Human Endogenous Retrovirus-K (HML-2)-Related Genetic Variation: Human Genome Diversity and Disease
Abstract
1. Introduction
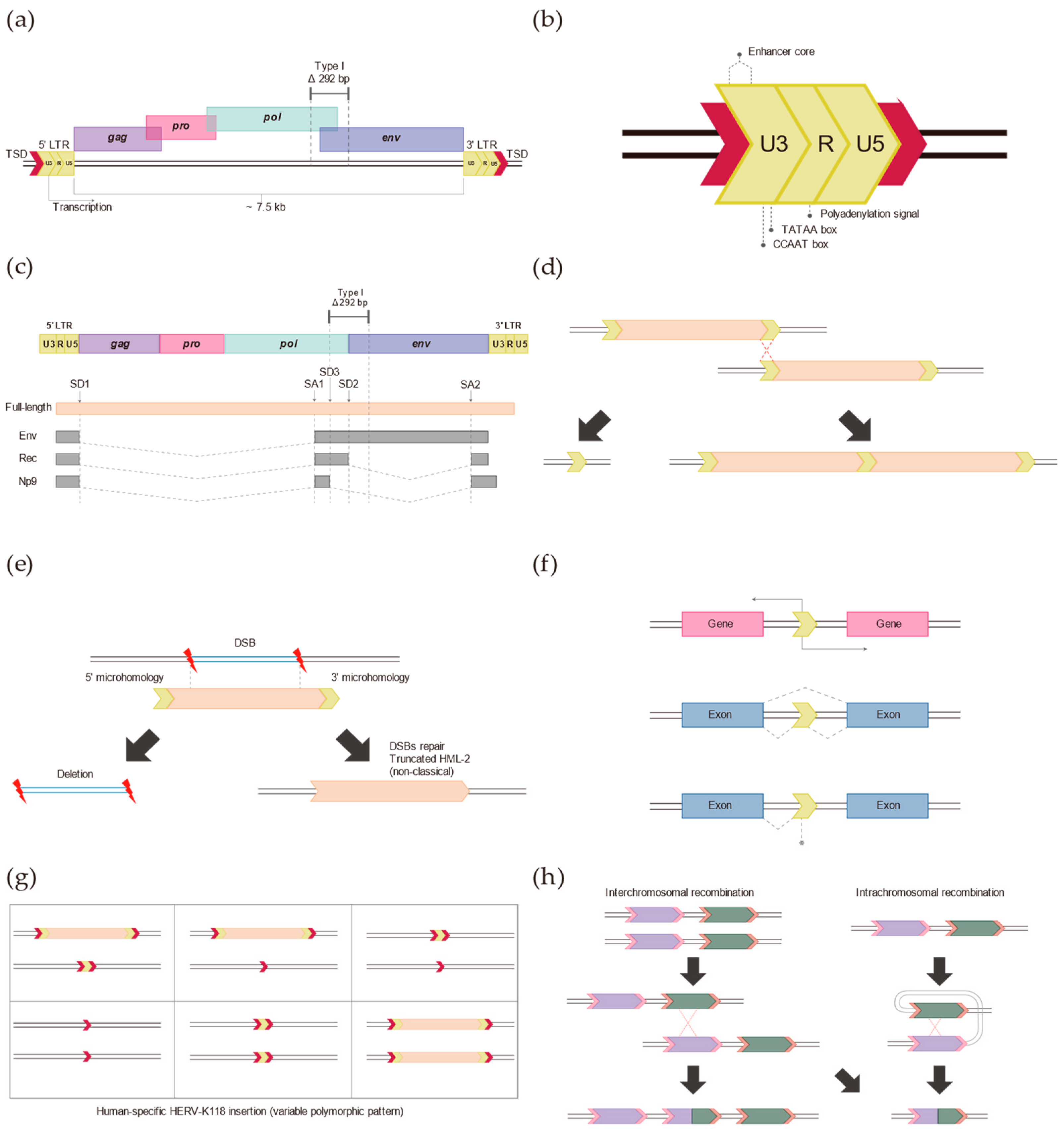
2. Typical HERV-K (HML-2)
3. Long Terminal Repeats (LTRs)
4. Insertion of HERV-K (HML-2) in the Human Genome
5. Discovery of Non-Reference HERV-K (HML-2)
6. Role of Non-Classical HERV-K (HML-2) Insertions in the Human Genome
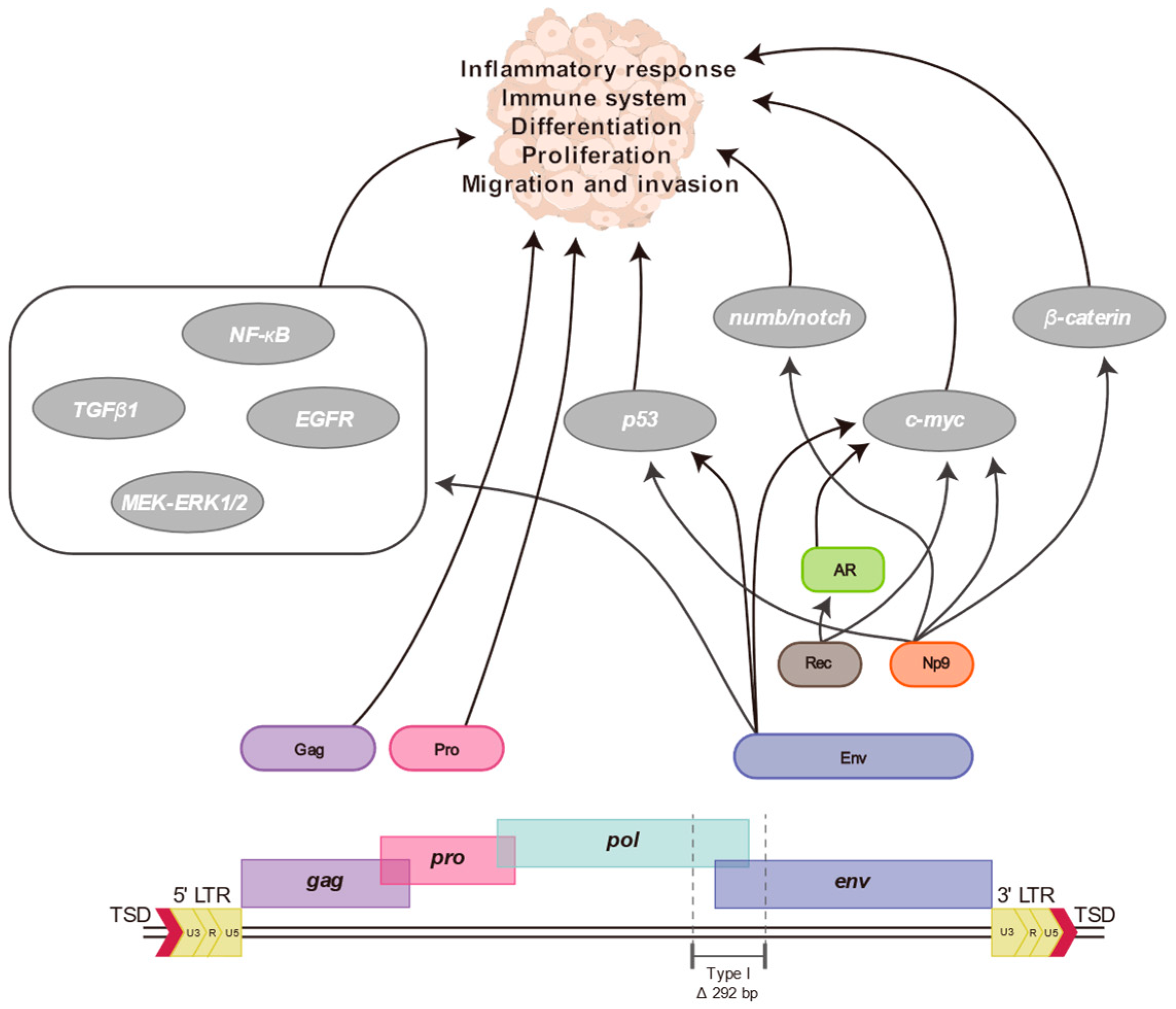
7. Proteins and Particles of HERV-K (HML-2)
8. Association between HERV-K (HML-2) and Human Diseases
9. HERV-K (HML-2) Transcriptome and Human Diseases
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.A.; DePaola, R.V. Human endogenous retroviruses: Our genomic fossils and companions. Physiol. Genom. 2023, 55, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Sechi, L.A.; Kelvin, D.J. Human Endogenous Retrovirus K (HML-2) in Health and Disease. Front. Microbiol. 2020, 11, 1690. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Curty, G.; Marston, J.L.; Rougvie, M.D.; Leal, F.E.; Nixon, D.F.; Soares, M.A. Human Endogenous Retrovirus K in Cancer: A Potential Biomarker and Immunotherapeutic Target. Viruses 2020, 12, 726. [Google Scholar] [CrossRef] [PubMed]
- Blikstad, V.; Benachenhou, F.; Sperber, G.O.; Blomberg, J. Evolution of human endogenous retroviral sequences: A conceptual account. Cell. Mol. Life Sci. 2008, 65, 3348–3365. [Google Scholar] [CrossRef]
- Dervan, E.; Bhattacharyya, D.D.; McAuliffe, J.D.; Khan, F.H.; Glynn, S.A. Ancient Adversary—HERV-K (HML-2) in Cancer. Front. Oncol. 2021, 11, 658489. [Google Scholar] [CrossRef]
- Alcazer, V.; Bonaventura, P.; Depil, S. Human Endogenous Retroviruses (HERVs): Shaping the Innate Immune Response in Cancers. Cancers 2020, 12, 610. [Google Scholar] [CrossRef]
- Gifford, R.; Tristem, M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 2003, 26, 291–315. [Google Scholar] [CrossRef]
- Greenwood, A.D.; Ishida, Y.; O’Brien, S.P.; Roca, A.L.; Eiden, M.V. Transmission, Evolution, and Endogenization: Lessons Learned from Recent Retroviral Invasions. Microbiol. Mol. Biol. Rev. 2018, 82, e00044-17. [Google Scholar] [CrossRef]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Trono, D. Dynamic control of endogenous retroviruses during development. Virology 2011, 411, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.C.; Lorincz, M.C. Silencing of endogenous retroviruses: When and why do histone marks predominate? Trends Biochem. Sci. 2012, 37, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Ayarpadikannan, S.; Kim, H.S. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genom. Inform. 2014, 12, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhu, W.; Zhou, J.; Li, H.; Xu, X.; Zhang, B.; Gao, X. Repetitive DNA sequence detection and its role in the human genome. Commun. Biol. 2023, 6, 954. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef]
- Yan, Y.; Buckler-White, A.; Wollenberg, K.; Kozak, C.A. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. USA 2009, 106, 3259–3263. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Gifford, R.J.; Bieniasz, P.D. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. eLife 2017, 6, e22519. [Google Scholar] [CrossRef]
- Mortelmans, K.; Wang-Johanning, F.; Johanning, G.L. The role of human endogenous retroviruses in brain development and function. APMIS 2016, 124, 105–115. [Google Scholar] [CrossRef]
- Bhat, R.K.; Rudnick, W.; Antony, J.M.; Maingat, F.; Ellestad, K.K.; Wheatley, B.M.; Tonjes, R.R.; Power, C. Human endogenous retrovirus-K(II) envelope induction protects neurons during HIV/AIDS. PLoS ONE 2014, 9, e97984. [Google Scholar] [CrossRef] [PubMed]
- Dembny, P.; Newman, A.G.; Singh, M.; Hinz, M.; Szczepek, M.; Kruger, C.; Adalbert, R.; Dzaye, O.; Trimbuch, T.; Wallach, T.; et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight 2020, 5, e131093. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, Q.; Cong, Y.S. Human endogenous retroviruses in development and disease. Comput. Struct. Biotechnol. J. 2021, 19, 5978–5986. [Google Scholar] [CrossRef] [PubMed]
