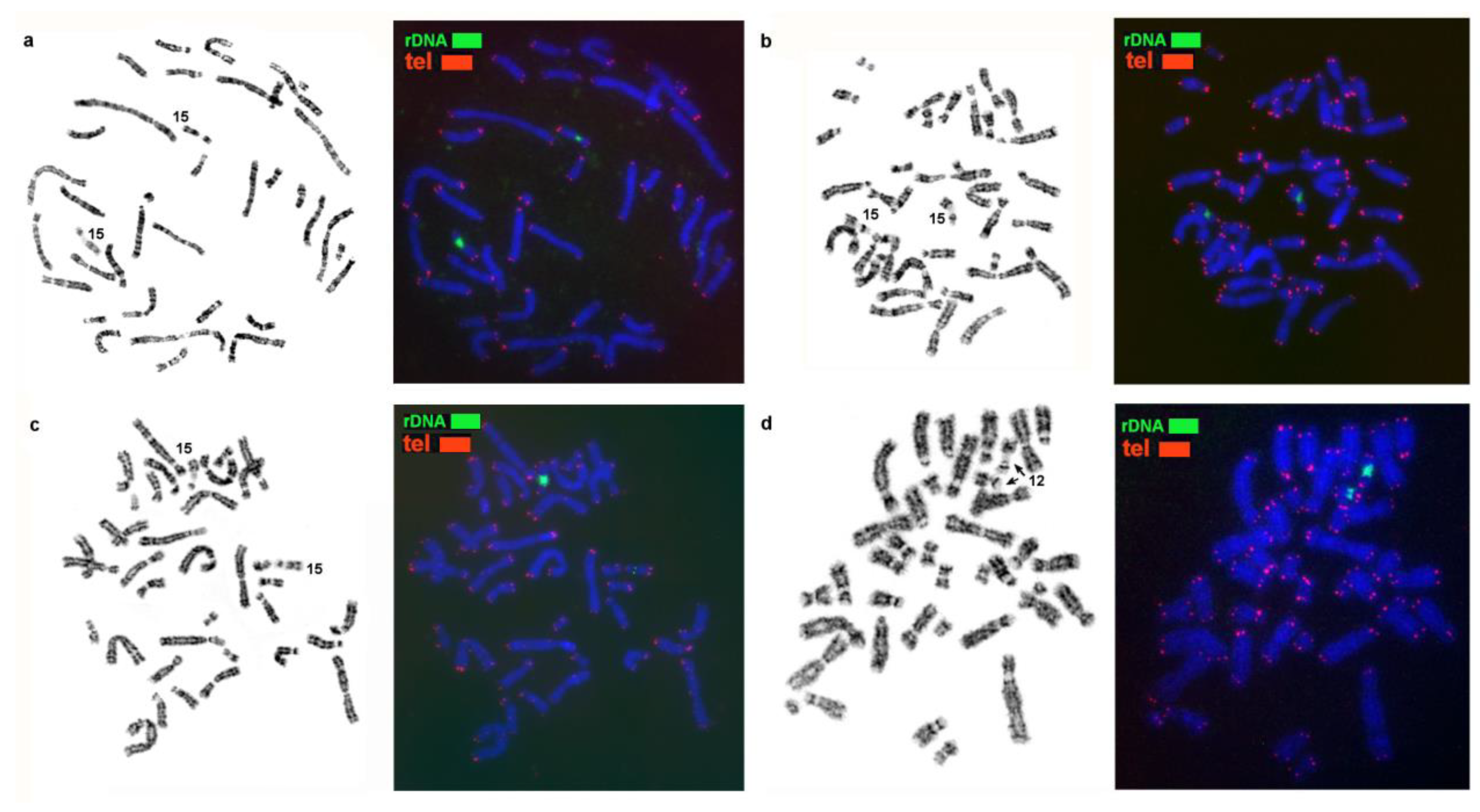
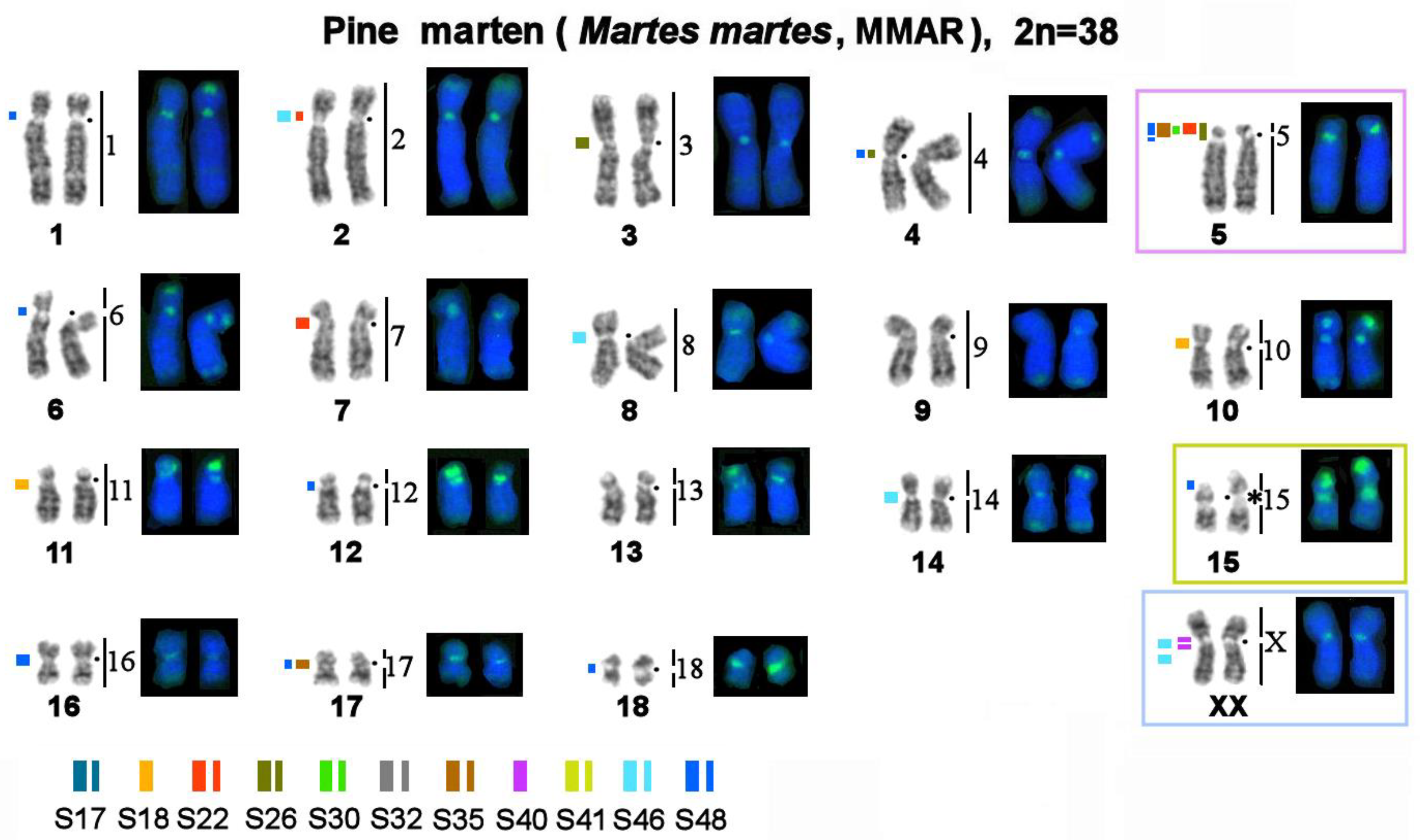
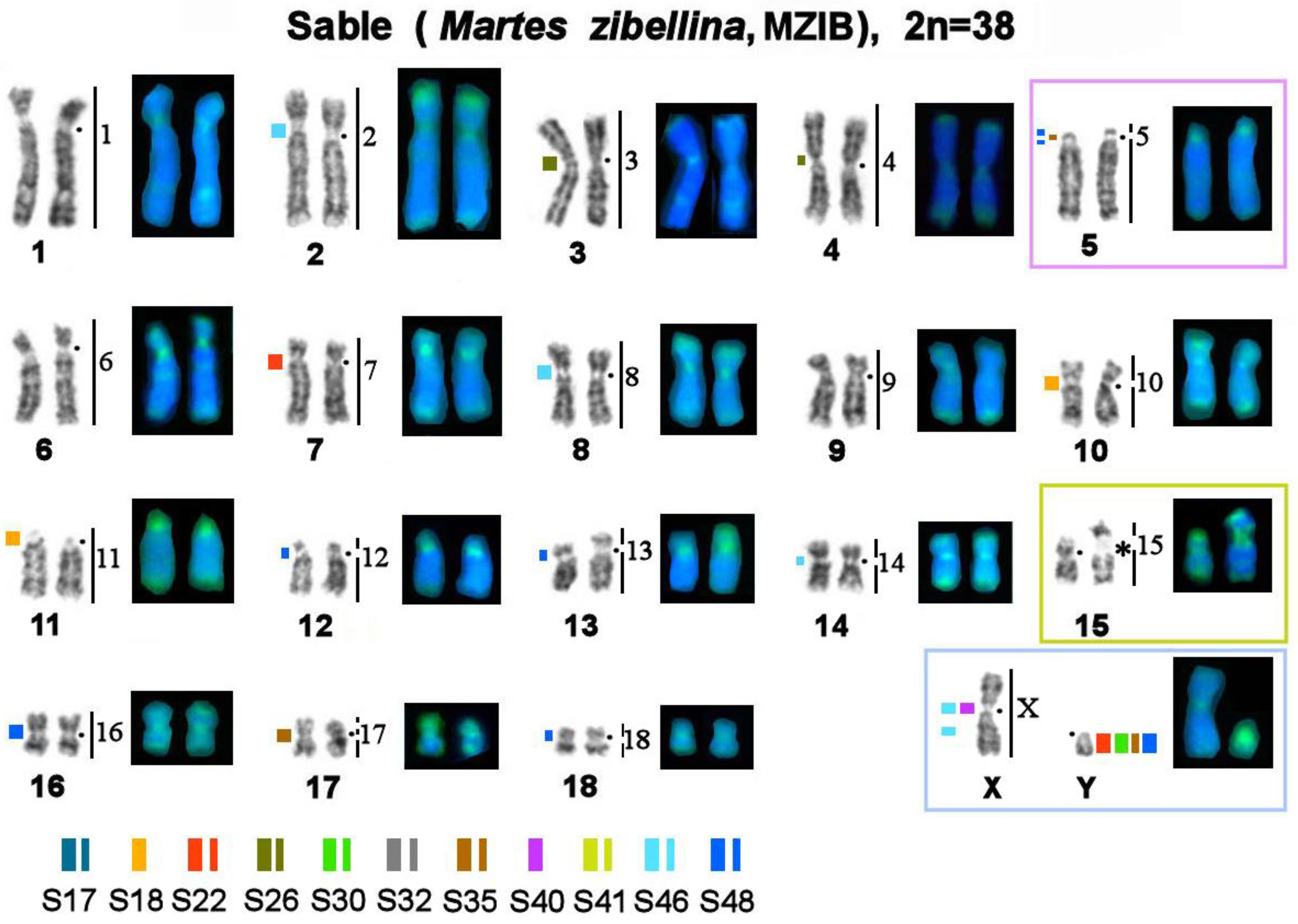
| No. | Scientific Name | Code | 2n | Sex | Common Name | Reference for FISH Data |
|---|---|---|---|---|---|---|
| 1 | M. foina | MFO | 38 | M | stone (beach) marten | [43] |
| 2 | M. flavigula | MFLA | 40 | M | yellow-throated marten | [39] |
| 3 | M. martes | MMAR | 38 | F | pine marten | this article |
| 4 | M. zibellina | MZIB | 38 | M | sable | this article |
| Repeat Name | Genome Proportion | Monomer Length, bp | GC Content | Homology to Known satDNA | Probe Length, bp |
|---|---|---|---|---|---|
| S17H | 0.3% | 714 | 60.22% | Mustela vison clone I225 microsatellite | 410 |
| S18H | 0.28% | 1041 | 70.61% | Mustela putorius 1080 bp Bam HI repeat | 449 |
| S22H | 0.25% | 986 | 69.68% | N/A | 269 |
| S26L | 0.140% | 1157 | 57.30% | N/A | 425 |
| S30H | 0.12% | 1580 | 68.35% | N/A | 320 |
| S32H | 0.11% | 1010 | 60.50% | N/A | 440 |
| S35H | 0.11% | 1148 | 68.12% | N/A | 447 |
