Large Deletions, Cleavage of the Telomeric Repeat Sequence, and Reverse Transcriptase-Mediated DNA Damage Response Associated with Long Interspersed Element-1 ORF2p Enzymatic Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of L1-, ORF2-, and EN-Containing Plasmids
2.2. Cell Culture
2.3. Generation of Doxycycline-Inducible 293T Cell Lines
2.4. Protein Harvest for Western Blot Analysis
2.5. Western Blot Analysis
2.6. LEAP Assay
2.7. Retrotransposition Assay
2.8. Analysis of L1-Induced Mutations
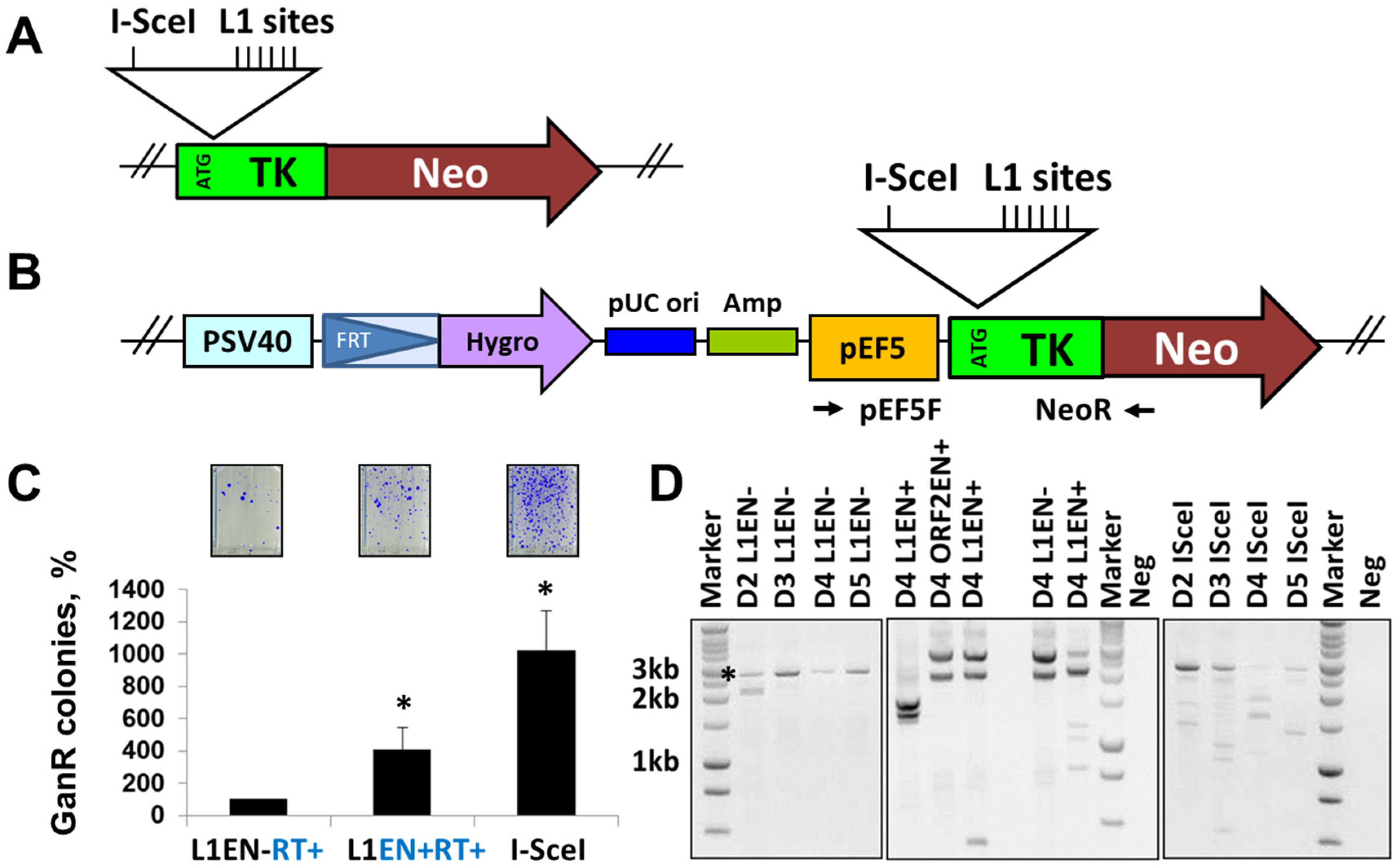
2.9. Site-Specific PCR Analysis
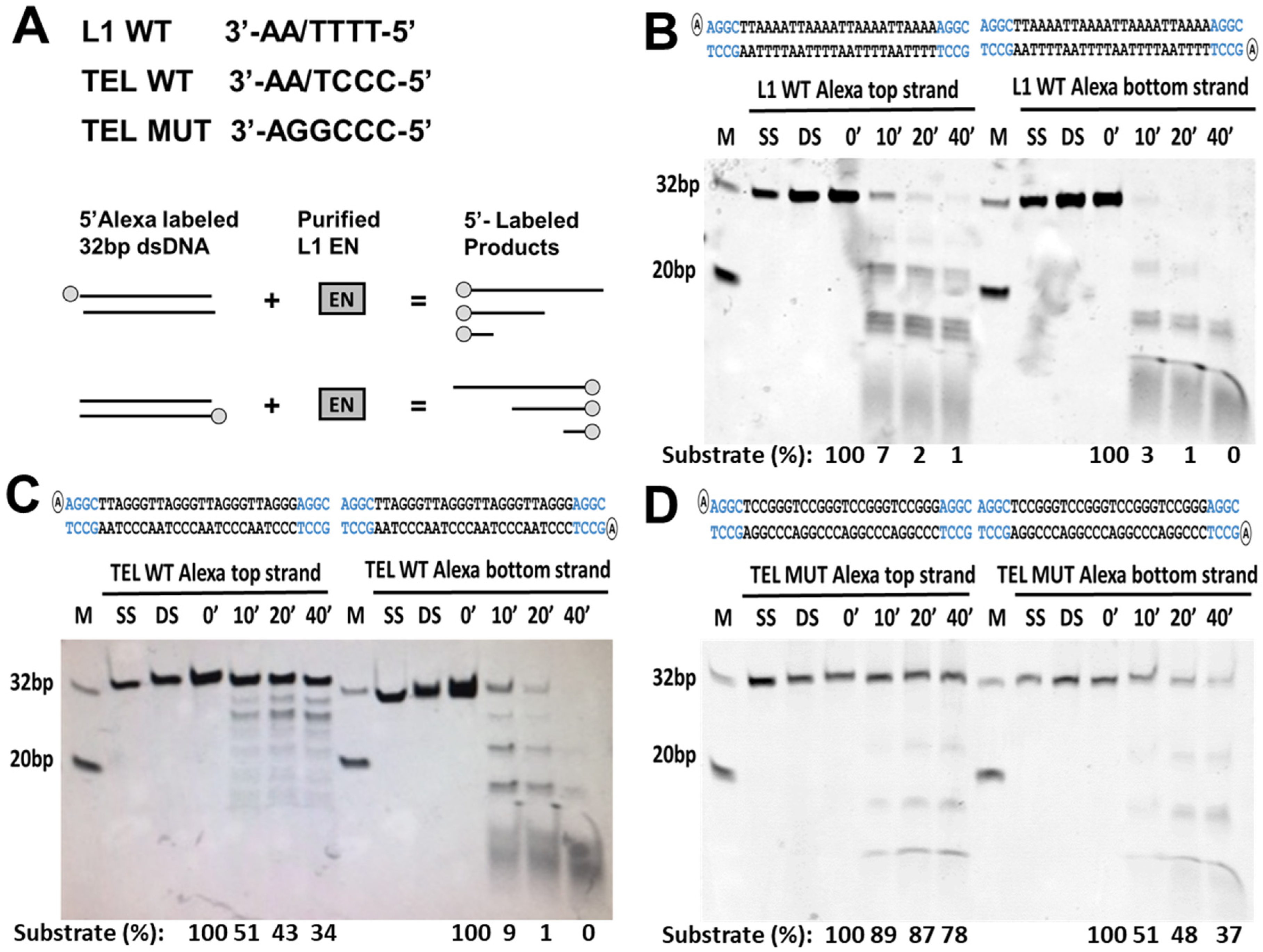
2.10. Endonuclease In Vitro Cleavage Assay
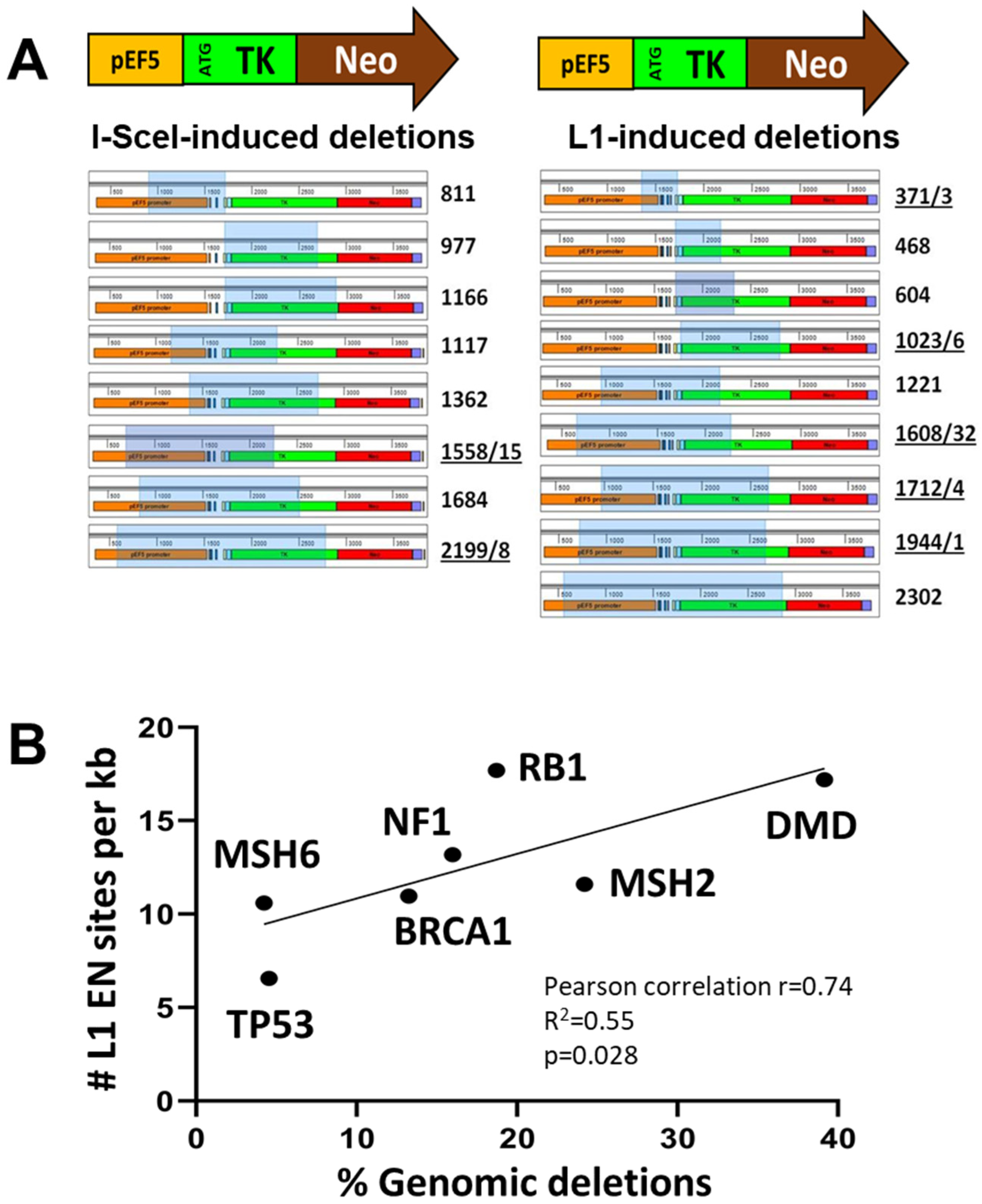
2.11. Assessment of Disease-Causing Mutations and the Number of L1 EN Sites in Human Genes
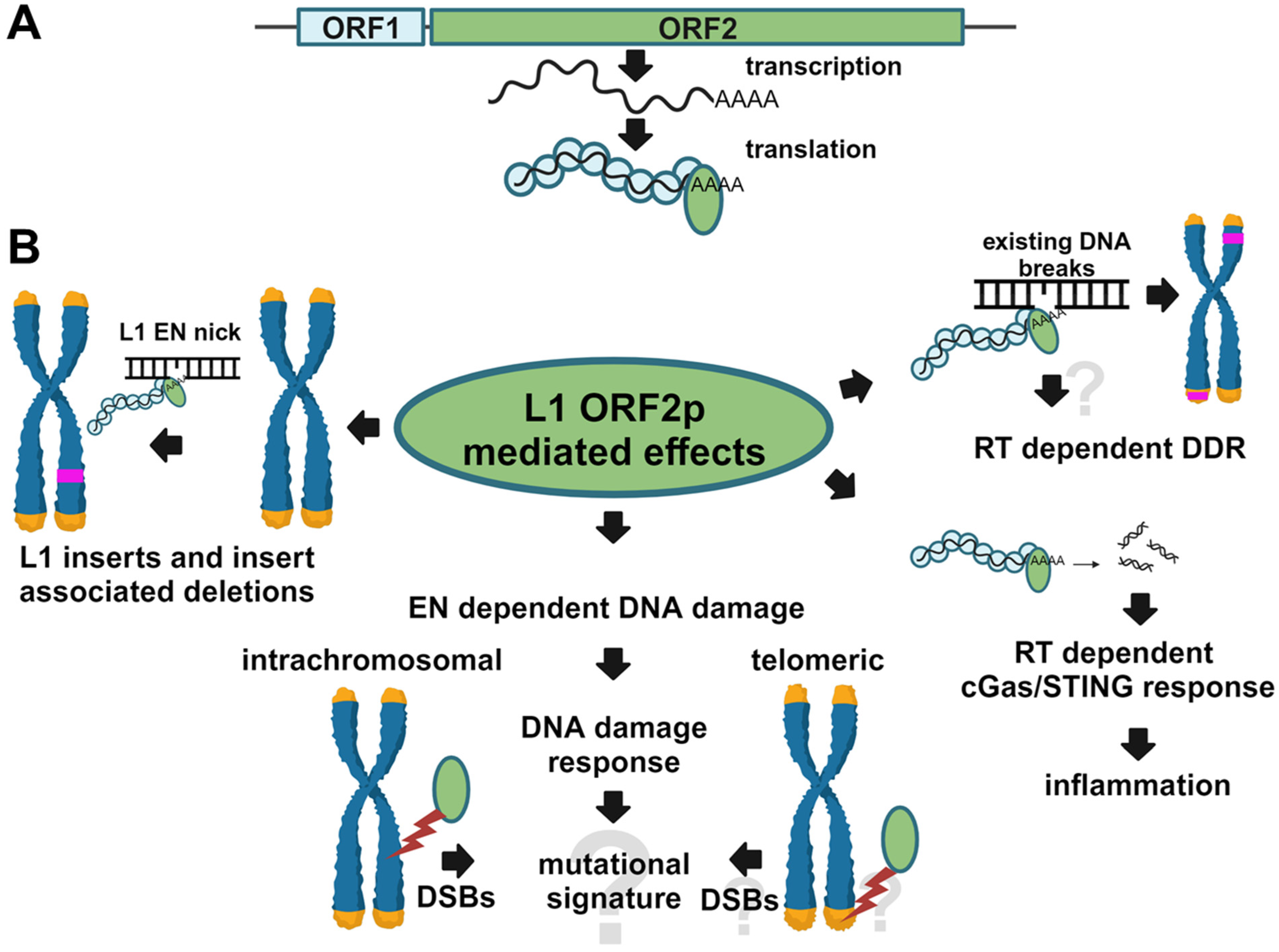
3. Results
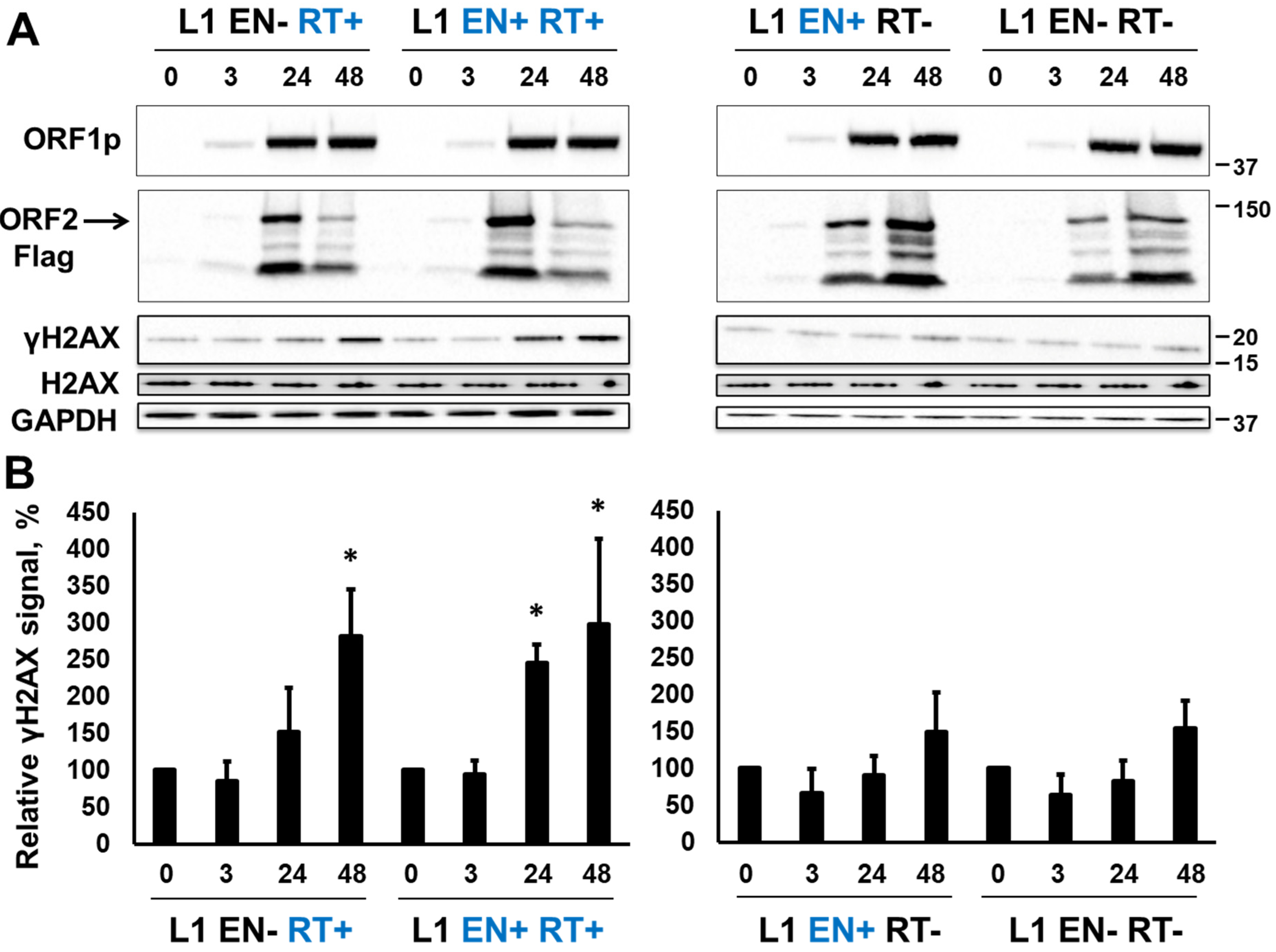
3.1. RT-Dependent Accumulation of γH2AX Signal upon Induction of L1 Expression
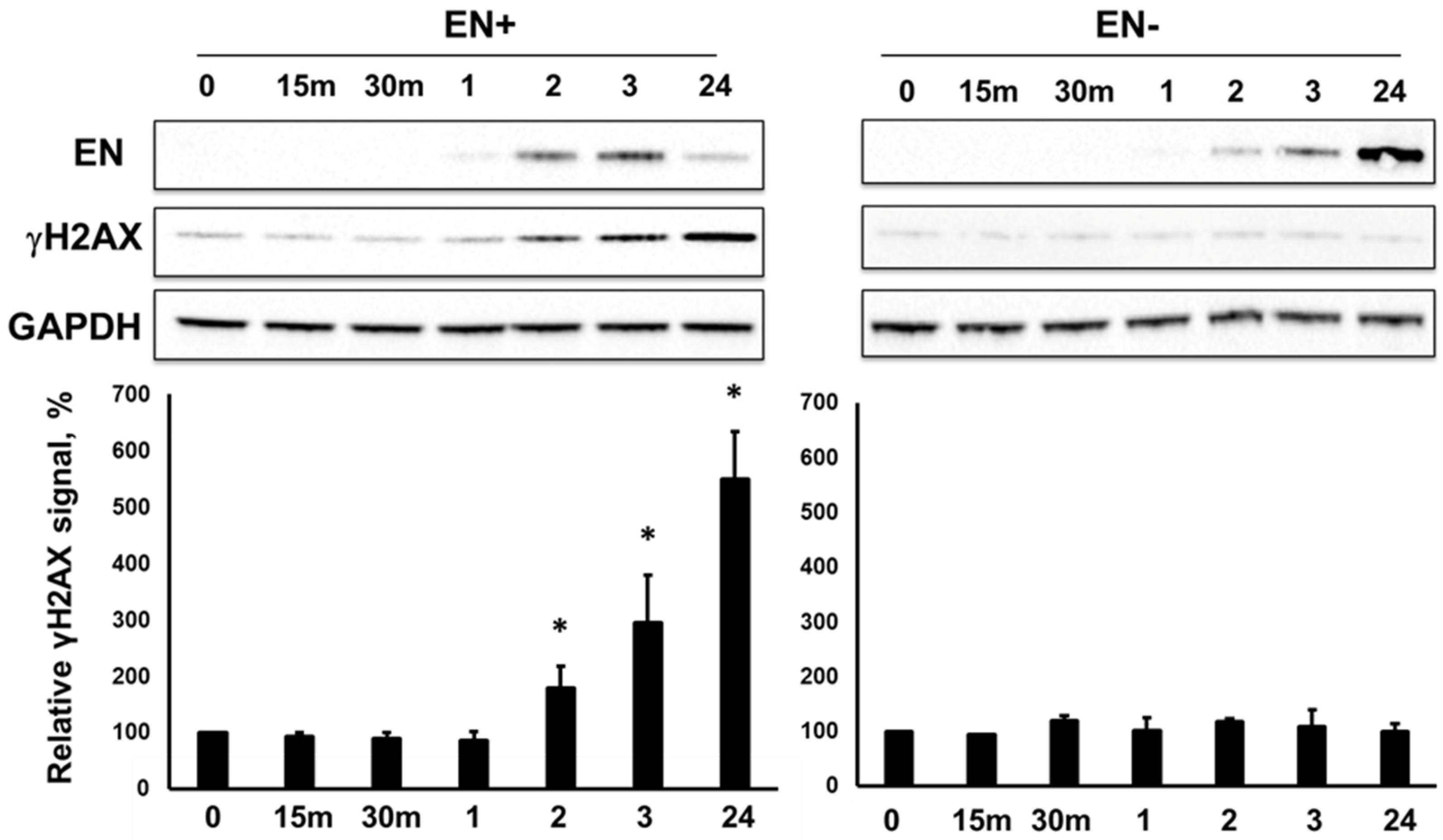
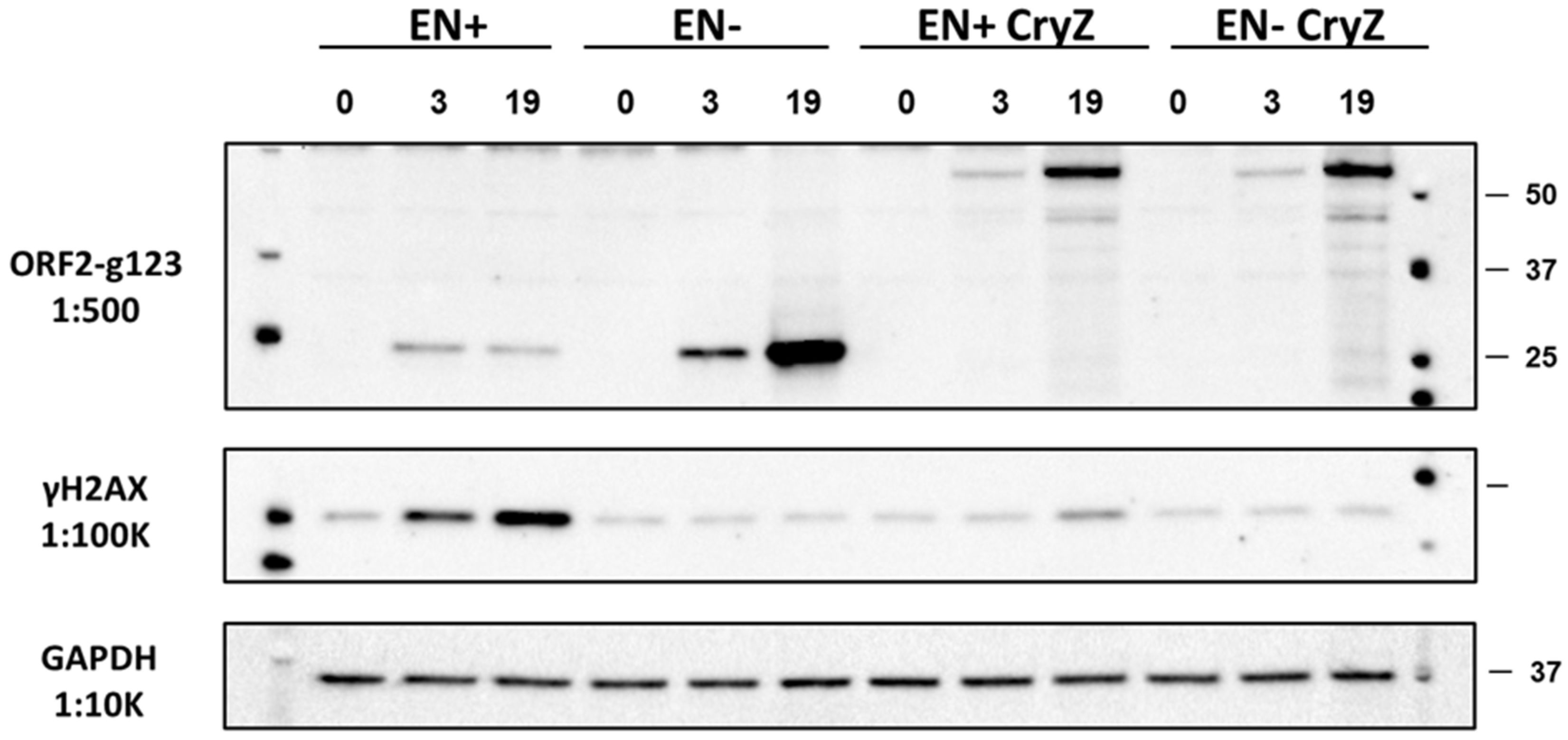
3.2. RT-Dependent Accumulation of γH2AX Signal upon Induction of ORF2p Expression
3.3. L1 Expression Causes Large Genomic Deletions That Do Not Contain Any L1 Sequences
3.4. L1 Endonuclease Cuts Telomeric Sequences In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [PubMed]
- Gardner, E.J.; Lam, V.K.; Harris, D.N.; Chuang, N.T.; Scott, E.C.; Pittard, W.S.; Mills, R.E.; Devine, S.E. The Mobile Element Locator Tool (MELT): Population-scale mobile element discovery and biology. Genome Res. 2017, 27, 1916–1929. [Google Scholar] [CrossRef] [PubMed]
- Evrony, G.D.; Cai, X.; Lee, E.; Hills, L.B.; Elhosary, P.C.; Lehmann, H.S.; Parker, J.J.; Atabay, K.D.; Gilmore, E.C.; Poduri, A. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 2012, 151, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.R.; Gerdes, P.; Gerhardt, D.J.; Sanchez-Luque, F.J.; Bodea, G.O.; Muñoz-Lopez, M.; Jesuadian, J.S.; Kempen, M.H.C.; Carreira, P.E.; Jeddeloh, J.A. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 2017, 27, 1395–1405. [Google Scholar] [CrossRef]
- Platt, R.N., 2nd; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018, 26, 25–43. [Google Scholar] [CrossRef]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Belancio, V.P.; Roy-Engel, A.M.; Pochampally, R.R.; Deininger, P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010, 38, 3909–3922. [Google Scholar] [CrossRef]
- Stow, E.C.; Kaul, T.; deHaro, D.L.; Dem, M.R.; Beletsky, A.G.; Morales, M.E.; Du, Q.; LaRosa, A.J.; Yang, H.; Smither, E. Organ-, sex- and age-dependent patterns of endogenous L1 mRNA expression at a single locus resolution. Nucleic Acids Res. 2021, 49, 5813–5831. [Google Scholar] [CrossRef]