- Buzdin, A.; Ustyugova, S.; Khodosevich, K.; Mamedov, I.; Lebedev, Y.; Hunsmann, G.; Sverdlov, E. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: Three master genes were active simultaneously during branching of hominoid lineages. Genomics 2003, 81, 149–156. [Google Scholar] [CrossRef]
- Gallahan, D.; Callahan, R. Mammary tumorigenesis in feral mice: Identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J. Virol. 1987, 61, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Kahyo, T.; Yamada, H.; Tao, H.; Kurabe, N.; Sugimura, H. Insertionally polymorphic sites of human endogenous retrovirus-K (HML-2) with long target site duplications. BMC Genom. 2017, 18, 487. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. S2), 14572–14579. [Google Scholar] [CrossRef]
- Montesion, M.; Bhardwaj, N.; Williams, Z.H.; Kuperwasser, C.; Coffin, J.M. Mechanisms of HERV-K (HML-2) Transcription during Human Mammary Epithelial Cell Transformation. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Contreras-Galindo, R.; Kaplan, M.H.; Dube, D.; Gonzalez-Hernandez, M.J.; Chan, S.; Meng, F.; Dai, M.; Omenn, G.S.; Gitlin, S.D.; Markovitz, D.M. Human Endogenous Retrovirus Type K (HERV-K) Particles Package and Transmit HERV-K-Related Sequences. J. Virol. 2015, 89, 7187–7201. [Google Scholar] [CrossRef]
- Shin, W.; Lee, J.; Son, S.Y.; Ahn, K.; Kim, H.S.; Han, K. Human-specific HERV-K insertion causes genomic variations in the human genome. PLoS ONE 2013, 8, e60605. [Google Scholar] [CrossRef] [PubMed]
- Hohn, O.; Hanke, K.; Bannert, N. HERV-K(HML-2), the Best Preserved Family of HERVs: Endogenization, Expression, and Implications in Health and Disease. Front. Oncol. 2013, 3, 246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Li, M.; Wei, Y.; Lin, K.; Lu, Y.; Shen, J.; Johanning, G.L.; Wang-Johanning, F. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 2016, 7, 84093–84117. [Google Scholar] [CrossRef] [PubMed]
- Agoni, L.; Guha, C.; Lenz, J. Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines. Front. Oncol. 2013, 3, 180. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.A.; Downey, R.F.; Seufert, C.J.; Schetter, A.; Dorsey, T.H.; Johnson, C.A.; Goldman, R.; Loffredo, C.A.; Yan, P.; Sullivan, F.J.; et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 2014, 35, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Buscher, K.; Trefzer, U.; Hofmann, M.; Sterry, W.; Kurth, R.; Denner, J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005, 65, 4172–4180. [Google Scholar] [CrossRef]
- Bergallo, M.; Montanari, P.; Mareschi, K.; Merlino, C.; Berger, M.; Bini, I.; Dapra, V.; Galliano, I.; Fagioli, F. Expression of the pol gene of human endogenous retroviruses HERV-K and -W in leukemia patients. Arch. Virol. 2017, 162, 3639–3644. [Google Scholar] [CrossRef]
- Wildschutte, J.H.; Williams, Z.H.; Montesion, M.; Subramanian, R.P.; Kidd, J.M.; Coffin, J.M. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. USA 2016, 113, E2326–E2334. [Google Scholar] [CrossRef]
- Moyes, D.; Griffiths, D.J.; Venables, P.J. Insertional polymorphisms: A new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007, 23, 326–333. [Google Scholar] [CrossRef]
- Macfarlane, C.; Simmonds, P. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 2004, 59, 642–656. [Google Scholar] [CrossRef]
- Martin, M.A.; Bryan, T.; Rasheed, S.; Khan, A.S. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 4892–4896. [Google Scholar] [CrossRef]
- Nelson, P.N.; Carnegie, P.R.; Martin, J.; Davari Ejtehadi, H.; Hooley, P.; Roden, D.; Rowland-Jones, S.; Warren, P.; Astley, J.; Murray, P.G. Demystified. Human endogenous retroviruses. Mol. Pathol. 2003, 56, 11–18. [Google Scholar] [CrossRef]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 8, 90. [Google Scholar] [CrossRef]
- Medstrand, P.; Mager, D.L. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 1998, 72, 9782–9787. [Google Scholar] [CrossRef]
- Montesion, M.; Williams, Z.H.; Subramanian, R.P.; Kuperwasser, C.; Coffin, J.M. Promoter expression of HERV-K (HML-2) provirus-derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology 2018, 15, 57. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The impact of transposable elements in adaptive evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, D.J. Retrotransposons. Curr. Biol. 2012, 22, R432–R437. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Tempel, S.; Jurka, J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene 2009, 448, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, D.J. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001, 2, REVIEWS1017. [Google Scholar] [CrossRef] [PubMed]
