Severe COVID-19 May Impact Hepatic Fibrosis /Hepatic Stellate Cells Activation as Indicated by a Pathway and Population Genetic Study
Abstract
1. Introduction
2. Methods
2.1. Identification of Candidate Genes for Analysis
2.2. Canonical Signalling Pathway Analysis, Protein Interactions and GO Enrichment Pathway Analysis
2.3. Mortality Rate Frequency Calculation
2.4. Principal Component (PC) Analysis of Genomic Variants in Genes Identified by Pathway Analysis in European Populations
2.5. Differential Allele Frequencies Analysis and Functional Annotations of SNPs
3. Results
3.1. Shortlisted Candidate Genes and Their Role during Viral Infection
3.2. Signalling Pathways Involved in SARS Infection
3.3. Protein Interactions and Signalling Pathways Associated with COVID-19 Aggressiveness
3.4. Mortality Rate and Common COVID-19 Comorbidities Data in European Populations
3.5. PC Analysis Reveals Genetic Variants in Hepatic Fibrosis/HSC Activation Pathway Segregates Differently in European Populations
3.6. Functional SNPs within Genes Associated with Severe COVID-19
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cherry, J.D. The chronology of the 2002–2003 SARS mini pandemic. Paediatr. Respir. Rev. 2004, 5, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Middle East Respiratory Syndrome; World Health Organisation: Geneva, Switzerland, 2021. [Google Scholar]
- Josset, L.; Engelmann, F.; Haberthur, K.; Kelly, S.; Park, B.; Kawoaka, Y.; García-Sastre, A.; Katze, M.G.; Messaoudi, I. Increased Viral Loads and Exacerbated Innate Host Responses in Aged Macaques Infected with the 2009 Pandemic H1N1 Influenza A Virus. J. Virol. 2012, 86, 11115–11127. [Google Scholar] [CrossRef] [PubMed]
- Diallo, M.S.K.; Toure, A.; Sow, M.S.; Kpamou, C.; Keita, A.K.; Taverne, B.; Peeters, M.; Msellati, P.; Barry, T.A.; Etard, J.F.; et al. Understanding the long-term evolution and predictors of sequelae of Ebola virus disease survivors in Guinea: A 48-month prospective, longitudinal cohort study (PostEboGui). Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Brake, S.J.; Barnsley, K.; Lu, W.; McAlinden, K.D.; Eapen, M.S.; Sohal, S.S. Smoking Upregulates Angiotensin-Converting Enzyme-2 Receptor: A Potential Adhesion Site for Novel Coronavirus SARS-CoV-2 (Covid-19). J. Clin. Med. 2020, 9, 841. [Google Scholar] [CrossRef]
- Chakravarty, D.; Nair, S.S.; Hammouda, N.; Ratnani, P.; Gharib, Y.; Wagaskar, V.; Mohamed, N.; Lundon, D.; Dovey, Z.; Kyprianou, N.; et al. Sex differences in SARS-CoV-2 infection rates and the potential link to prostate cancer. Commun. Biol. 2020, 3, 374. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.; Wang, Y.; Yuen, T.T.; Chai, Y.; Hou, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 1400–1409. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Mercatelli, D.; Ceraolo, C.; Giorgi, F.M. Master Regulator Analysis of the SARS-CoV-2/Human Interactome. J. Clin. Med. 2020, 9, 982. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 Hijacking of Host Mitochondria in Pathogenesis of COVID-19. Am. J. Physiol. Cell Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Cui, H.; Gao, Z.; Liu, M.; Lu, S.; Mkandawire, W.; Narykov, O.; Sun, M.; Korkin, D. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses 2020, 12, 360. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhou, Y.-S.; Lian, J.-Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.-Y.; Geng, J.-J.; et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Fernández-Rhodes, L.; Young, K.L.; Lilly, A.G.; Raffield, L.M.; Highland, H.M.; Wojcik, G.L.; Agler, C.; Love, S.-A.M.; Okello, S.; Petty, L.E.; et al. Importance of Genetic Studies of Cardiometabolic Disease in Diverse Populations. Circ. Res. 2020, 126, 1816–1840. [Google Scholar] [CrossRef]
- Nikoghosyan, M.; Hakobyan, S.; Hovhannisyan, A.; Loeffler-Wirth, H.; Binder, H.; Arakelyan, A. Population Levels Assessment of the Distribution of Disease-Associated Variants With Emphasis on Armenians—A Machine Learning Approach. Front. Genet. 2019, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Deka, R.; Courtney, P.T.; Parsons, J.K.; Nelson, T.J.; Nalawade, V.; Luterstein, E.; Cherry, D.R.; Simpson, D.R.; Mundt, A.J.; Murphy, J.D.; et al. Association Between African American Race and Clinical Outcomes in Men Treated for Low-Risk Prostate Cancer With Active Surveillance. JAMA 2020, 324, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Chan, V.S.F.; Chan, K.Y.K.; Chen, Y.; Poon, L.L.M.; Cheung, A.N.Y.; Zheng, B.; Chan, K.-H.; Mak, W.; Ngan, H.Y.S.; Xu, X.; et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 2006, 38, 38–46. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Coronavirus Disease (COVID-2019) Situation Reports. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 20 December 2020).