- Ergün, S.; Buschmann, C.; Heukeshoven, J.; Dammann, K.; Schnieders, F.; Lauke, H.; Chalajour, F.; Kilic, N.; Strätling, W.H.; Schumann, G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004, 279, 27753–27763. [Google Scholar] [CrossRef]
- Martin, S.L. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell Biol. 1991, 11, 4804–4807. [Google Scholar] [PubMed]
- Branciforte, D.; Martin, S.L. Developmental and cell type specificity of LINE-1 expression in mouse testis: Implications for transposition. Mol. Cell Biol. 1994, 14, 2584–2592. [Google Scholar] [PubMed]
- Harris, C.R.; Normart, R.; Yang, Q.; Stevenson, E.; Haffty, B.G.; Ganesan, S.; Cordon-Cardo, C.; Levine, A.J.; Tang, L.H. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes. Cancer 2010, 1, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., 3rd; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Iskow, R.C.; McCabe, M.T.; Mills, R.E.; Torene, S.; Pittard, W.S.; Neuwald, A.F.; Van Meir, E.G.; Vertino, P.M.; Devine, S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010, 141, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Rodić, N.; Sharma, R.; Sharma, R.; Zampella, J.; Dai, L.; Taylor, M.S.; Hruban, R.H.; Iacobuzio-Donahue, C.A.; Maitra, A.; Torbenson, M.S. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 2014, 184, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Rodić, N.; Steranka, J.P.; Makohon-Moore, A.; Moyer, A.; Shen, P.; Sharma, R.; Kohutek, Z.A.; Huang, C.R.; Ahn, D.; Mita, P. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 2015, 21, 1060–1064. [Google Scholar] [CrossRef]
- Solyom, S.; Ewing, A.D.; Rahrmann, E.P.; Doucet, T.; Nelson, H.H.; Burns, M.B.; Harris, R.S.; Sigmon, D.F.; Casella, A.; Erlanger, B. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012, 22, 2328–2338. [Google Scholar] [CrossRef]
- Tubio, J.M.C.; Li, Y.; Ju, Y.S.; Martincorena, I.; Cooke, S.L.; Tojo, M.; Gundem, G.; Pipinikas, C.P.; Zamora, J.; Raine, K. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014, 345, 1251343. [Google Scholar] [CrossRef]
- Taylor, M.S.; Wu, C.; Fridy, P.C.; Zhang, S.J.; Senussi, Y.; Wolters, J.C.; Cajuso, T.; Cheng, W.C.; Heaps, J.D.; Miller, B.D. Ultrasensitive detection of circulating LINE-1 ORF1p as a specific multi-cancer biomarker. Cancer Discov. 2023, 13, 2532–2547. [Google Scholar] [CrossRef]
- Swergold, G.D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990, 10, 6718–6729. [Google Scholar] [PubMed]
- Hata, K.; Sakaki, Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 1997, 189, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Chan, M.F.; Tomigahara, Y.; Tsai, Y.C.; Gonzales, F.A.; Li, E.; Laird, P.W.; Jones, P.A. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell Biol. 2002, 22, 480–491. [Google Scholar] [CrossRef]