- Petrizzo, A.; Ragone, C.; Cavalluzzo, B.; Mauriello, A.; Manolio, C.; Tagliamonte, M.; Buonaguro, L. Human Endogenous Retrovirus Reactivation: Implications for Cancer Immunotherapy. Cancers 2021, 13, 1999. [Google Scholar] [CrossRef] [PubMed]
- Lower, R.; Lower, J.; Kurth, R. The viruses in all of us: Characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 1996, 93, 5177–5184. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Zeng, T.; Jia, L.; Yang, D.; Lin, S.L.; Sechi, L.A.; Kelvin, D.J. Identification of the distribution of human endogenous retroviruses K (HML-2) by PCR-based target enrichment sequencing. Retrovirology 2020, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Reus, K.; Mayer, J.; Sauter, M.; Zischler, H.; Muller-Lantzsch, N.; Meese, E. HERV-K(OLD): Ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 2001, 75, 8917–8926. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef]
- Holloway, J.R.; Williams, Z.H.; Freeman, M.M.; Bulow, U.; Coffin, J.M. Gorillas have been infected with the HERV-K (HML-2) endogenous retrovirus much more recently than humans and chimpanzees. Proc. Natl. Acad. Sci. USA 2019, 116, 1337–1346. [Google Scholar] [CrossRef]
- Fuentes, D.R.; Swigut, T.; Wysocka, J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife 2018, 7, e35989. [Google Scholar] [CrossRef]
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 2018, 26, 5–23. [Google Scholar] [CrossRef]
- Mustelin, T.; Ukadike, K.C. How Retroviruses and Retrotransposons in Our Genome May Contribute to Autoimmunity in Rheumatological Conditions. Front. Immunol. 2020, 11, 593891. [Google Scholar] [CrossRef]
- Groger, V.; Wieland, L.; Naumann, M.; Meinecke, A.C.; Meinhardt, B.; Rossner, S.; Ihling, C.; Emmer, A.; Staege, M.S.; Cynis, H. Formation of HERV-K and HERV-Fc1 Envelope Family Members is Suppressed on Transcriptional and Translational Level. Int. J. Mol. Sci. 2020, 21, 7855. [Google Scholar] [CrossRef] [PubMed]
- Balada, E.; Ordi-Ros, J.; Vilardell-Tarres, M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev. Med. Virol. 2009, 19, 273–286. [Google Scholar] [CrossRef]
- Magin, C.; Lower, R.; Lower, J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 1999, 73, 9496–9507. [Google Scholar] [CrossRef]
- Ryan, F.P. Human endogenous retroviruses in health and disease: A symbiotic perspective. J. R. Soc. Med. 2004, 97, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Tongyoo, P.; Avihingsanon, Y.; Prom-On, S.; Mutirangura, A.; Mhuantong, W.; Hirankarn, N. EnHERV: Enrichment analysis of specific human endogenous retrovirus patterns and their neighboring genes. PLoS ONE 2017, 12, e0177119. [Google Scholar] [CrossRef]
- Conley, A.B.; Piriyapongsa, J.; Jordan, I.K. Retroviral promoters in the human genome. Bioinformatics 2008, 24, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Reiss, D.; Zhang, Y.; Mager, D.L. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 2007, 35, 4743–4754. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, L.; Jiang, J.; Wang, Y.; Jiang, Y.; Yan, T.; Cao, Y. The structure and retrotransposition mechanism of LTR-retrotransposons in the asexual yeast Candida albicans. Virulence 2014, 5, 655–664. [Google Scholar] [CrossRef][Green Version]
- Manghera, M.; Douville, R.N. Endogenous retrovirus-K promoter: A landing strip for inflammatory transcription factors? Retrovirology 2013, 10, 16. [Google Scholar] [CrossRef]
- Trubetskoy, A.M.; Okenquist, S.A.; Lenz, J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J. Virol. 1999, 73, 3477–3483. [Google Scholar] [CrossRef]