- Univeristy of Oxford; Blavatnik School of Government. Coronavirus Government Response Tracker; Blavatnik School of Government: Oxford, UK, 2020. [Google Scholar]
- Healthcare, D.-G.F.P. FAQ—COVID-19; Sallute, M.D.L., Ed.; 2021. [Google Scholar]
- Publica, S. Desarrollo del Estado de Alarma en las Comunidades Autónomas; Ministerio de Sanidad: Madrid, Spain, 2021. [Google Scholar]
- Modi, C.; Böhm, V.; Ferraro, S.; Stein, G.; Seljak, U. Estimating COVID-19 mortality in Italy early in the COVID-19 pandemic. Nat. Commun. 2021, 12, 2729. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformation 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Brohée, S.; van Helden, J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinform. 2006, 7, 488. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformation 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformation 2013, 29, 661–663. [Google Scholar] [CrossRef]
- CDC. Mortality Frequency Measures. Available online: https://www.cdc.gov/csels/dsepd/ss1978/lesson3/section3.html (accessed on 4 June 2020).
- World Health Organisation. International Guidelines for Certification and Classification (Coding) Of COVID-19 as Cause of Death; WHO: Geneva, Switzerland, 2020; p. 14. [Google Scholar]
- Population 1st January 2020. In Eurostat Data Browser. Available online: https://ec.europa.eu/eurostat/databrowser/view/tps00001/default/table?lang=en (accessed on 20 June 2020).
- Guan, W.-J.; Liang, W.-H.; Zhao, Y.; Liang, H.-R.; Chen, Z.-S.; Li, Y.-M.; Liu, X.-Q.; Chen, R.-C.; Tang, C.-L.; Wang, T.; et al. Comorbidity and its impact on 1590 patients with Covid-19 in China: A Nationwide Analysis. Eur. Respir. J. 2020, 55, 2000547. [Google Scholar] [CrossRef]
- Caramelo, F.; Ferreira, N.; Oliveiros, B. Estimation of risk factors for COVID-19 mortality—Preliminary results. MedRxiv 2020. MedRxiv: 2020.02.24.20027268. [Google Scholar]
- Shahid, Z.; Kalayanamitra, R.; McClafferty, B.; Kepko, D.; Ramgobin, D.; Patel, R.; Aggarwal, C.S.; Vunnam, R.R.; Sahu, N.; Bhatt, D.; et al. COVID-19 And Older Adults: What We Know. J. Am. Geriatr. Soc. 2020, 68, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Worldometer Age, Sex, Existing Conditions of COVID-19 Cases and Deaths. Available online: https://www.worldometers.info/coronavirus/coronavirus-age-sex-demographics/ (accessed on 20 April 2020).
