Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell and Culture Conditions
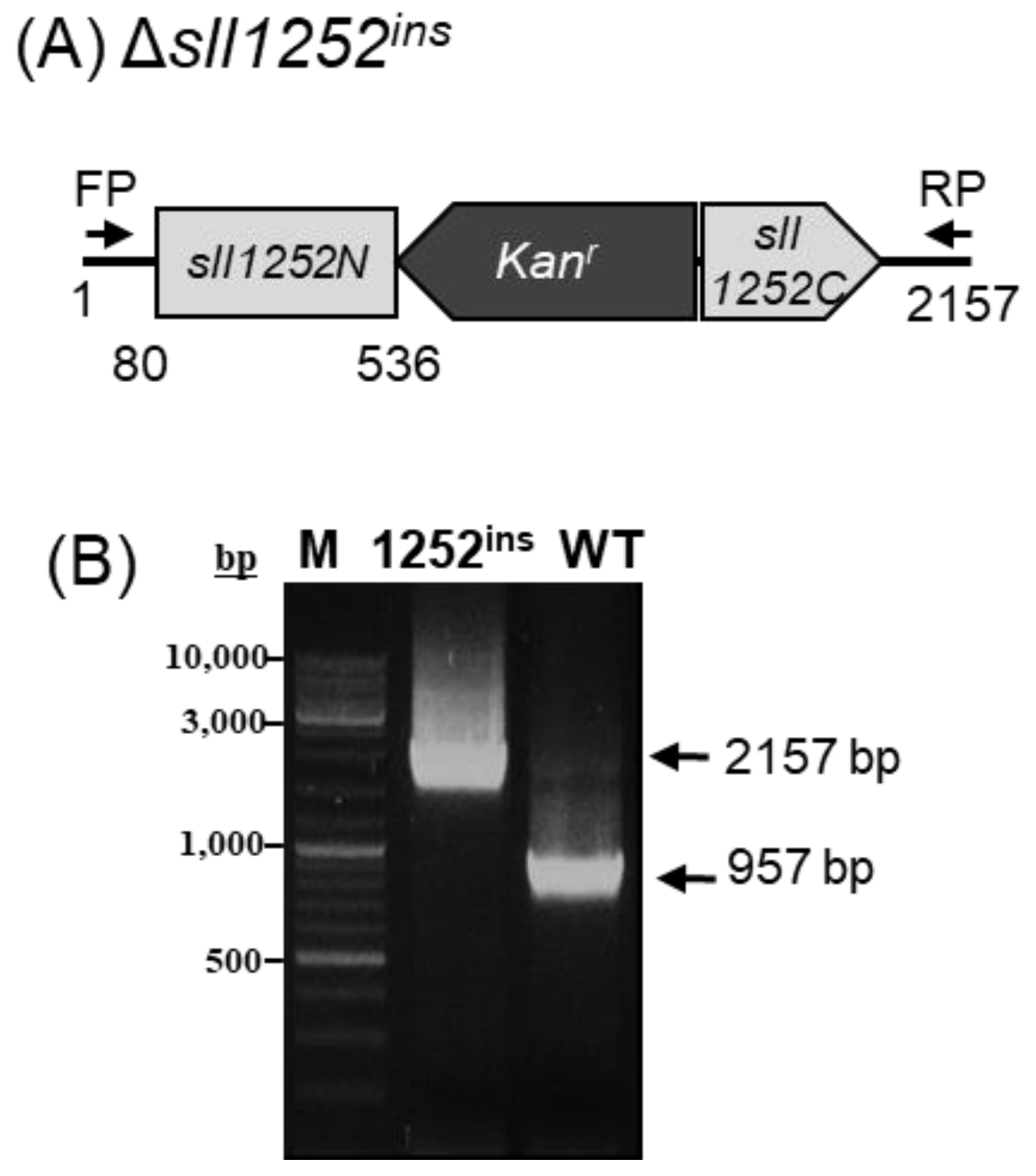
2.2. Insertional Inactivation of sll1252 by Transposon Mutagenesis (Δsll1252ins)
2.3. Screening for Spontaneous Suppressor Mutants of Δsll1252ins
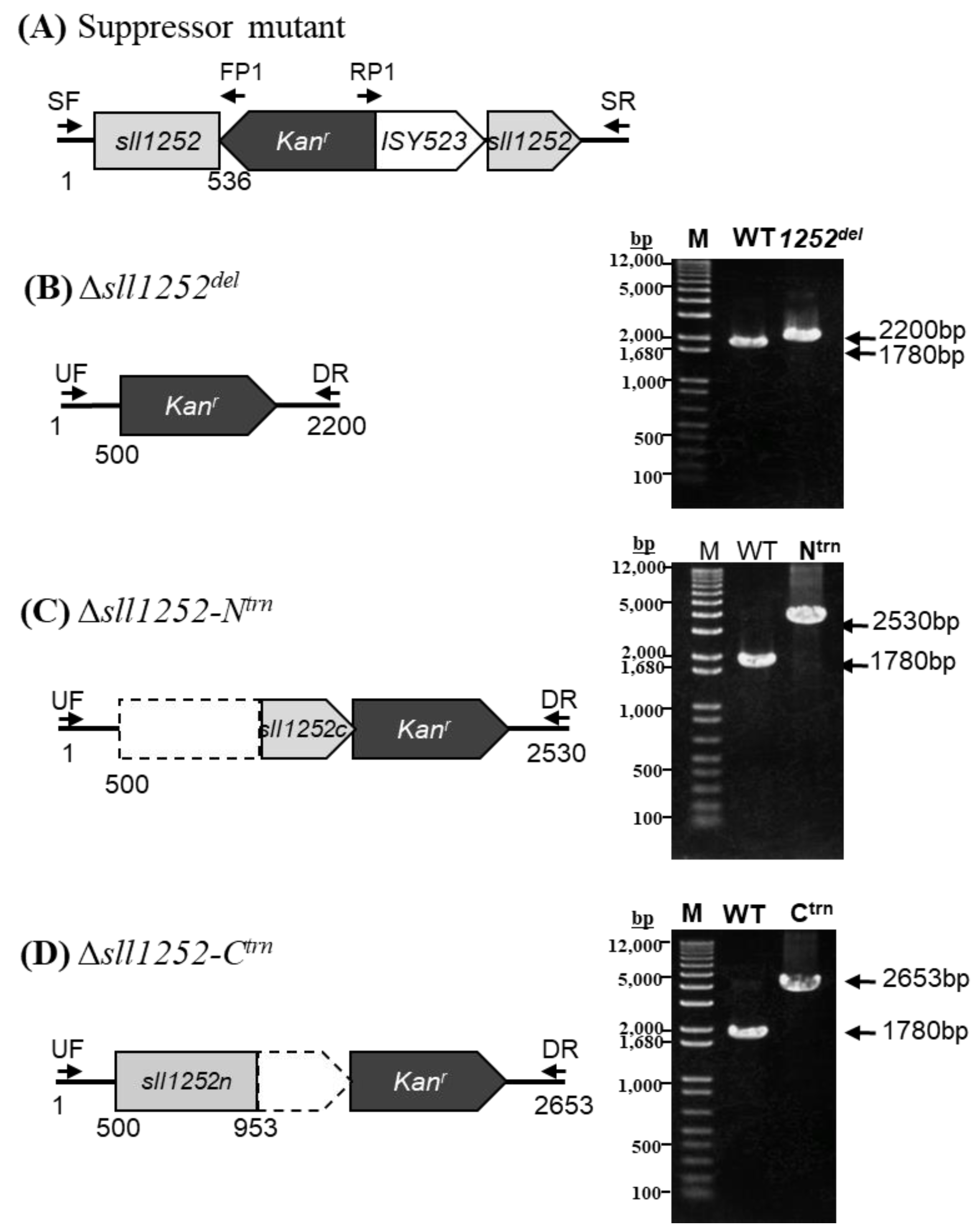
2.4. Deletional Mutagenesis of the sll1252 Gene (Δsll1252del)
2.5. Construction of sll1252 N-Terminal Truncated Mutant (Δsll1252-Ntrn)
2.6. Construction of sll1252 C-Terminal Truncated Mutant (Δsll1252-Ctrn)
2.7. Measurement of Photosynthetic Evolution of Oxygen
2.8. Preparation of cDNA for DNA Microarray Analysis
2.9. DNA Microarray Analysis
2.10. Transmission Electron Microscopy
3. Results
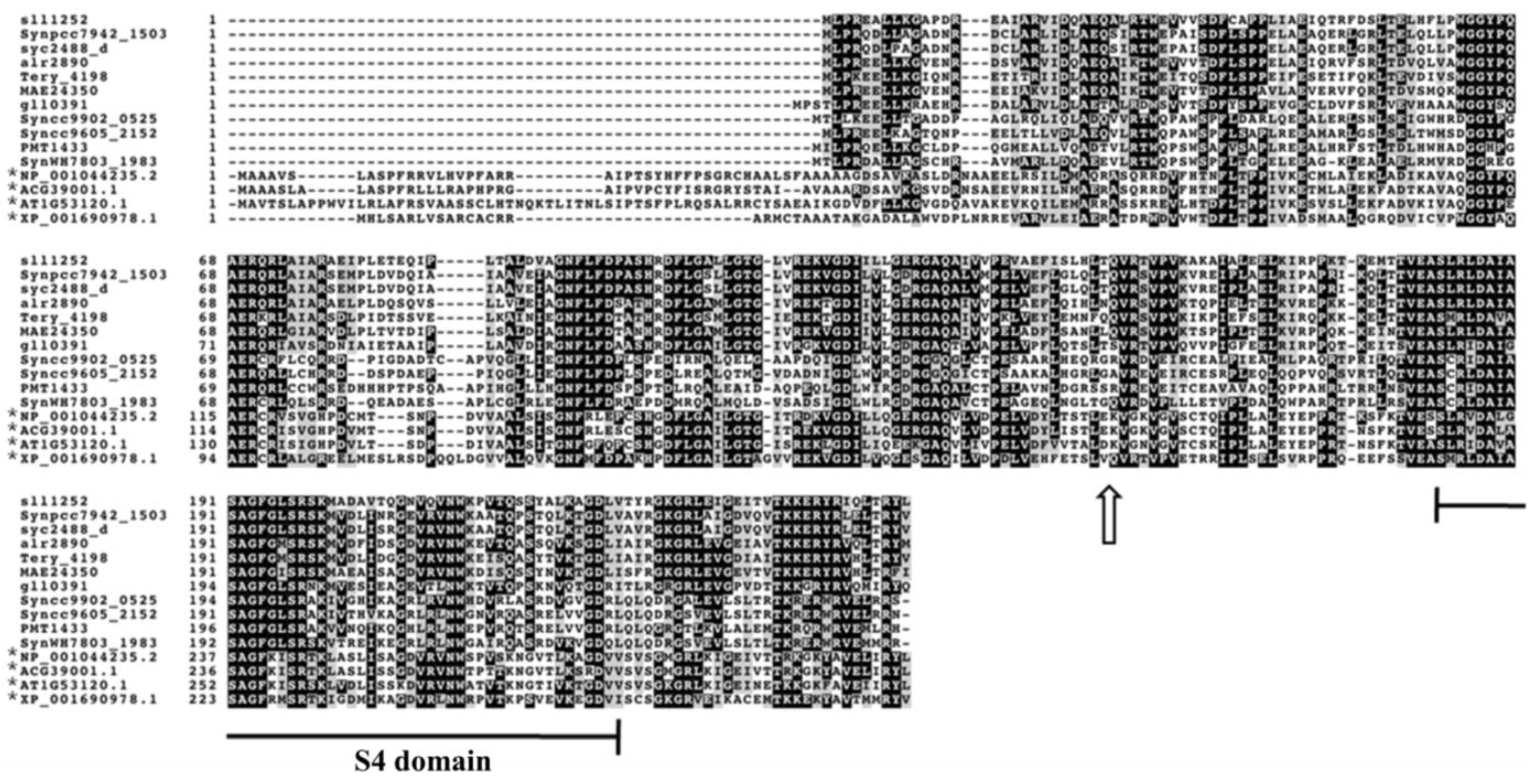
3.1. Sll1252 Is a Conserved S4-Domain Containing Protein
3.2. Mutagenesis of sll1252 in Synechocystis
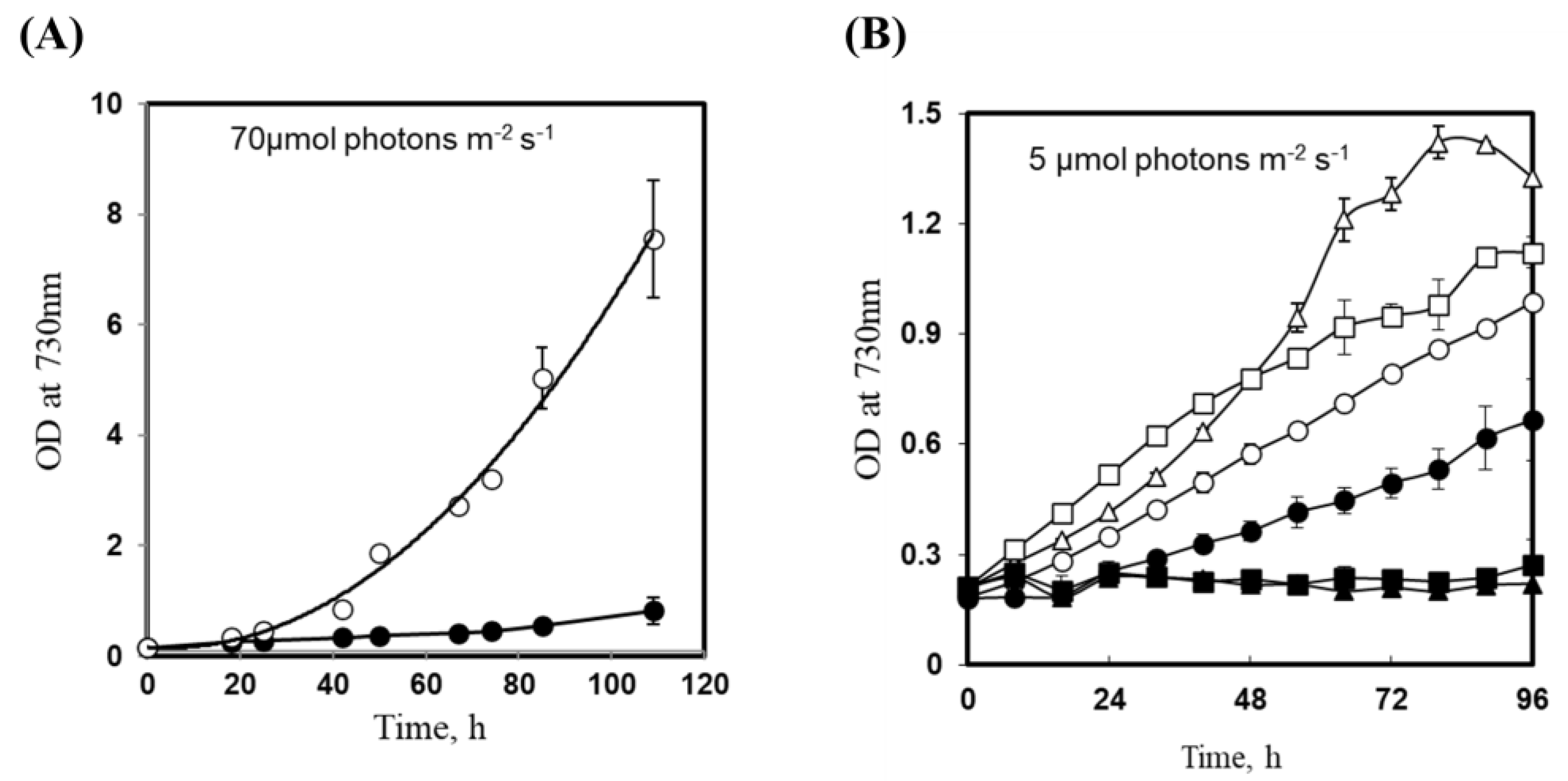
3.3. Insertional Inactivation of sll1252 Influences Growth at Optimal Light
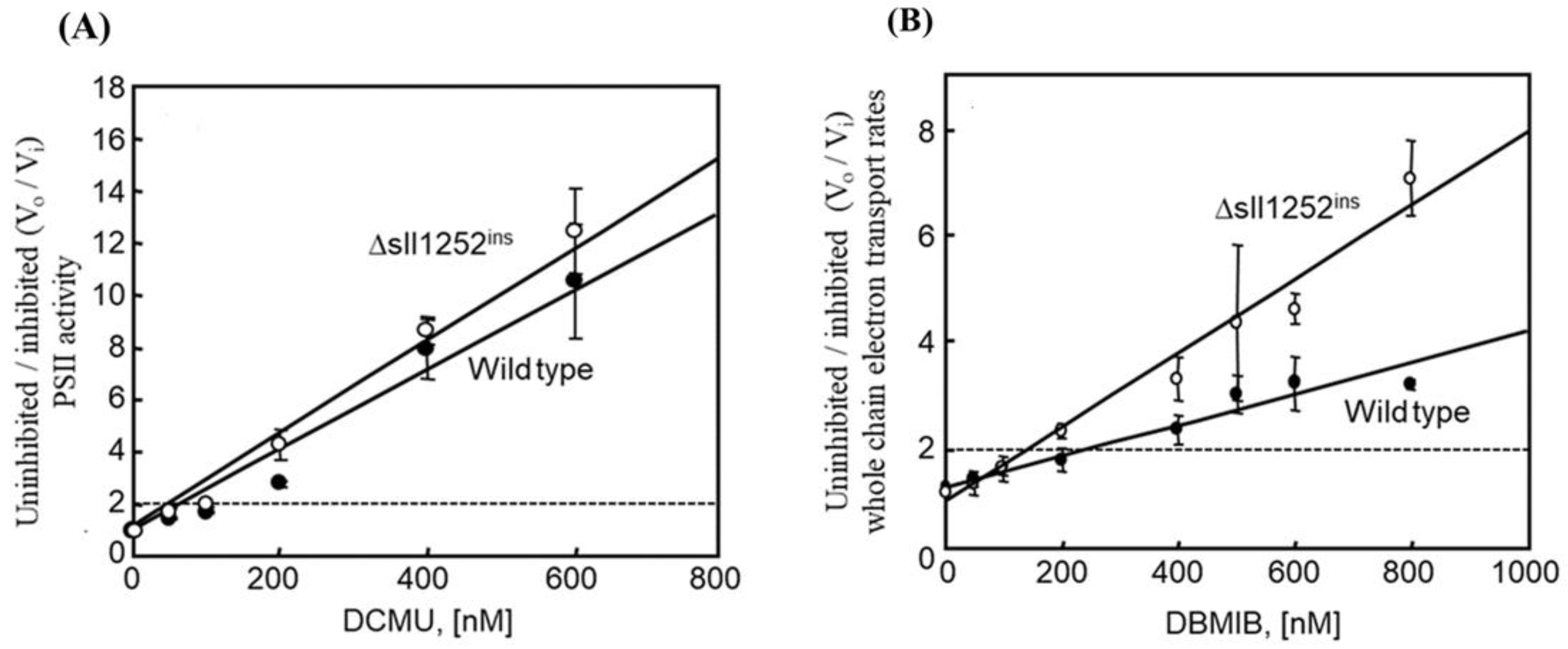
3.4. Insertional Inactivation of sll1252 Alters the Rate of Whole Chain Electron Transport
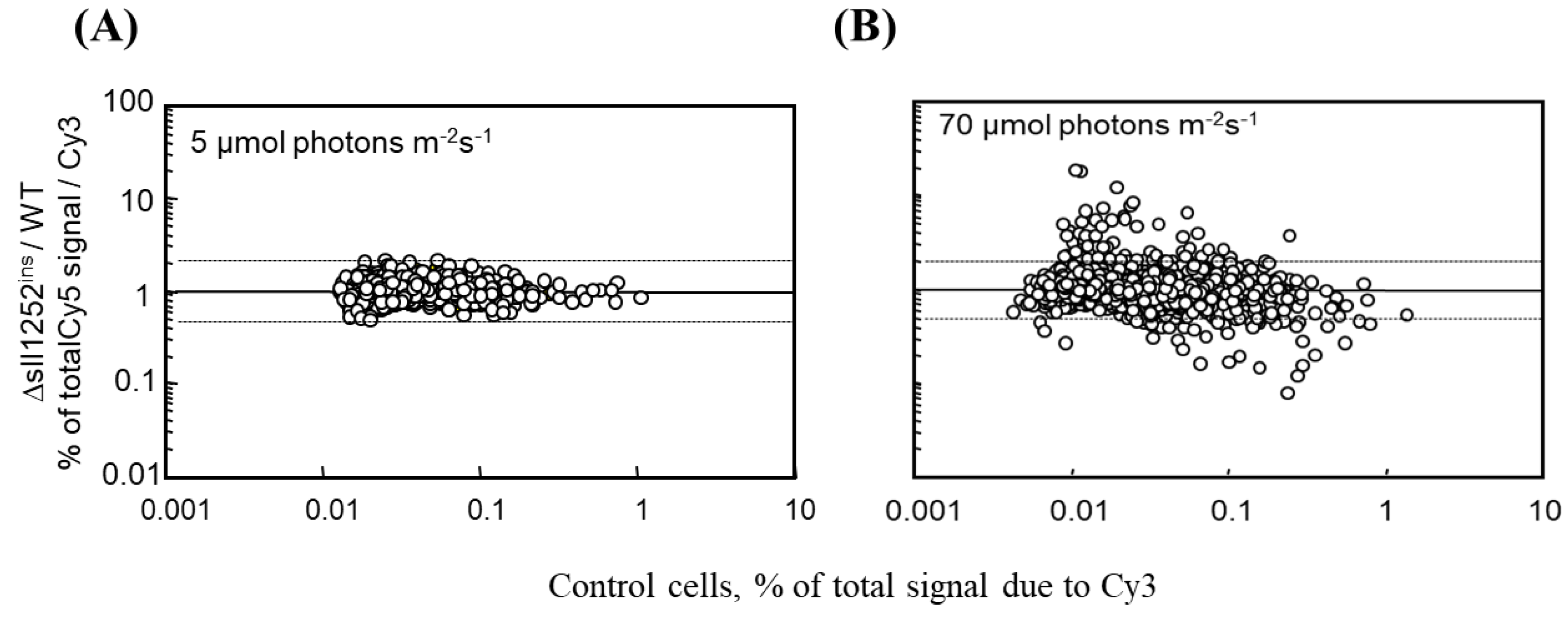
3.5. The Global Gene Expression Profiles of DBMIB-Treated Wild-Type and Δsll1252ins Cells Grown under Optimal Light Are Similar
3.6. Δsll1252ins Cells Are More Sensitive to Inhibition by DBMIB Than Wild-Type Cells
3.7. Suppressor Mutants of Δsll1252ins (Δsll1252ins-Rev)
3.8. Truncated and Deletional Mutants of sll1252 (Δsll1252Ntrn, Δsll1252Ctrn, and Δsll1252del)
3.9. Expression of Only C-Terminal Region of sll1252 Influences Growth of the Cells
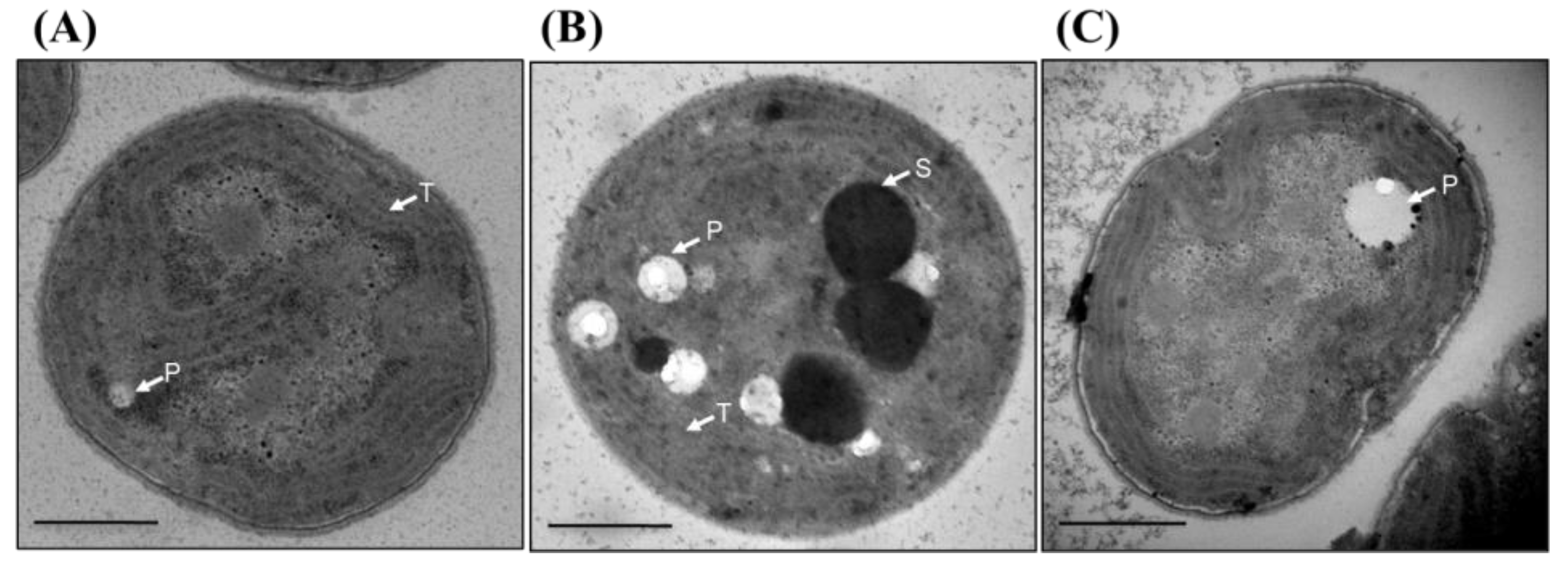
3.10. Thylakoid Membrane Is Disorganized in Δsll1252Ntrn
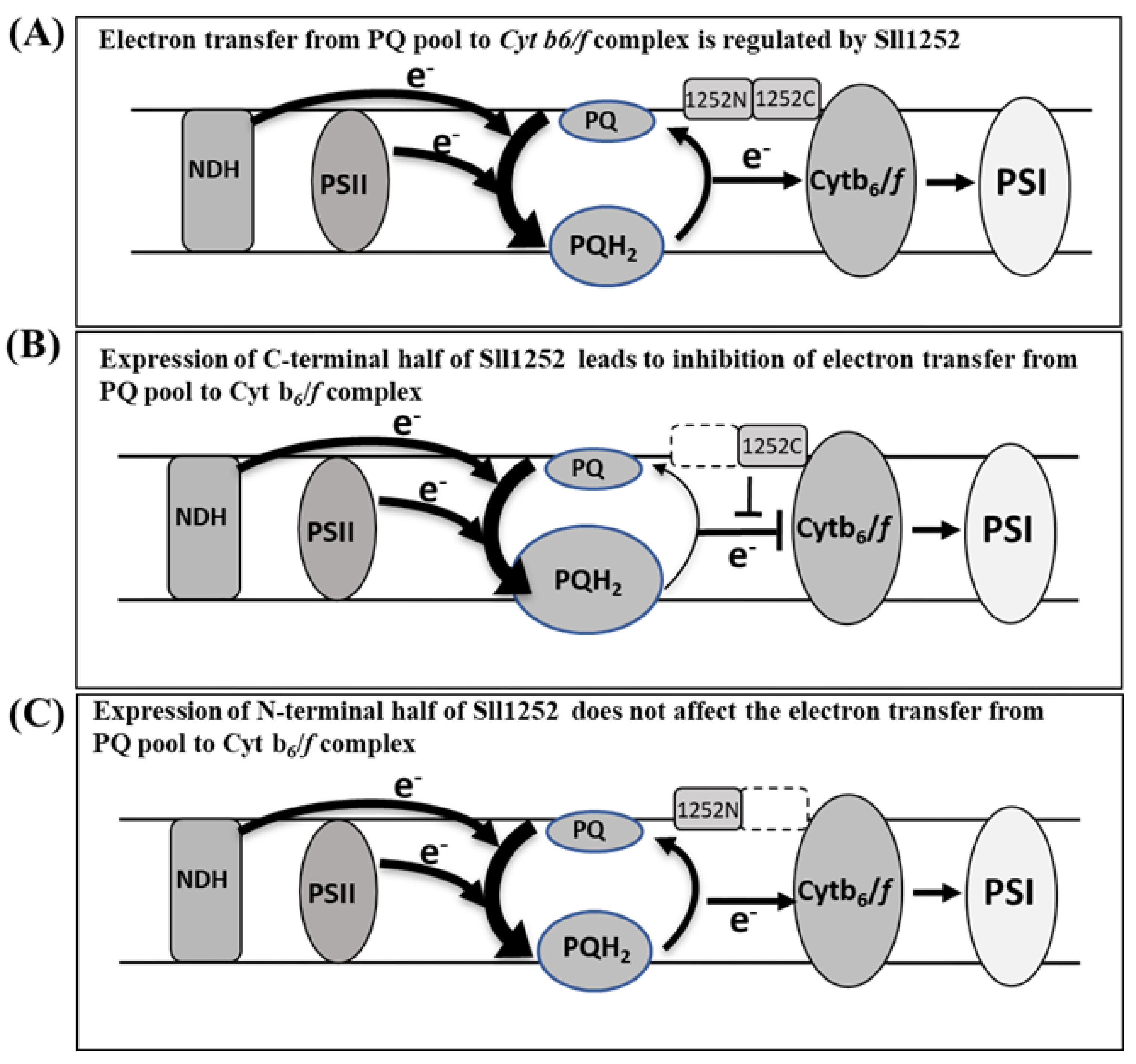
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sl. No | Primer Name | Sequence |
|---|---|---|
| 1 | sll1252-F | 5′-AAC CAT AGG CGC ATT GTA GCT CCT TG-3′ |
| 2 | sll1252-R | 5′-GAT ATC AAC ATT ATC CTT TGC CTC GAA CG-3′ |