- Liu, M.; Jia, L.; Li, H.; Liu, Y.; Han, J.; Wang, X.; Li, T.; Li, J.; Zhang, B.; Zhai, X.; et al. p53 Binding Sites in Long Terminal Repeat 5Hs (LTR5Hs) of Human Endogenous Retrovirus K Family (HML-2 Subgroup) Play Important Roles in the Regulation of LTR5Hs Transcriptional Activity. Microbiol. Spectr. 2022, 10, e0048522. [Google Scholar] [CrossRef] [PubMed]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Chen, T.; Meng, Z.; Gan, Y.; Wang, X.; Xu, F.; Gu, Y.; Xu, X.; Tang, J.; Zhou, H.; Zhang, X.; et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 2013, 27, 1469–1478. [Google Scholar] [CrossRef]
- Manghera, M.; Magnusson, A.; Douville, R.N. The sense behind retroviral anti-sense transcription. Virol. J. 2017, 14, 9. [Google Scholar] [CrossRef]
- Domansky, A.N.; Kopantzev, E.P.; Snezhkov, E.V.; Lebedev, Y.B.; Leib-Mosch, C.; Sverdlov, E.D. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 2000, 472, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Gebrie, A. Transposable elements as essential elements in the control of gene expression. Mob. DNA 2023, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Lenz, J. HERV-K HML-2 diversity among humans. Proc. Natl. Acad. Sci. USA 2016, 113, 4240–4242. [Google Scholar] [CrossRef]
- Rivas, S.R.; Valdez, M.J.M.; Govindarajan, V.; Seetharam, D.; Doucet-O’Hare, T.T.; Heiss, J.D.; Shah, A.H. The Role of HERV-K in Cancer Stemness. Viruses 2022, 14, 2019. [Google Scholar] [CrossRef]
- Ono, M.; Kawakami, M.; Ushikubo, H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J. Virol. 1987, 61, 2059–2062. [Google Scholar] [CrossRef]
- Goering, W.; Ribarska, T.; Schulz, W.A. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 2011, 32, 1484–1492. [Google Scholar] [CrossRef]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., 3rd; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Hong, Z.; Liu, H.; Chen, X.; Ding, L.; Liu, Z.; Zhou, F.; Yuan, Y. Human Endogenous Retroviruses-K (HML-2) Expression Is Correlated with Prognosis and Progress of Hepatocellular Carcinoma. Biomed. Res. Int. 2016, 2016, 8201642. [Google Scholar] [CrossRef] [PubMed]
- Hosseiniporgham, S.; Sechi, L.A. Anti-HERV-K Drugs and Vaccines, Possible Therapies against Tumors. Vaccines 2023, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.A.; van de Lagemaat, L.N.; Baillie, G.J.; Mager, D.L. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: The case of primate beta3GAL-T5. Gene 2005, 364, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, N.V.; Kraft, M.; Tondera, C.; Hanschmann, K.M.; Lower, J.; Lower, R. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J. Virol. 2011, 85, 3436–3448. [Google Scholar] [CrossRef] [PubMed]
- Manghera, M.; Ferguson-Parry, J.; Lin, R.; Douville, R.N. NF-kappaB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5′ Long Terminal Repeat. J. Virol. 2016, 90, 9338–9349. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Kusko, R.; Xiao, W.; Zheng, Y.; Liu, Z.; Xiao, C.; Sakkiah, S.; Guo, W.; Gong, P.; Zhang, C.; et al. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinform. 2019, 20, 101. [Google Scholar] [CrossRef]
- Shin, W.; Mun, S.; Kim, J.; Lee, W.; Park, D.G.; Choi, S.; Lee, T.Y.; Cha, S.; Han, K. Novel Discovery of LINE-1 in a Korean Individual by a Target Enrichment Method. Mol. Cells 2019, 42, 87–95. [Google Scholar] [CrossRef]
- Hughes, J.F.; Coffin, J.M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 2004, 101, 1668–1672. [Google Scholar] [CrossRef]
- Turner, G.; Barbulescu, M.; Su, M.; Jensen-Seaman, M.I.; Kidd, K.K.; Lenz, J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 2001, 11, 1531–1535. [Google Scholar] [CrossRef]
- Boissinot, S.; Chevret, P.; Furano, A.V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000, 17, 915–928. [Google Scholar] [CrossRef]
- Kahyo, T.; Tao, H.; Shinmura, K.; Yamada, H.; Mori, H.; Funai, K.; Kurabe, N.; Suzuki, M.; Tanahashi, M.; Niwa, H.; et al. Identification and association study with lung cancer for novel insertion polymorphisms of human endogenous retrovirus. Carcinogenesis 2013, 34, 2531–2538. [Google Scholar] [CrossRef]
- Wang, L.; Norris, E.T.; Jordan, I.K. Human Retrotransposon Insertion Polymorphisms Are Associated with Health and Disease via Gene Regulatory Phenotypes. Front. Microbiol. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Kang, Q.; Guo, X.; Li, T.; Yang, C.; Han, J.; Jia, L.; Liu, Y.; Wang, X.; Zhang, B.; Li, J.; et al. Identification of differentially expressed HERV-K(HML-2) loci in colorectal cancer. Front. Microbiol. 2023, 14, 1192900. [Google Scholar] [CrossRef]