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. New Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Dong, S.; Boyle, A.P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 2019, 40, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.; Yamamoto, N.; Nakai-Murakami, C.; Osawa, Y.; Tokunaga, K.; Sata, T.; Yamamoto, N.; Sasazuki, T.; Ishizaka, Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc. Natl. Acad. Sci. USA 2008, 105, 7809–7814. [Google Scholar] [CrossRef]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef]
- Muramatsu, T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem 2016, 159, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, W.D.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef] [PubMed]
- Lozach, P.-Y.; Burleigh, L.; Staropoli, I.; Amara, A. The C type lectins DC-SIGN and L-SIGN: Receptors for viral glycoproteins. Methods Mol. Biol. 2007, 379, 51–68. [Google Scholar] [PubMed]
- Alvarez, C.P.; Lasala, F.; Carrillo, J.; Muñiz, O.; Corbí, A.L.; Delgado, R. C-Type Lectins DC-SIGN and L-SIGN Mediate Cellular Entry by Ebola Virus in cis and in trans. J. Virol. 2002, 76, 6841–6844. [Google Scholar] [CrossRef] [PubMed]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef]
- Kamitani, W.; Huang, C.; Narayanan, K.; Lokugamage, K.G.; Makino, S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009, 16, 1134–1140. [Google Scholar] [CrossRef]
- Tanaka, T.; Kamitani, W.; DeDiego, M.L.; Enjuanes, L.; Matsuura, Y. Severe Acute Respiratory Syndrome Coronavirus nsp1 Facilitates Efficient Propagation in Cells through a Specific Translational Shutoff of Host mRNA. J. Virol. 2012, 86, 11128–11137. [Google Scholar] [CrossRef]
- Law, A.H.Y.; Lee, D.C.W.; Cheung, B.K.W.; Yim, H.C.H.; Lau, A.S.Y. Role for Nonstructural Protein 1 of Severe Acute Respiratory Syndrome Coronavirus in Chemokine Dysregulation. J. Virol. 2007, 81, 416–422. [Google Scholar] [CrossRef]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef]
- Saikatendu, K.S.; Joseph, J.S.; Subramanian, V.; Clayton, T.; Griffith, M.; Moy, K.; Velasquez, J.; Neuman, B.W.; Buchmeier, M.J.; Stevens, R.C.; et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1’’-phosphate dephosphorylation by a conserved domain of nsP3. Structure 2005, 13, 1665–1675. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Mozzi, A.; Pozzoli, U.; Al-Daghri, N.; Clerici, M.; Sironi, M. Extensive Positive Selection Drives the Evolution of Nonstructural Proteins in Lineage C Betacoronaviruses. J. Virol. 2016, 90, 3627–3639. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Hauer, D.; McPherson, R.L.; Utt, A.; Kirby, I.T.; Cohen, M.S.; Merits, A.; Leung, A.K.L.; Griffin, D.E. ADP-ribosyl–binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. 2018, 115, E10457–E10466. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Blet, A.; Smyth, D.; Li, H. The Science Underlying COVID-19: Implications for the Cardiovascular System. Circulation 2020, 142, 68–78. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, H.-T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M.; et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Ingenuity Pathway Analysis. Caveolar-mediated Endocytosis Signaling; Ingenuity Target Explorer, 2020. [Google Scholar]
- Ingenuity Pathway Analysis. MSP-RON Signaling Pathway; Ingenuity Target Explorer, 2020. [Google Scholar]
- Ingenuity Pathway Analysis. The Communication between Innate and Adaptive Immune Cells; Ingenuity Target Explorer, 2020. [Google Scholar]
- European Union. Available online: Europa.eu (accessed on 20 June 2020).
- Population Reference Bureau, P. Countries With the Oldest Populations in the World. Available online: https://www.prb.org/countries-with-the-oldest-populations/ (accessed on 20 June 2020).