- Takai, D.; Yagi, Y.; Habib, N.; Sugimura, T.; Ushijima, T. Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Jpn. J. Clin. Oncol. 2000, 30, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.; Morales, M.E.; White, T.B.; Baddoo, M.; Hedges, D.J.; Servant, G.; Srivastav, S.; Smither, M.E.; Concha, M.; DeHaro, D.L. A comprehensive approach to expression of L1 loci. Nucleic Acids Res. 2017, 45, e31. [Google Scholar] [CrossRef]
- Philippe, C.; Vargas-Landin, D.B.; Doucet, A.J.; van Essen, D.; Vera-Otarola, J.; Kuciak, M.; Corbin, A.; Nigumann, P.; Cristofari, G. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. elife 2016, 5, e13926. [Google Scholar] [CrossRef]
- Stow, E.C.; Baddoo, M.; LaRosa, A.J.; LaCoste, D.; Deininger, P.; Belancio, V. SCIFER: Approach for analysis of LINE-1 mRNA expression in single cells at a single locus resolution. Mob. DNA 2022, 13, 21. [Google Scholar] [CrossRef]
- Freeman, B.; White, T.; Kaul, T.; Stow, E.C.; Baddoo, M.; Ungerleider, N.; Morales, M.; Yang, H.; Deharo, D.; Deininger, P. Analysis of epigenetic features characteristic of L1 loci expressed in human cells. Nucleic Acids Res. 2022, 50, 1888–1907. [Google Scholar] [CrossRef]
- Taylor, D.; Lowe, R.; Philippe, C.; Cheng, K.C.L.; Grant, O.A.; Zabet, N.R.; Cristofari, G.; Branco, M.R. Locus-specific chromatin profiling of evolutionarily young transposable elements. Nucleic Acids Res. 2021, 50, e33. [Google Scholar] [CrossRef]
- Morrish, T.A.; Garcia-Perez, J.L.; Stamato, T.D.; Taccioli, G.E.; Sekiguchi, J.; Moran, J.V. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 2007, 446, 208–212. [Google Scholar] [CrossRef]
- Morrish, T.A.; Gilbert, N.; Myers, J.S.; Vincent, B.J.; Stamato, T.D.; Taccioli, G.E.; Batzer, M.A.; Moran, J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002, 31, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Kines, K.J.; Sokolowski, M.; deHaro, D.L.; Christian, C.M.; Belancio, V.P. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 2014, 42, 10488–10502. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, M.; Chynces, M.; deHaro, D.; Christian, C.M.; Belancio, V.P. Truncated ORF1 proteins can suppress LINE-1 retrotransposition in trans. Nucleic Acids Res. 2017, 45, 5294–5308. [Google Scholar] [CrossRef][Green Version]