| 3 | KAN-2 FP-1 | 5′-ACC TAC AAC AAA GCT CTC ATC AAC C-3′ |
| 4 | KAN-2 RP-1 | 5′-GCA ATG TAA CAT CAG AGA TTT TGA G-3′ |
| 5 | Supp-F | 5′-AGA TCT GCC AAG ATA ATT GAT TG-3′ |
| 6 | Supp-R | 5′-AGA TCT GTC GAC ATT CCA CTG GCA AAA-3′ |
| 7 | UF | 5′-GCA TCA ATT TGT TCT GTC AC-3′ |
| 8 | UR | 5′-AGA TCT CGA GCA ACG GCA TTA GCT-3′ |
| 9 | DF | 5′-CGT TGC TCG AGA TCT CCA GTT TCA AGG CTT GCC-3′ |
| 10 | DR | 5′-CCA CAA TTA AAG TTG TAC TG-3′ |
| 11 | KAN-2 SalI-F | 5′-TCG TCG ACC AAC CAT CAT CGA TGA ATT-3′ |
| 12 | KAN-2 SalI-R | 5′-TCG TCG ACA AAG CCG CCG TCC CGT CAA-3′ |
| 13 | sll1252-trn-UR | 5′-CGA GCA ACG GCA TTA GCT AA-3′ |
| 14 | sll1252-Ntrn-F | 5′-TAA TGC CGT TGC TCG ATG GTG CGT ACT GTA-3′ |
| 15 | sll1252-Ntrn-R | 5′-CCC GGG TTA AAG ATA TCG GGT-3′ |
| 16 | sll1252-Ntrn-DF | 5′-TAT CTT TAA CCC GGG CCA GTT TCA AGG CTT-3′ |
| 17 | sll1252-trn-DR | 5′-CCC GGG TTA CTG GGT TAA ATG AAG ACT-3′ |
| 18 | sll1252-Ctrn-F | 5′-TAA CCC AGT AAC CCG GGC CAG TTT CAA GGC TT-3′ |
References
- Delwiche, C.F.; Kuhsel, M.; Palmer, J.D. Phylogenetic Analysis of tufA Sequences Indicates a Cyanobacterial Origin of All Plastids. Mol. Phylogenet. Evol. 1995, 4, 110–128. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Medlin, L. Phylogeny of plastids A review based on comparisons of small subunit ribosomal RNA coding regions. J. Phycol. 1995, 31, 487–496. [Google Scholar] [CrossRef]
- Brocks, J.J.; Logan, G.A.; Buick, R.; Summons, R.E. Archean molecular fossils and the early rise of eukaryotes. Science 1999, 285, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Allen, J.F. Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 2003, 4, 209. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef] [PubMed]
- Kashino, Y.; Lauber, W.M.; Carroll, J.A.; Wang, Q.; Whitmarsh, J.; Satoh, K.; Pakrasi, H.B. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 2002, 41, 8004–8012. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, K.N.; Iverson, T.M.; Maghlaoui, K.; Barber, J.; Iwata, S. Architecture of the photosynthetic oxygen-evolving center. Science 2004, 303, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef]