- Townsend, N.; Wilson, L.; Bhatnagar, P.; Wickramasinghe, K.; Rayner, M.; Nichols, M. Cardiovascular disease in Europe: Epidemiological update 2016. Eur. Heart J. 2016, 37, 3232–3245. [Google Scholar] [CrossRef]
- Patterson, C.C.; Harjutsalo, V.; Rosenbauer, J.; Neu, A.; Cinek, O.; Skrivarhaug, T.; Rami-Merhar, B.; Soltesz, G.; Svensson, J.; Parslow, R.C.; et al. Trends and cyclical variation in the incidence of childhood type 1 diabetes in 26 European centres in the 25 year period 1989–2013: A multicentre prospective registration study. Diabetologia 2019, 62, 408–417. [Google Scholar] [CrossRef]
- Khan, M.A.B.; Hashim, M.J.; King, J.K.; Govender, R.D.; Mustafa, H.; Al Kaabi, J. Epidemiology of Type 2 Diabetes—Global Burden of Disease and Forecasted Trends. J. Epidemiol. Glob. Health 2020, 10, 107–111. [Google Scholar] [CrossRef]
- Gallus, S.; Lugo, A.; Liu, X.; Behrakis, P.; Boffi, R.; Bosetti, C.; Carreras, G.; Chatenoud, L.; Clancy, L.; Continente, X.; et al. Who smokes in Europe? Data from 12 European countries in the TackSHS survey (2017–2018). J. Epidemiol. 2020, 31, 145–151. [Google Scholar] [CrossRef]
- Chheda, H.; Palta, P.; Pirinen, M.; McCarthy, S.; Walter, K.; Koskinen, S.; Salomaa, V.; Daly, M.; Durbin, R.; Palotie, A.; et al. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur. J. Hum. Genet. EJHG 2017, 25, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Locke, A.E.; Steinberg, K.M.; Chiang, C.W.K.; Service, S.K.; Havulinna, A.S.; Stell, L.; Pirinen, M.; Abel, H.J.; Chiang, C.C.; Fulton, R.S.; et al. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 2019, 572, 323–328. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Vankadari, N.; Wilce, J.A. Emerging COVID-19 coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020, 9, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Prokscha, A.; Naim, H.Y.; Müller, M.A.; Drosten, C.; Pöhlmann, S.; Hoffmann, M. Polymorphisms in dipeptidyl peptidase 4 reduce host cell entry of Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2020, 9, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.J.; Liu, C.Y.; Chiang, B.L.; Chao, Y.C.; Chen, C.C. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: Identification of two functional regions. J. Immunol. 2004, 173, 7602–7614. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef]
- Wathelet, M.G.; Orr, M.; Frieman, M.B.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus Evades Antiviral Signaling: Role of nsp1 and Rational Design of an Attenuated Strain. J. Virol. 2007, 81, 11620–11633. [Google Scholar] [CrossRef]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.D.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease Ubiquitin-Like Domain and Catalytic Domain Regulate Antagonism of IRF3 and NF-κB Signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef] [PubMed]
- Siu, K.-L.; Yuen, K.-S.; Castano-Rodriguez, C.; Ye, Z.-W.; Yeung, M.-L.; Fung, S.-Y.; Yuan, S.; Chan, C.-P.; Yuen, K.-Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef]
- Sakai, Y.; Kawachi, K.; Terada, Y.; Omori, H.; Matsuura, Y.; Kamitani, W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology 2017, 510, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.-Q.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct. Target. Ther. 2020, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Meng, M.; Kumar, R.; Wu, Y.; Huang, J.; Deng, Y.; Weng, Z.; Yang, L. Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A systemic review and meta-analysis. Int. J. Infect. Dis. 2020, 96, 131–135. [Google Scholar] [CrossRef]
- Sungur, C.M.; Murphy, W.J. Positive and negative regulation by NK cells in cancer. Crit Rev Oncog 2014, 19, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Radujkovic, A.; Hippchen, T.; Tiwari-Heckler, S.; Dreher, S.; Boxberger, M.; Merle, U. Vitamin D Deficiency and Outcome of COVID-19 Patients. Nutrients 2020, 12, 2757. [Google Scholar] [CrossRef]