- Kines, K.J.; Sokolowski, M.; deHaro, D.L.; Christian, C.M.; Baddoo, M.; Smither, M.E.; Belancio, V.P. The endonuclease domain of the LINE-1 ORF2 protein can tolerate multiple mutations. Mob. DNA 2016, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Belancio, V.P.; Hedges, D.J.; Deininger, P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008, 18, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- Alisch, R.S.; Garcia-Perez, J.L.; Muotri, A.R.; Gage, F.H.; Moran, J.V. Unconventional translation of mammalian LINE-1 retrotransposons. Genes. Dev. 2006, 20, 210–224. [Google Scholar] [CrossRef]
- Callahan, K.E.; Hickman, A.B.; Jones, C.E.; Ghirlando, R.; Furano, A.V. Polymerization and nucleic acid-binding properties of human L1 ORF1 protein. Nucleic Acids Res. 2012, 40, 813–827. [Google Scholar] [CrossRef]
- Christian, C.M.; Sokolowski, M.; deHaro, D.; Kines, K.J.; Belancio, V.P. Involvement of Conserved Amino Acids in the C-Terminal Region of LINE-1 ORF2p in Retrotransposition. Genetics 2017, 205, 1139–1149. [Google Scholar] [CrossRef]
- Christian, C.M.; deHaro, D.; Kines, K.J.; Sokolowski, M.; Belancio, V.P. Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR). Nucleic Acids Res. 2016, 44, 4818–4834. [Google Scholar] [CrossRef]
- Adney, E.M.; Ochmann, M.T.; Sil, S.; Truong, D.M.; Mita, P.; Wang, X.; Kahler, D.J.; Fenyö, D.; Holt, L.J.; Boeke, J.D. Comprehensive Scanning Mutagenesis of Human Retrotransposon LINE-1 Identifies Motifs Essential for Function. Genetics 2019, 213, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Li, J.; Epperson, L.E.; Lieberman, B. Functional reverse transcriptases encoded by A-type mouse LINE-1: Defining the minimal domain by deletion analysis. Gene 1998, 215, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Cost, G.J.; Boeke, J.D. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 1998, 37, 18081–18093. [Google Scholar] [CrossRef] [PubMed]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H.; Boeke, J.D.; Gabriel, A. Reverse transcriptase encoded by a human transposable element. Science 1991, 254, 1808. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, M.; deHaro, D.; Christian, C.M.; Kines, K.J.; Belancio, V.P. Characterization of L1 ORF1p self-interaction and cellular localization using a mammalian two-hybrid system. PLoS ONE 2013, 8, e82021. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.V.; Holmes, S.E.; Naas, T.P.; DeBerardinis, R.J.; Boeke, J.D.; Kazazian, H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell 1996, 87, 917–927. [Google Scholar] [CrossRef]
- Martin, S.L.; Bushman, F.D. Nucleic Acid Chaperone Activity of the ORF1 Protein from the Mouse LINE-1 Retrotransposon. Mol. Cell. Biol. 2001, 21, 467. [Google Scholar] [CrossRef]
- Martin, S.L.; Li, J.; Weisz, J.A. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 2000, 304, 11–20. [Google Scholar] [CrossRef]
- Basame, S.; Wai-lun Li, P.; Howard, G.; Branciforte, D.; Keller, D.; Martin, S.L. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Biol. 2006, 357, 351–357. [Google Scholar] [CrossRef]