- Suga, M.; Akita, F.; Hirata, K.; Ueno, G.; Murakami, H.; Nakajima, Y.; Shimizu, T.; Yamashita, K.; Yamamoto, M.; Ago, H. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 2015, 517, 99–103. [Google Scholar] [CrossRef]
- Gisriel, C.J.; Wang, J.; Liu, J.; Flesher, D.A.; Reiss, K.M.; Huang, H.-L.; Yang, K.R.; Armstrong, W.H.; Gunner, M.; Batista, V.S. High-resolution cryo-electron microscopy structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2022, 119, e2116765118. [Google Scholar] [CrossRef]
- Srikumar, A.; Krishna, P.S.; Sivaramakrishna, D.; Kopfmann, S.; Hess, W.R.; Swamy, M.J.; Lin-Chao, S.; Prakash, J.S. The Ssl2245-Sll1130 toxin-antitoxin system mediates heat-induced programmed cell death in Synechocystis sp. PCC6803. J. Biol. Chem. 2017, 292, 4222–4234. [Google Scholar] [CrossRef]
- Roose, J.L.; Kashino, Y.; Pakrasi, H.B. The PsbQ protein defines cyanobacterial Photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. USA 2007, 104, 2548–2553. [Google Scholar] [CrossRef]
- Wegener, K.M.; Bennewitz, S.; Oelmüller, R.; Pakrasi, H.B. The Psb32 protein aids in repairing photodamaged photosystem II in the cyanobacterium Synechocystis 6803. Mol. Plant 2011, 4, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Keren, N.; Ohkawa, H.; Welsh, E.A.; Liberton, M.; Pakrasi, H.B. Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 2005, 17, 2768–2781. [Google Scholar] [CrossRef] [PubMed]
- Calderone, V.; Trabucco, M.; Vujičić, A.; Battistutta, R.; Giacometti, G.M.; Andreucci, F.; Barbato, R.; Zanotti, G. Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep. 2003, 4, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Inoue-Kashino, N.; Kashino, Y.; Orii, H.; Satoh, K.; Terashima, I.; Pakrasi, H.B. S4 protein Sll1252 is necessary for energy balancing in photosynthetic electron transport in Synechocystis sp. PCC 6803. Biochemistry 2011, 50, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Sireesha, K.; Radharani, B.; Krishna, P.S.; Sreedhar, N.; Subramanyam, R.; Mohanty, P.; Prakash, J.S. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp. PCC6803. Biochim. Biophys. Acta (BBA)-Bioenerg. 2012, 1817, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Hihara, Y.; Sonoike, K.; Kanehisa, M.; Ikeuchi, M. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003, 185, 1719–1725. [Google Scholar] [CrossRef]
- Hihara, Y.; Kamei, A.; Kanehisa, M.; Kaplan, A.; Ikeuchi, M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 2001, 13, 793–806. [Google Scholar] [CrossRef]
- Georg, J.; Dienst, D.; Schürgers, N.; Wallner, T.; Kopp, D.; Stazic, D.; Kuchmina, E.; Klähn, S.; Lokstein, H.; Hess, W.R. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 2014, 26, 3661–3679. [Google Scholar] [CrossRef]
- Li, D.; Xie, J.; Zhao, J.; Xia, A.; Li, D.; Gong, Y. Light-induced excitation energy redistribution in Spirulina platensis cells: “spillover” or “mobile PBSs”? Biochim. Biophys. Acta (BBA)-Bioenerg. 2004, 1608, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Joshua, S.; Mullineaux, C.W. Phycobilisome diffusion is required for light-state transitions in cyanobacteria. Plant Physiol. 2004, 135, 2112–2119. [Google Scholar] [CrossRef] [PubMed]
- Karapetyan, N.V. Protective dissipation of excess absorbed energy by photosynthetic apparatus of cyanobacteria: Role of antenna terminal emitters. Photosynth. Res. 2008, 97, 195–204. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, H.; Niedzwiedzki, D.M.; Prado, M.; He, G.; Gross, M.L.; Blankenship, R.E. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 2013, 342, 1104–1107. [Google Scholar] [CrossRef]