- Douville, R.; Liu, J.; Rothstein, J.; Nath, A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 2011, 69, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, T.; Ebert, A.D.; Pritze, W.; Loddenkemper, C.; Schwartz, S.; Thiel, E. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res. Hum. Retroviruses 2004, 20, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.M.; van Marle, G.; Opii, W.; Butterfield, D.A.; Mallet, F.; Yong, V.W.; Wallace, J.L.; Deacon, R.M.; Warren, K.; Power, C. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 2004, 7, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Depil, S.; Roche, C.; Dussart, P.; Prin, L. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 2002, 16, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lee, M.H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef]
- Boller, K.; Schonfeld, K.; Lischer, S.; Fischer, N.; Hoffmann, A.; Kurth, R.; Tonjes, R.R. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 2008, 89, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.N.; Hooley, P.; Roden, D.; Davari Ejtehadi, H.; Rylance, P.; Warren, P.; Martin, J.; Murray, P.G.; Molecular Immunology Research, G. Human endogenous retroviruses: Transposable elements with potential? Clin. Exp. Immunol. 2004, 138, 1–9. [Google Scholar] [CrossRef]
- Bray, S.; Turnbull, M.; Hebert, S.; Douville, R.N. Insight into the ERVK Integrase—Propensity for DNA Damage. Front. Microbiol. 2016, 7, 1941. [Google Scholar] [CrossRef] [PubMed]
- Burn, A.; Roy, F.; Freeman, M.; Coffin, J.M. Widespread expression of the ancient HERV-K (HML-2) provirus group in normal human tissues. PLoS Biol. 2022, 20, e3001826. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Kim, S.; Lee, W.; Kang, K.; Meyer, T.J.; Han, B.G.; Han, K.; Kim, H.S. A study of transposable element-associated structural variations (TASVs) using a de novo-assembled Korean genome. Exp. Mol. Med. 2021, 53, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Witherspoon, D.J.; Xing, J.; Zhang, Y.; Watkins, W.S.; Batzer, M.A.; Jorde, L.B. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genom. 2010, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Badge, R.M.; Alisch, R.S.; Moran, J.V. ATLAS: A system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 2003, 72, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Streva, V.A.; Jordan, V.E.; Linker, S.; Hedges, D.J.; Batzer, M.A.; Deininger, P.L. Sequencing, identification and mapping of primed L1 elements (SIMPLE) reveals significant variation in full length L1 elements between individuals. BMC Genom. 2015, 16, 220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef]
- Sfeir, A.; Symington, L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef]
- Onozawa, M.; Zhang, Z.; Kim, Y.J.; Goldberg, L.; Varga, T.; Bergsagel, P.L.; Kuehl, W.M.; Aplan, P.D. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. USA 2014, 111, 7729–7734. [Google Scholar] [CrossRef]
- Mameli, G.; Erre, G.L.; Caggiu, E.; Mura, S.; Cossu, D.; Bo, M.; Cadoni, M.L.; Piras, A.; Mundula, N.; Colombo, E.; et al. Identification of a HERV-K env surface peptide highly recognized in Rheumatoid Arthritis (RA) patients: A cross-sectional case-control study. Clin. Exp. Immunol. 2017, 189, 127–131. [Google Scholar] [CrossRef]
- Tai, A.K.; O’Reilly, E.J.; Alroy, K.A.; Simon, K.C.; Munger, K.L.; Huber, B.T.; Ascherio, A. Human endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult. Scler. 2008, 14, 1175–1180. [Google Scholar] [CrossRef]
- Huang, G.; Li, Z.; Wan, X.; Wang, Y.; Dong, J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J. Carcinog. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Liu, J.; Rycaj, K.; Huang, M.; Tsai, K.; Rosen, D.G.; Chen, D.T.; Lu, D.W.; Barnhart, K.F.; Johanning, G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 2007, 120, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Dao Thi, V.L.; Denner, J. The transmembrane protein of the human endogenous retrovirus—K (HERV-K) modulates cytokine release and gene expression. PLoS ONE 2013, 8, e70399. [Google Scholar] [CrossRef] [PubMed]