| S40H | 0.098% | 2949 | 61.99% | N/A | 400 |
| S41H | 0.096% | 356 | 53.37% | M. vison clone I225 microsatellite | 181 |
| S46L | 0.079% | 1210 | 44.88% | N/A | 267 |
| S48H | 0.073% | 1146 | 65.27% | N/A | 371 |
| Repeat Name | MFO, Male ♂ | MMAR, Female ♀ | MZIB, Male ♂ | MFLA, Male ♂ |
|---|---|---|---|---|
| S17H | 2c, 8c | n/a | n/a | 2c |
| S18H | 10c, 11c | 10c, 11c | 10c, 11c | 3c, 10c, 11c |
| S22H | 1c, 5p, 9c, Yq | 2c, 5p, 7 | 7c, Yqprox | 5p, 5q, 7q-p, 11p, 11c, 15c, Yq |
| S26L | 3c, 4c, 6c | 3c, 4c, 5p, 5c | 3c, 4c | 3c, 4c, 5c, 7q-c, 12p, 12c, 14c, 15p, 15c |
| S30H | 5p, Yp, Yqprox | 5p | Yqprox | 5p, 7qp, 7p-p, Xc, Yq |
| S32H | 9c | n/a | n/a | n/a |
| S35H | 5p, Ypq | 5p, 7c | 5c, 17c, Yqprox | 5c, 7q-c, 15p, 16c, Yq |
| S40H | Xc | Xc | Xc | Xc |
| S41H | 2c, 8c, 14c | n/a | n/a | n/a |
| S46L | 1c, 2c, 8c, Xc, Xqint | 2c, 8c, 14c, Xc, Xqint | 2c, 8c, 14c, Xc, Xqint | Xc |