- Bhatti, A.F.; Choubeh, R.R.; Kirilovsky, D.; Wientjes, E.; van Amerongen, H. State transitions in cyanobacteria studied with picosecond fluorescence at room temperature. Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148255. [Google Scholar] [CrossRef] [PubMed]
- Pfannschmidt, T.; Bräutigam, K.; Wagner, R.; Dietzel, L.; Schröter, Y.; Steiner, S.; Nykytenko, A. Potential regulation of gene expression in photosynthetic cells by redox and energy state: Approaches towards better understanding. Ann. Bot. 2009, 103, 599–607. [Google Scholar] [CrossRef]
- Rochaix, J.D. Regulation of photosynthetic electron transport. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 375–383. [Google Scholar] [CrossRef]
- Foyer, C.H.; Neukermans, J.; Queval, G.; Noctor, G.; Harbinson, J. Photosynthetic control of electron transport and the regulation of gene expression. J. Exp. Bot. 2012, 63, 1637–1661. [Google Scholar] [CrossRef]
- Williams, J.G. [85] Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1988; Volume 167, pp. 766–778. [Google Scholar] [CrossRef]
- Prakash, J.S.; Krishna, P.S.; Sirisha, K.; Kanesaki, Y.; Suzuki, I.; Shivaji, S.; Murata, N. An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology 2010, 156, 442–451. [Google Scholar] [CrossRef]
- Krishna, P.S.; Rani, B.R.; Mohan, M.K.; Suzuki, I.; Shivaji, S.; Prakash, J.S. A novel transcriptional regulator, Sll1130, negatively regulates heat-responsive genes in Synechocystis sp. PCC6803. Biochem. J. 2013, 449, 751–760. [Google Scholar] [CrossRef]
- Sendersky, E.; Simkovsky, R.; Golden, S.S.; Schwarz, R. Quantification of chlorophyll as a proxy for biofilm formation in the cyanobacterium Synechococcus elongatus. Bio-Protocol 2017, 7, e2406. [Google Scholar] [CrossRef] [PubMed]
- Yubuki, N.; Inagaki, Y.; Nakayama, T.; Inouye, I. Ultrastructure and ribosomal RNA phylogeny of the free-living heterotrophic flagellate Dysnectes brevis n. gen., n. sp., a new member of the Fornicata. J. Eukaryot. Microbiol. 2007, 54, 191–200. [Google Scholar] [CrossRef]