- Greenig, M. HERVs, immunity, and autoimmunity: Understanding the connection. PeerJ 2019, 7, e6711. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, K.; Heyne, K.; Roemer, K.; Meese, E.; Mayer, J. HERV-K(HML-2) rec and np9 transcripts not restricted to disease but present in many normal human tissues. Mob. DNA 2015, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Hanke, K.; Chudak, C.; Kurth, R.; Bannert, N. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int. J. Cancer. 2013, 132, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Rigogliuso, G.; Biniossek, M.L.; Goodier, J.L.; Mayer, B.; Pereira, G.C.; Schilling, O.; Meese, E.; Mayer, J. A human endogenous retrovirus encoded protease potentially cleaves numerous cellular proteins. Mob. DNA 2019, 10, 36. [Google Scholar] [CrossRef]
- Boller, K.; Konig, H.; Sauter, M.; Mueller-Lantzsch, N.; Lower, R.; Lower, J.; Kurth, R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 1993, 196, 349–353. [Google Scholar] [CrossRef]
- Ruprecht, K.; Ferreira, H.; Flockerzi, A.; Wahl, S.; Sauter, M.; Mayer, J.; Mueller-Lantzsch, N. Human endogenous retrovirus family HERV-K(HML-2) RNA transcripts are selectively packaged into retroviral particles produced by the human germ cell tumor line Tera-1 and originate mainly from a provirus on chromosome 22q11.21. J. Virol. 2008, 82, 10008–10016. [Google Scholar] [CrossRef] [PubMed]
- Serafino, A.; Balestrieri, E.; Pierimarchi, P.; Matteucci, C.; Moroni, G.; Oricchio, E.; Rasi, G.; Mastino, A.; Spadafora, C.; Garaci, E.; et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell. Res. 2009, 315, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachar Badarinarayan, S.; Shcherbakova, I.; Langer, S.; Koepke, L.; Preising, A.; Hotter, D.; Kirchhoff, F.; Sparrer, K.M.J.; Schotta, G.; Sauter, D. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res. 2020, 48, 10890–10908. [Google Scholar] [CrossRef]
- Freimanis, G.; Hooley, P.; Ejtehadi, H.D.; Ali, H.A.; Veitch, A.; Rylance, P.B.; Alawi, A.; Axford, J.; Nevill, A.; Murray, P.G.; et al. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: Investigating mechanisms of pathogenesis. Clin. Exp. Immunol. 2010, 160, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Bergallo, M.; Marozio, L.; Botta, G.; Tancredi, A.; Dapra, V.; Galliano, I.; Montanari, P.; Coscia, A.; Benedetto, C.; Tovo, P.A. Human Endogenous Retroviruses Are Preferentially Expressed in Mononuclear Cells from Cord Blood Than From Maternal Blood and in the Fetal Part of Placenta. Front. Pediatr. 2020, 8, 244. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachar Badarinarayan, S.; Sauter, D. Not all viruses cause disease: HERV-K(HML-2) in healthy human tissues. PLoS Biol. 2022, 20, e3001884. [Google Scholar] [CrossRef]
- Muller, M.D.; Holst, P.J.; Nielsen, K.N. A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 1330. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Frost, A.R.; Johanning, G.L.; Khazaeli, M.B.; LoBuglio, A.F.; Shaw, D.R.; Strong, T.V. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin. Cancer Res. 2001, 7, 1553–1560. [Google Scholar]
- Kleiman, A.; Senyuta, N.; Tryakin, A.; Sauter, M.; Karseladze, A.; Tjulandin, S.; Gurtsevitch, V.; Mueller-Lantzsch, N. HERV-K(HML-2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int. J. Cancer 2004, 110, 459–461. [Google Scholar] [CrossRef]
- Sauter, M.; Schommer, S.; Kremmer, E.; Remberger, K.; Dolken, G.; Lemm, I.; Buck, M.; Best, B.; Neumann-Haefelin, D.; Mueller-Lantzsch, N. Human endogenous retrovirus K10: Expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 1995, 69, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Dolci, M.; Favero, C.; Toumi, W.; Favi, E.; Tarantini, L.; Signorini, L.; Basile, G.; Bollati, V.; D’Alessandro, S.; Bagnoli, P.; et al. Human Endogenous Retroviruses Long Terminal Repeat Methylation, Transcription, and Protein Expression in Human Colon Cancer. Front. Oncol. 2020, 10, 569015. [Google Scholar] [CrossRef]