- Keenan, B.P.; Fong, L.; Kelley, R.K. Immunotherapy in hepatocellular carcinoma: The complex interface between inflammation, fibrosis, and the immune response. J. ImmunoTherapy Cancer 2019, 7, 267. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef]
- Liu, R.; Li, X.; Zhu, W.; Wang, Y.; Zhao, D.; Wang, X.; Gurley, E.C.; Liang, G.; Chen, W.; Lai, G.; et al. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 2019, 70, 1317–1335. [Google Scholar] [CrossRef] [PubMed]
- Weiskirchen, R.; Tacke, F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg. Nutr. 2014, 3, 344–363. [Google Scholar] [PubMed]
- Wang, Y.; Li, J.; Wang, X.; Sang, M.; Ho, W. Hepatic stellate cells, liver innate immunity, and hepatitis C virus. J. Gastroenterol. Hepatol. 2013, 28 (Suppl. 1), 112–115. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.; Jouve, T.; Malvezzi, P.; Rostaing, L. Renal complications of liver diseases. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, L.; Wang, F.S. Liver injury in COVID-19: Management and challenges. Lancet. Gastroenterol. Hepatol. 2020, 5, 428–430. [Google Scholar] [CrossRef]
- Marjot, T.; Moon, A.M.; Cook, J.A.; Abd-Elsalam, S.; Aloman, C.; Armstrong, M.J.; Pose, E.; Brenner, E.J.; Cargill, T.; Catana, M.-A.; et al. Outcomes following SARS-CoV-2 infection in patients with chronic liver disease: An international registry study. J. Hepatol. 2021, 74, 567–577. [Google Scholar] [CrossRef]
- Téllez, L.; Martín Mateos, R.M. COVID-19 and liver disease: An update. Gastroenterol. Y Hepatol. (Engl. Ed. ) 2020, 43, 472–480. [Google Scholar] [CrossRef]
- Oyelade, T.; Alqahtani, J.; Canciani, G. Prognosis of COVID-19 in Patients with Liver and Kidney Diseases: An Early Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2020, 5, 80. [Google Scholar] [CrossRef]
- Wang, Q.; Davis, P.B.; Xu, R. COVID-19 risk, disparities and outcomes in patients with chronic liver disease in the United States. EClinicalMedicine 2021, 31, 100688. [Google Scholar] [CrossRef]
- Mokhtari, T.; Hassani, F.; Ghaffari, N.; Ebrahimi, B.; Yarahmadi, A.; Hassanzadeh, G. COVID-19 and multiorgan failure: A narrative review on potential mechanisms. J. Mol. Histol. 2020, 51, 613–628. [Google Scholar] [CrossRef]
- Testino, G. Covid-19 infection, liver injury and prognosis: A suggestion. Eur. J. Gastroenterol. Hepatol. 2021, 33, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Pimpin, L.; Cortez-Pinto, H.; Negro, F.; Corbould, E.; Lazarus, J.V.; Webber, L.; Sheron, N. Hepahealth Project Report; EASL, EU; 2018; p. 177. [Google Scholar]
- Jiménez, E.; Fontán-Vela, M.; Valencia, J.; Fernandez-Jimenez, I.; Álvaro-Alonso, E.A.; Izquierdo-García, E.; Lazaro Cebas, A.; Gallego Ruiz-Elvira, E.; Troya, J.; Tebar-Martinez, A.J.; et al. Characteristics, complications and outcomes among 1549 patients hospitalised with COVID-19 in a secondary hospital in Madrid, Spain: A retrospective case series study. BMJ Open 2020, 10, e042398. [Google Scholar] [CrossRef] [PubMed]
- Lobo-Valbuena, B.; García-Arias, M.; Pérez, R.B.; Delgado, D.V.; Gordo, F. Characteristics of critical patients with COVID-19 in a Spanish second-level hospital. Med. Intensiv. 2021, 45, 56–58. [Google Scholar] [CrossRef]
- Plus, M. Lactate Dehydrogenase (LDH) Isoenzymes Test. Available online: https://medlineplus.gov/lab-tests/lactate-dehydrogenase-ldh-isoenzymes-test/ (accessed on 20 June 2020).
- University of Rochester. Lactate Dehydrogenase Isoenzymes. Available online: https://www.urmc.rochester.edu/encyclopedia/content.aspx?contenttypeid=167&contentid=lactate_dehydrogenase_isoenzymes (accessed on 1 September 2020).