- Kaneko, T.; Sato, S.; Kotani, H.; Tanaka, A.; Asamizu, E.; Nakamura, Y.; Miyajima, N.; Hirosawa, M.; Sugiura, M.; Sasamoto, S.; et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996, 3, 109–136. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shen, G.; Wang, T.; Bryant, D.A.; Golbeck, J.H.; McIntosh, L. Suppressor mutations in the study of photosystem I biogenesis: sll0088 is a previously unidentified gene involved in reaction center accumulation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003, 185, 3878–3887. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, S.; Kurkela, J.; Linhartová, M.; Tyystjärvi, T. The genome sequence of Synechocystis sp. PCC 6803 substrain GT-T and its implications for the evolution of PCC 6803 substrains. FEBS Open Bio 2023, 13, 701–712. [Google Scholar] [CrossRef]
- Zhang, C.-C.; Jeanjean, R.; Joset, F. Obligate phototrophy in cyanobacteria: More than a lack of sugar transport. FEMS Microbiol. Lett. 1998, 161, 285–292. [Google Scholar] [CrossRef][Green Version]
- Hihara, Y.; Sonoike, K.; Ikeuchi, M. A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis species PCC 6803 in response to high light. Plant Physiol. 1998, 117, 1205–1216. [Google Scholar] [CrossRef]
- de Porcellinis, A.J.; Klähn, S.; Rosgaard, L.; Kirsch, R.; Gutekunst, K.; Georg, J.; Hess, W.R.; Sakuragi, Y. The non-coding RNA Ncr0700/PmgR1 is required for photomixotrophic growth and the regulation of glycogen accumulation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016, 57, 2091–2103. [Google Scholar] [CrossRef]
- Stiehl, H.; Witt, H.T. Quantitative treatment of the function of plastoquinone in photosynthesis. Z. Für Nat. B 1969, 24, 1588–1598. [Google Scholar] [CrossRef]
| Strain | Mean Generation Time (h) | PSII Activity (H2O to PBQ) | WCE Activity (H2O to MV) |
|---|---|---|---|
| Wild-type | 8.3 ± 0.3 | 1 ± 0.11 | 1 ± 0.13 |