- Freeman, B.T.; Sokolowski, M.; Roy-Engel, A.M.; Smither, M.E.; Belancio, V.P. Identification of charged amino acids required for nuclear localization of human L1 ORF1 protein. Mob. DNA 2019, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Cost, G.J.; Golding, A.; Schlissel, M.S.; Boeke, J.D. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001, 29, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.T.; van Eeuwen, T.; Hoyos, D.; Zalevsky, A.; Tchesnokov, E.P.; Sánchez, R.; Miller, B.D.; Di Stefano, L.H.; Ruiz, F.X.; Hancock, M. Structures, functions, and adaptations of the human LINE-1 ORF2 protein. Nature 2023. [Google Scholar] [CrossRef] [PubMed]
- Dombroski, B.A.; Feng, Q.; Mathias, S.L.; Sassaman, D.M.; Scott, A.F.; Kazazian, H.H., Jr.; Boeke, J.D. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol. Cell Biol. 1994, 14, 4485–4492. [Google Scholar] [PubMed]
- Piskareva, O.; Denmukhametova, S.; Schmatchenko, V. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expr. Purif. 2003, 28, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Weichenrieder, O.; Repanas, K.; Perrakis, A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 2004, 12, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.S.; LaCava, J.; Mita, P.; Molloy, K.R.; Huang, C.R.; Li, D.; Adney, E.M.; Jiang, H.; Burns, K.H.; Chait, B.T. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 2013, 155, 1034–1048. [Google Scholar] [CrossRef]
- Clements, A.P.; Singer, M.F. The human LINE-1 reverse transcriptase:effect of deletions outside the common reverse transcriptase domain. Nucleic Acids Res. 1998, 26, 3528–3535. [Google Scholar] [CrossRef]
- Burke, W.D.; Malik, H.S.; Jones, J.P.; Eickbush, T.H. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 1999, 16, 502–511. [Google Scholar] [CrossRef]
- Thawani, A.; Ariza, A.J.F.; Nogales, E.; Collins, K. Template and target site recognition by human LINE-1 in retrotransposition. Nature 2023, 81. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Jamburuthugoda, V.K. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008, 134, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Viollet, S.; Monot, C.; Cristofari, G. L1 retrotransposition: The snap-velcro model and its consequences. Mob. Genet. Elem. 2014, 4, e28907. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martin, B.; Alvarez, E.G.; Baez-Ortega, A.; Zamora, J.; Supek, F.; Demeulemeester, J.; Santamarina, M.; Ju, Y.S.; Temes, J.; Garcia-Souto, D. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020, 52, 306–319. [Google Scholar] [CrossRef]