- Bellan, M.; Patti, G.; Hayden, E.; Azzolina, D.; Pirisi, M.; Acquaviva, A.; Aimaretti, G.; Aluffi Valletti, P.; Angilletta, R.; Arioli, R.; et al. Fatality rate and predictors of mortality in an Italian cohort of hospitalized COVID-19 patients. Sci. Rep. 2020, 10, 20731. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Greco, M.; Zanella, A.; Albano, G.; Antonelli, M.; Bellani, G.; Bonanomi, E.; Cabrini, L.; Carlesso, E.; Castelli, G.; et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020, 180, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Niu, Z.; Jiang, X.; Zhang, Z.; Zheng, Y.; Wang, Z.; Zhu, Y.; Gao, L.; Huang, H.; Wang, X.; et al. SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. CellPress 2020, 23, 101744. [Google Scholar] [CrossRef]
- Marjot, T.; Webb, G.J.; Barritt, A.S.; Ginès, P.; Lohse, A.W.; Moon, A.M.; Pose, E.; Trivedi, P.; Barnes, E. SARS-CoV-2 vaccination in patients with liver disease: Responding to the next big question. Lancet Gastroenterol. Hepatol. 2021, 6, 156–158. [Google Scholar] [CrossRef] [PubMed]
- The, C.-H.G.I. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 2020, 28, 715–718. [Google Scholar]
- Scherger, S.; Henao-Martínez, A.; Franco-Paredes, C.; Shapiro, L. Rethinking interleukin-6 blockade for treatment of COVID-19. Med. Hypotheses 2020, 144, 110053. [Google Scholar] [CrossRef]
- Ghosn, L.; Chaimani, A.; Evrenoglou, T.; Davidson, M.; Graña, C.; Schmucker, C.; Bollig, C.; Henschke, N.; Sguassero, Y.; Nejstgaard, C.H.; et al. Interleukin-6 blocking agents for treating COVID-19: A living systematic review. Cochrane Database Syst. Rev. 2021, 2021, CD013881. [Google Scholar]