| Δsll1252ins | Not detected | 0.7 ± 0.11 | 0.55 ± 0.06 |
| Δsll1252del | 9.8 ± 0.2 | 0.78 ± 0.23 | 0.82 ± 0.06 |
| Δsll1252inv-Rev | 8.1 ± 0.2 | 0.89 ± 0.16 | 0.91 ± 0.09 |
| Δsll1252-Ntrn | 22.0 ± 2.5 | 0.75 ± 0.04 | 0.55 ± 0.06 |
| Δsll1252-Ctrn | 11.5 ± 0.7 | 0.59 ± 0.03 | 0.91 ± 0.06 |
| ORF and Description | (Δsll1252ins/Wild-Type) | Effect of Inhibitor on Expression a | ||
|---|---|---|---|---|
| 5 µmol/m−2 s−1 | 70 µmol/m−2 s−1 | DBMIB | DCMU | |
| sll0330, sepiapterine reductase | 1.3 ± 0.4 | 18.7 ± 5.5 | ||
| slr1544 | 1.1 ± 0.1 | 17.8 ± 1.7 | I | |
| slr1704 | 1.3 ± 0.1 | 11.9 ± 0.2 | ||
| slr1674 | 1.1 ± 0.1 | 8.3 ± 0.6 | I | |
| sll0528 | 1.1 ± 0.1 | 7.6 ± 0.2 | I | |
| sll1483, periplasmic protein | 1.1 ± 0.0 | 7.2 ± 0.7 | I | I |
| sll1652 | 1.2 ± 0.1 | 6.6 ± 3.6 | ||
| sll0306, sigB, rpoD, RNA polymerase group 2 sigma factor | 1.5 ± 0.1 | 6.5 ± 0.4 | I | |
| sll1862 | 1.2 ± 0.1 | 5.8 ± 0.8 | ||
| sll1515, gifB, glutamine synthetase inactivating factor IF17 | 1.2 ± 0.1 | 5.6 ± 1.0 | I | I |
| sll1653 menG, probable phylloquinone biosynthesis methlytransferase | 0.9 ± 0.1 | 5.5 ± 0.9 | ||
| sll1514, hspA, hsp1, 16.6 kDa small heat shock protein | 1.1 ± 0.1 | 5.5 ± 1.3 | I | |
| ssr2194 | 1.1 ± 0.1 | 5.2 ± 0.1 | ||
| slr1516, sodB, superoxide dismutase | 0.8 ± 0.1 | 4.9 ± 0.4 | ||
| sll1236 | 1.0 ± 0.1 | 4.9 ± 0.0 | ||
| slr1738 | 1.1 ± 0.1 | 4.7 ± 0.2 | I | |
| slr0581 | 1.0 ± 0.1 | 4.5 ± 0.5 | I | I |
| ssr2016 | 1.1 ± 0.0 | 4.1 ± 0.2 | ||
| slr2075, groES, 10 kDa chaperonin, GroES protein | 0.7 ± 0.0 | 3.9 ± 0.5 | I | |
| ssr2595, hilB, scpD, high light-inducible protein | 1.1 ± 0.1 | 3.9 ± 0.2 | I | I |
| sll1598, mntC, ABC-type manganese transport system substrate-binding protein | 1.1 ± 0.2 | 3.7 ± 0.8 | ||
| slr1675, hypA1, putative hydrogenase expression/formation protein HypA1 | 0.7 ± 0.1 | 3.7 ± 0.5 | I | |
| slr2076, groEL1, cpn60, 60 kDa chaperonin 1, GroEL1, molecular chaperone | 0.9 ± 0.0 | 3.7 ± 0.1 | I | R |