- Gasior, S.L.; Wakeman, T.P.; Xu, B.; Deininger, P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006, 357, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Cajuso, T.; Sulo, P.; Tanskanen, T.; Katainen, R.; Taira, A.; Hänninen, U.A.; Kondelin, J.; Forsström, L.; Välimäki, N.; Aavikko, M. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat. Commun. 2019, 10, 4022. [Google Scholar] [CrossRef]
- Gilbert, N.; Lutz-Prigge, S.; Moran, J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell 2002, 110, 315–325. [Google Scholar] [CrossRef]
- Sokolowski, M.; DeFreece, C.B.; Servant, G.; Kines, K.J.; deHaro, D.L.; Belancio, V.P. Development of a monoclonal antibody specific to the endonuclease domain of the human LINE-1 ORF2 protein. Mob. DNA 2014, 5, 29. [Google Scholar] [CrossRef]
- Christian, C.M.; Kines, K.J.; Belancio, V.P. The importance of L1 ORF2p cryptic sequence to ORF2p fragment-mediated cytotoxicity. Mob. Genet. Elem. 2016, 6, e1198300. [Google Scholar] [CrossRef]
- Wallace, N.A.; Belancio, V.P.; Deininger, P.L. L1 mobile element expression causes multiple types of toxicity. Gene 2008, 419, 75–81. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Capetillo, O.; Celeste, A.; Nussenzweig, A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle 2003, 2, 426–427. [Google Scholar] [CrossRef] [PubMed]
- Celeste, A.; Petersen, S.; Romanienko, P.J.; Fernandez-Capetillo, O.; Chen, H.T.; Sedelnikova, O.A.; Reina-San-Martin, B.; Coppola, V.; Meffre, E.; Difilippantonio, M.J. Genomic instability in mice lacking histone H2AX. Science 2002, 296, 922–927. [Google Scholar] [CrossRef]
- Sedelnikova, O.A.; Rogakou, E.P.; Panyutin, I.G.; Bonner, W.M. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat. Res. 2002, 158, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Valdiglesias, V.; Giunta, S.; Fenech, M.; Neri, M.; Bonassi, S. γH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat. Res. 2013, 753, 24–40. [Google Scholar] [CrossRef]
- Kuo, L.J.; Yang, L.X. Gamma-H2AX—A novel biomarker for DNA double-strand breaks. Vivo 2008, 22, 305–309. [Google Scholar]
- Cisneros-Aguirre, M.; Ping, X.; Stark, J.M. To indel or not to indel: Factors influencing mutagenesis during chromosomal break end joining. DNA Repair. 2022, 118, 103380. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Kopera, H.C.; Flasch, D.A.; Nakamura, M.; Miyoshi, T.; Doucet, A.J.; Moran, J.V. LEAP: L1 Element Amplification Protocol. Methods Mol. Biol. 2016, 1400, 339–355. [Google Scholar]
- Igoucheva, O.; Alexeev, V.; Yoon, K. Differential cellular responses to exogenous DNA in mammalian cells and its effect on oligonucleotide-directed gene modification. Gene Ther. 2006, 13, 266–275. [Google Scholar] [CrossRef][Green Version]