- Cavalli, G.; Larcher, A.; Tomelleri, A.; Campochiaro, C.; Della-Torre, E.; De Luca, G.; Farina, N.; Boffini, N.; Ruggeri, A.; Poli, A.; et al. Interleukin-1 and interleukin-6 inhibition compared with standard management in patients with COVID-19 and hyperinflammation: A cohort study. Lancet Rheumatol. 2021, 3, e253–e261. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.C.; Liew, D.F.L.; Liew, J.W.; Monaco, C.; Richards, D.; Shivakumar, S.; Tanner, H.L.; Feldmann, M. The Potential for Repurposing Anti-TNF as a Therapy for the Treatment of COVID-19. Med 2020, 1, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; Shao, R.; Han, X.; Su, C.; Lu, W. Risk and clinical outcomes of COVID-19 in patients with rheumatic diseases compared with the general population: A systematic review and meta-analysis. Rheumatol Int. 2021, 41, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Repurposed Antiviral Drugs for Covid-19—Interim WHO Solidarity Trial Results. New Engl. J. Med. 2020, 384, 497–511.
- Wang, B.; Li, D.; Liu, T.; Wang, H.; Luo, F.; Liu, Y. Subcutaneous injection of IFN alpha-2b for COVID-19: An observational study. BMC Infect. Dis. 2020, 20, 723. [Google Scholar] [CrossRef]
- Ricardo Pereda, D.G.; Rivero, H.B.; Rivero, J.C.; Pérez, A.; Lopez, L.d.; Mezquia, N.; Venegas, R.; Betancourt, J.R.; Domínguez, R.E.; Nodarse, H. Therapeutic Effectiveness of Interferon Alpha 2b Treatment for COVID-19 Patient Recovery. J. Interferon Cytokine Res. 2020, 40, 578–588. [Google Scholar] [CrossRef]
- Pandit, A.; Bhalani, N.; Bhushan, B.L.S.; Koradia, P.; Gargiya, S.; Bhomia, V.; Kansagra, K. Efficacy and safety of pegylated interferon alfa-2b in moderate COVID-19: A phase II, randomized, controlled, open-label study. Int. J. Infect. Dis. 2021, 105, 516–521. [Google Scholar] [CrossRef]