| slr0967 | 0.8 ± 0.2 | 3.6 ± 0.4 | I | I |
| ssl3044, hydrogenase component | 1.0 ± 0.1 | 3.5 ± 0.4 | ||
| sl0416, groEL2, cpn 60, 60 kDa chaperonin 2, GroEL2, | 1.1 ± 0.0 | 3.5 ± 1.1 | R | |
| sll0846 | 1.0 ± 0.0 | 3.2 ± 0.3 | I | |
| sll1863 | 1.0 ± 0.1 | 3.2 ± 1.3 | ||
| slr0093, dnaJ, DnaJ protein, heat shock protein 40, molecular chaperone | 1.1 ± 0.0 | 3.1 ± 0.2 | I | |
| ssl1911, gifA, glutamine synthetase inactivating factor IF7 | 1.3 ± 0.1 | 2.9 ± 0.5 | I | I |
| slr1687 | 1.1 ± 0.2 | 2.8 ± 0.1 | I | I |
| sll1620 | 1.2 ± 0.0 | 2.8 ± 0.6 | I | |
| slr1204, htrA, serine protease Htra | 1.1 ± 0.2 | 2.8 ± 1.2 | ||
| sll1621, membrane protein | 0.8 ± 0.1 | 2.7 ± 0.4 | I | |
| slr1963, water-soluble carotenoid protein | 1.1 ± 0.1 | 2.6 ± 0.8 | I | R |
| ssl3364, cp12 | 1.3 ± 0.0 | 2.6 ± 0.7 | ||
| ssl2542, hliA, scpC, high light-inducible protein | 0.8 ± 0.1 | 2.6 ± 0.5 | I | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaga, R.R.; Itoh, F.; Chauhan, S.; Mandal, M.; Krishna, P.S.; Suzuki, I.; Prakash, J.S.S. Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803. Genes 2023, 14, 2151. https://doi.org/10.3390/genes14122151
Balaga RR, Itoh F, Chauhan S, Mandal M, Krishna PS, Suzuki I, Prakash JSS. Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803. Genes. 2023; 14(12):2151. https://doi.org/10.3390/genes14122151
Chicago/Turabian StyleBalaga, Radha Rani, Fumihiro Itoh, Suraj Chauhan, Mukulika Mandal, Pilla Sankara Krishna, Iwane Suzuki, and Jogadhenu S. S. Prakash. 2023. "Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803" Genes 14, no. 12: 2151. https://doi.org/10.3390/genes14122151
APA StyleBalaga, R. R., Itoh, F., Chauhan, S., Mandal, M., Krishna, P. S., Suzuki, I., & Prakash, J. S. S. (2023). Sll1252 Coordinates Electron Transport between Plastoquinone and Cytochrome b6/f Complex in Synechocystis PCC 6803. Genes, 14(12), 2151. https://doi.org/10.3390/genes14122151