- Huerfano, S.; Ryabchenko, B.; Forstová, J. Nucleofection of expression vectors induces a robust interferon response and inhibition of cell proliferation. DNA Cell Biol. 2013, 32, 467–479. [Google Scholar] [CrossRef]
- Dewannieux, M.; Esnault, C.; Heidmann, T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003, 35, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Honma, M.; Sakuraba, M.; Koizumi, T.; Takashima, Y.; Sakamoto, H.; Hayashi, M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair. 2007, 6, 781–788. [Google Scholar] [CrossRef]
- Repanas, K.; Zingler, N.; Layer, L.E.; Schumann, G.G.; Perrakis, A.; Weichenrieder, O. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007, 35, 4914–4926. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kazazian, H.H., Jr.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef]
- Kirilyuk, A.; Tolstonog, G.V.; Damert, A.; Held, U.; Hahn, S.; Löwer, R.; Buschmann, C.; Horn, A.V.; Traub, P.; Schumann, G.G. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008, 36, 648–665. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, Y.; Payer, L.M.; Lords, H.; Steranka, J.P.; Burns, K.H.; Xing, J. Polymorphic mobile element insertions contribute to gene expression and alternative splicing in human tissues. Genome Biol. 2020, 21, 185. [Google Scholar] [CrossRef]
- Belancio, V.P.; Roy-Engel, A.M.; Deininger, P. The impact of multiple splice sites in human L1 elements. Gene 2008, 411, 38–45. [Google Scholar] [CrossRef]
- Gasior, S.L.; Roy-Engel, A.M.; Deininger, P.L. ERCC1/XPF limits L1 retrotransposition. DNA Repair. 2008, 7, 983–989. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Kajikawa, M.; Okada, N. LINE retrotransposition and host DNA repair machinery. Mob. Genet. Elem. 2015, 5, 92–97. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suzuki, J.; Yamaguchi, K.; Kajikawa, M.; Ichiyanagi, K.; Adachi, N.; Koyama, H.; Takeda, S.; Okada, N. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009, 5, e1000461. [Google Scholar] [CrossRef] [PubMed]
- Pezone, A.; Olivieri, F.; Napoli, M.V.; Procopio, A.; Avvedimento, E.V.; Gabrielli, A. Inflammation and DNA damage: Cause, effect or both. Nat. Rev. Rheumatol. 2023, 19, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Han, K.; Meyer, T.J.; Kim, H.S.; Batzer, M.A. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE 2008, 3, e4047. [Google Scholar] [CrossRef]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Cai, Y.; Sablina, A.A. Cancer-associated chromosomal deletions: Size makes a difference. Cell Cycle 2016, 15, 2850–2851. [Google Scholar] [CrossRef][Green Version]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M. Punctuated evolution of prostate cancer genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef]