| S48H | 4c, 5p, 6c, 16c, Yp, Yqprox | 1c, 4c, 5p, 5c, 6c, 12c, 15p 16c, 17c, 18c | 5p, 5c, 12c, 13c, 16c, 18c, Yqprox | 1c, 4c, 5c, 6c, 7q-c, 9c, 12c, 14c, 15c, 16c, 17c, 18c, Yq |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beklemisheva, V.R.; Lemskaya, N.A.; Prokopov, D.Y.; Perelman, P.L.; Romanenko, S.A.; Proskuryakova, A.A.; Serdyukova, N.A.; Utkin, Y.A.; Nie, W.; Ferguson-Smith, M.A.; et al. Maps of Constitutive-Heterochromatin Distribution for Four Martes Species (Mustelidae, Carnivora, Mammalia) Show the Formative Role of Macrosatellite Repeats in Interspecific Variation of Chromosome Structure. Genes 2023, 14, 489. https://doi.org/10.3390/genes14020489
Beklemisheva VR, Lemskaya NA, Prokopov DY, Perelman PL, Romanenko SA, Proskuryakova AA, Serdyukova NA, Utkin YA, Nie W, Ferguson-Smith MA, et al. Maps of Constitutive-Heterochromatin Distribution for Four Martes Species (Mustelidae, Carnivora, Mammalia) Show the Formative Role of Macrosatellite Repeats in Interspecific Variation of Chromosome Structure. Genes. 2023; 14(2):489. https://doi.org/10.3390/genes14020489