- Li, M.; Radvanyi, L.; Yin, B.N.; Li, J.; Chivukula, R.; Lin, K.; Lu, Y.; Shen, J.J.; Chang, D.Z.; Li, D.H.; et al. Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral. RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth. Clin. Cancer Res. 2017, 23, 5892–5911. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Galindo, R.; Kaplan, M.H.; Leissner, P.; Verjat, T.; Ferlenghi, I.; Bagnoli, F.; Giusti, F.; Dosik, M.H.; Hayes, D.F.; Gitlin, S.D.; et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 2008, 82, 9329–9336. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Yang, Y.; Zhang, N.; Soto, C.; Jiang, X.; An, Z.; Zheng, W.J. Human Endogenous Retroviruses in Glioblastoma Multiforme. Microorganisms 2021, 9, 764. [Google Scholar] [CrossRef]
- Bo, M.; Niegowska, M.; Erre, G.L.; Piras, M.; Longu, M.G.; Manchia, P.; Manca, M.; Passiu, G.; Sechi, L.A. Rheumatoid arthritis patient antibodies highly recognize IL-2 in the immune response pathway involving IRF5 and EBV antigens. Sci. Rep. 2018, 8, 1789. [Google Scholar] [CrossRef]
- Posso-Osorio, I.; Tobón, G.J.; Cañas, C.A. Human endogenous retroviruses (HERV) and non-HERV viruses incorporated into the human genome and their role in the development of autoimmune diseases. J. Transl. Autoimmun. 2021, 4, 100137. [Google Scholar] [CrossRef]
- Papiri, G.; D’Andreamatteo, G.; Cacchiò, G.; Alia, S.; Silvestrini, M.; Paci, C.; Luzzi, S.; Vignini, A. Multiple Sclerosis: Inflammatory and Neuroglial Aspects. Curr. Issues Mol. Biol. 2023, 45, 1443–1470. [Google Scholar] [CrossRef]
- Costas, J. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J. Mol. Evol. 2001, 53, 237–243. [Google Scholar] [CrossRef]
- Keon, M.; Musrie, B.; Dinger, M.; Brennan, S.E.; Santos, J.; Saksena, N.K. Destination Amyotrophic Lateral Sclerosis. Front. Neurol. 2021, 12, 596006. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Simula, E.R.; Fathi, S.; McMahan, C.; Ghosal, A.; Berry, J.D.; Cudkowicz, M.; Elkahloun, A.; Johnson, K.; Norato, G.; et al. Antibody Response to HML-2 May Be Protective in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2022, 92, 782–792. [Google Scholar] [CrossRef]
- Mayer, J.; Harz, C.; Sanchez, L.; Pereira, G.C.; Maldener, E.; Heras, S.R.; Ostrow, L.W.; Ravits, J.; Batra, R.; Meese, E.; et al. Transcriptional profiling of HERV-K(HML-2) in amyotrophic lateral sclerosis and potential implications for expression of HML-2 proteins. Mol. Neurodegener. 2018, 13, 39. [Google Scholar] [CrossRef]
- Savage, A.L.; Schumann, G.G.; Breen, G.; Bubb, V.J.; Al-Chalabi, A.; Quinn, J.P. Retrotransposons in the development and progression of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 284–293. [Google Scholar] [CrossRef]
- Minchev, D.S.; Popov, N.T.; Naimov, S.I.; Minkov, I.N.; Vachev, T.I. Comparative Expression Analysis of Human Endogenous Retrovirus Elements in Peripheral Blood of Children with Specific Language Impairment. Balk. J. Med. Genet. 2019, 22, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Ko, E.J.; Song, K.S.; Ock, M.S.; Choi, Y.H.; Kim, S.; Kim, H.S.; Cha, H.J. Expression profiles of human endogenous retrovirus (HERV)-K and HERV-R Env proteins in various cancers. BMB Rep. 2021, 54, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Rhyu, D.W.; Kang, Y.J.; Ock, M.S.; Eo, J.W.; Choi, Y.H.; Kim, W.J.; Leem, S.H.; Yi, J.M.; Kim, H.S.; Cha, H.J. Expression of human endogenous retrovirus env genes in the blood of breast cancer patients. Int. J. Mol. Sci. 2014, 15, 9173–9183. [Google Scholar] [CrossRef]
- Zhao, J.; Rycaj, K.; Geng, S.; Li, M.; Plummer, J.B.; Yin, B.; Liu, H.; Xu, X.; Zhang, Y.; Yan, Y.; et al. Expression of Human Endogenous Retrovirus Type K Envelope Protein is a Novel Candidate Prognostic Marker for Human Breast Cancer. Genes Cancer 2011, 2, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Sci. Rep. 2017, 7, 41960. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Li, M.; Esteva, F.J.; Hess, K.R.; Yin, B.; Rycaj, K.; Plummer, J.B.; Garza, J.G.; Ambs, S.; Johanning, G.L. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int. J. Cancer 2014, 134, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Rycaj, K.; Plummer, J.B.; Li, M.; Yin, B.; Frerich, K.; Garza, J.G.; Shen, J.; Lin, K.; Yan, P.; et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl. Cancer Inst. 2012, 104, 189–210. [Google Scholar] [CrossRef]