- McKerrow, W.; Wang, X.; Mendez-Dorantes, C.; Mita, P.; Cao, S.; Grivainis, M.; Ding, L.; LaCava, J.; Burns, K.H.; Boeke, J.D. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc. Natl. Acad. Sci. USA 2022, 119, e2115999119. [Google Scholar] [CrossRef]
- Chuang, N.T.; Gardner, E.J.; Terry, D.M.; Crabtree, J.; Mahurkar, A.A.; Rivell, G.L.; Hong, C.C.; Perry, J.A.; Devine, S.E. Mutagenesis of human genomes by endogenous mobile elements on a population scale. Genome Res. 2021, 31, 2225–2235. [Google Scholar] [CrossRef]
- Stevens, C.; Hightower, A.; Buxbaum, S.G.; Falzarano, S.M.; Rhie, S.K. Genomic, epigenomic, and transcriptomic signatures of prostate cancer between African American and European American patients. Front. Oncol. 2023, 13, 1079037. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.A.; Browne, C.; Gregson, N.; Joyce, C.; White, H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J. Med. Genet. 1992, 29, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Ardeljan, D.; Taylor, M.S.; Ting, D.T.; Burns, K.H. The Human Long Interspersed Element-1 Retrotransposon: An Emerging Biomarker of Neoplasia. Clin. Chem. 2017, 63, 816–822. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kines, K.J.; Sokolowski, M.; DeFreece, C.; Shareef, A.; deHaro, D.L.; Belancio, V.P. Large Deletions, Cleavage of the Telomeric Repeat Sequence, and Reverse Transcriptase-Mediated DNA Damage Response Associated with Long Interspersed Element-1 ORF2p Enzymatic Activities. Genes 2024, 15, 143. https://doi.org/10.3390/genes15020143
Kines KJ, Sokolowski M, DeFreece C, Shareef A, deHaro DL, Belancio VP. Large Deletions, Cleavage of the Telomeric Repeat Sequence, and Reverse Transcriptase-Mediated DNA Damage Response Associated with Long Interspersed Element-1 ORF2p Enzymatic Activities. Genes. 2024; 15(2):143. https://doi.org/10.3390/genes15020143
Chicago/Turabian StyleKines, Kristine J., Mark Sokolowski, Cecily DeFreece, Afzaal Shareef, Dawn L. deHaro, and Victoria P. Belancio. 2024. "Large Deletions, Cleavage of the Telomeric Repeat Sequence, and Reverse Transcriptase-Mediated DNA Damage Response Associated with Long Interspersed Element-1 ORF2p Enzymatic Activities" Genes 15, no. 2: 143. https://doi.org/10.3390/genes15020143
APA StyleKines, K. J., Sokolowski, M., DeFreece, C., Shareef, A., deHaro, D. L., & Belancio, V. P. (2024). Large Deletions, Cleavage of the Telomeric Repeat Sequence, and Reverse Transcriptase-Mediated DNA Damage Response Associated with Long Interspersed Element-1 ORF2p Enzymatic Activities. Genes, 15(2), 143. https://doi.org/10.3390/genes15020143