Chicago/Turabian StyleBeklemisheva, Violetta R., Natalya A. Lemskaya, Dmitry Yu. Prokopov, Polina L. Perelman, Svetlana A. Romanenko, Anastasia A. Proskuryakova, Natalya A. Serdyukova, Yaroslav A. Utkin, Wenhui Nie, Malcolm A. Ferguson-Smith, and et al. 2023. "Maps of Constitutive-Heterochromatin Distribution for Four Martes Species (Mustelidae, Carnivora, Mammalia) Show the Formative Role of Macrosatellite Repeats in Interspecific Variation of Chromosome Structure" Genes 14, no. 2: 489. https://doi.org/10.3390/genes14020489
APA StyleBeklemisheva, V. R., Lemskaya, N. A., Prokopov, D. Y., Perelman, P. L., Romanenko, S. A., Proskuryakova, A. A., Serdyukova, N. A., Utkin, Y. A., Nie, W., Ferguson-Smith, M. A., Yang, F., & Graphodatsky, A. S. (2023). Maps of Constitutive-Heterochromatin Distribution for Four Martes Species (Mustelidae, Carnivora, Mammalia) Show the Formative Role of Macrosatellite Repeats in Interspecific Variation of Chromosome Structure. Genes, 14(2), 489. https://doi.org/10.3390/genes14020489