- Zare, M.; Mostafaei, S.; Ahmadi, A.; Azimzadeh Jamalkandi, S.; Abedini, A.; Esfahani-Monfared, Z.; Dorostkar, R.; Saadati, M. Human endogenous retrovirus env genes: Potential blood biomarkers in lung cancer. Microb. Pathog. 2018, 115, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Goering, W.; Schmitt, K.; Dostert, M.; Schaal, H.; Deenen, R.; Mayer, J.; Schulz, W.A. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate 2015, 75, 1958–1971. [Google Scholar] [CrossRef]
- Thompson, I.M.; Ankerst, D.P. Prostate-specific antigen in the early detection of prostate cancer. Can. Med. Assoc. J. 2007, 176, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Muster, T.; Waltenberger, A.; Grassauer, A.; Hirschl, S.; Caucig, P.; Romirer, I.; Fodinger, D.; Seppele, H.; Schanab, O.; Magin-Lachmann, C.; et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003, 63, 8735–8741. [Google Scholar] [PubMed]
- Schmitt, K.; Reichrath, J.; Roesch, A.; Meese, E.; Mayer, J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol. Evol. 2013, 5, 307–328. [Google Scholar] [CrossRef]
- Kitsou, K.; Lagiou, P.; Magiorkinis, G. Human endogenous retroviruses in cancer: Oncogenesis mechanisms and clinical implications. J. Med. Virol. 2023, 95, e28350. [Google Scholar] [CrossRef]
- Fischer, S.; Echeverria, N.; Moratorio, G.; Landoni, A.I.; Dighiero, G.; Cristina, J.; Oppezzo, P.; Moreno, P. Human endogenous retrovirus np9 gene is over expressed in chronic lymphocytic leukemia patients. Leuk. Res. Rep. 2014, 3, 70–72. [Google Scholar] [CrossRef]
- Giebler, M.; Staege, M.S.; Blauschmidt, S.; Ohm, L.I.; Kraus, M.; Wurl, P.; Taubert, H.; Greither, T. Elevated HERV-K Expression in Soft Tissue Sarcoma Is Associated with Worsened Relapse-Free Survival. Front. Microbiol. 2018, 9, 211. [Google Scholar] [CrossRef]
- Ryan, F.P. Human Endogenous Retroviruses in Multiple Sclerosis: Potential for Novel Neuro-Pharmacological Research. Curr. Neuropharmacol. 2011, 9, 360–369. [Google Scholar] [CrossRef]
- Morris, G.; Maes, M.; Murdjeva, M.; Puri, B.K. Do Human Endogenous Retroviruses Contribute to Multiple Sclerosis, and if So, How? Mol. Neurobiol. 2019, 56, 2590–2605. [Google Scholar] [CrossRef]
- Karimi, A.; Esmaili, N.; Ranjkesh, M.; Zolfaghari, M.A. Expression of human endogenous retroviruses in pemphigus vulgaris patients. Mol. Biol. Rep. 2019, 46, 6181–6186. [Google Scholar] [CrossRef] [PubMed]
- Sicat, J.; Sutkowski, N.; Huber, B.T. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J. Rheumatol. 2005, 32, 1821–1831. [Google Scholar] [PubMed]
- Reynier, F.; Verjat, T.; Turrel, F.; Imbert, P.E.; Marotte, H.; Mougin, B.; Miossec, P. Increase in human endogenous retrovirus HERV-K (HML-2) viral load in active rheumatoid arthritis. Scand. J. Immunol. 2009, 70, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Frank, O.; Giehl, M.; Zheng, C.; Hehlmann, R.; Leib-Mosch, C.; Seifarth, W. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 2005, 79, 10890–10901. [Google Scholar] [CrossRef]
| Class | Genus | Family | Subgroups | |
| HERVs | Class I | Gammaretrovirus-like and epsilonretrovirus-like | HERV-W, HERV-H, HERV-F, HERV-P, HERV-E, HERV-R, HERV-T, HERV-I, ERV-FRD, ERV-FTD | |
| Class II | Betaretrovirus-like | HERV-K | HML 1-11 | |
| Class III | Spumaretrovirus-like | HERV-L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, W.; Mun, S.; Han, K. Human Endogenous Retrovirus-K (HML-2)-Related Genetic Variation: Human Genome Diversity and Disease. Genes 2023, 14, 2150. https://doi.org/10.3390/genes14122150
Shin W, Mun S, Han K. Human Endogenous Retrovirus-K (HML-2)-Related Genetic Variation: Human Genome Diversity and Disease. Genes. 2023; 14(12):2150. https://doi.org/10.3390/genes14122150
Chicago/Turabian StyleShin, Wonseok, Seyoung Mun, and Kyudong Han. 2023. "Human Endogenous Retrovirus-K (HML-2)-Related Genetic Variation: Human Genome Diversity and Disease" Genes 14, no. 12: 2150. https://doi.org/10.3390/genes14122150
APA StyleShin, W., Mun, S., & Han, K. (2023). Human Endogenous Retrovirus-K (HML-2)-Related Genetic Variation: Human Genome Diversity and Disease. Genes, 14(12), 2150. https://doi.org/10.3390/genes14122150