- Group, T.S.C.-G. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. New Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar]
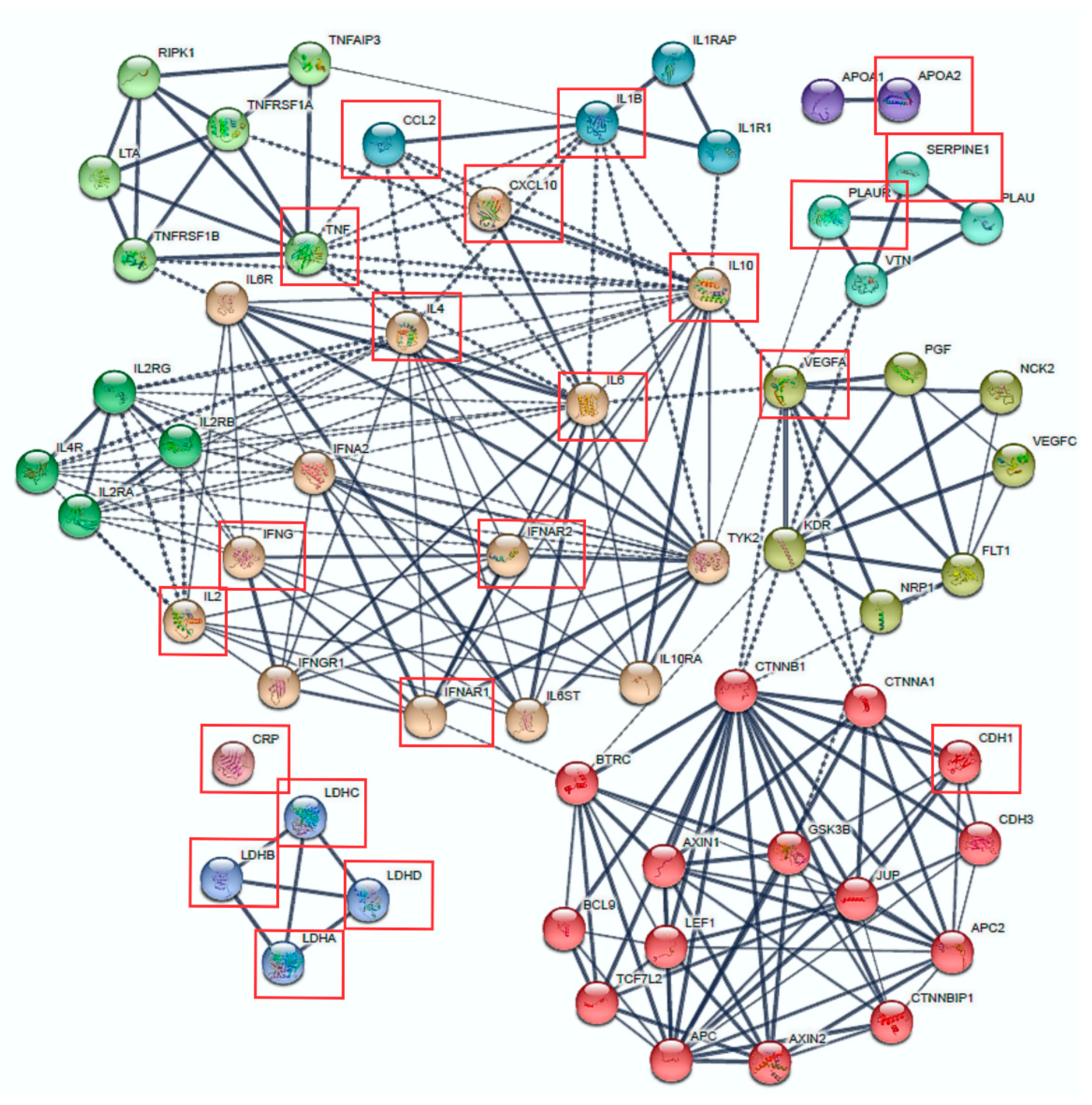

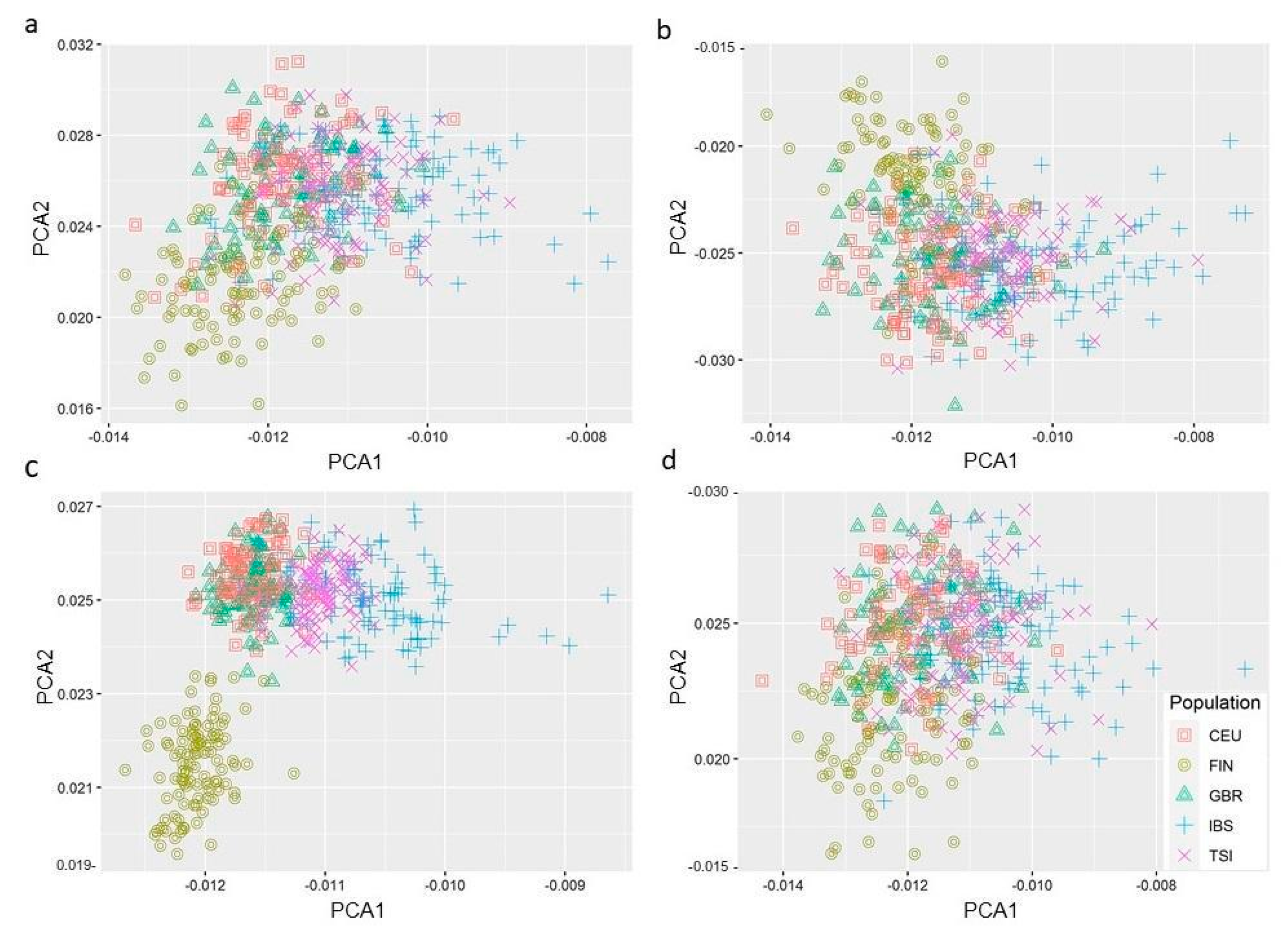
| Candidate Genes | Virus | Host Cell Response/Viral Mechanism | ASSAY Methods | Refs. |
|---|---|---|---|---|
| ACE2 | SARS-CoV-1, SARS-CoV-2 | High S binding affinity. Facilitating host cell recognition. | Multiple in vitro and in silico analysis. | [12,13,14,45,46,47] |
| TMPRSS2 | SARS-CoV-1, SARS-CoV-2 | S protein activator, leading to viral membrane conformational change and facilitating SARS virus. | Multiple in vitro and in silico analyses. | [13,46,48,49] |
| BSG | SARS-CoV-2, malaria, HIV, HepB and HHV | Basigin genes encode CD147 transmembrane glycoprotein recognised by several pathogens. CD147 directly binds to SARS-CoV-2 S protein affecting viral replication. | Review and in vitro. SARS-CoV-2 strain isolated from COVID-19 patients. Direct in vitro infection, Co-IP and ELISA. | [18,50] |
| HAT | SARS-CoV-1, HCoV (229E) | Histone acetyltransferases family, encoding for a family of cell nuclear enzymes. They contribute to SARS-CoV-1 entry, but are not essential for S protein activation. | In vitro. Both studies: gene cloning, lentiviral expression system, protein expression and cell-cell fusion analysis. | [46,48] |
| CLEC4M | SARS-CoV-1, EVD, Dengue, HCV, CMV, Sindbis, HIV | C-type lectin domain family 4 member M genes encode for L-SIGN membrane receptor, recognised by the S protein. Homozygous L-SIGN associated with SARS disease protective role. | In vitro. Infection of SARS-CoV-1 human cells, gene expression, cDNA library, IHC assays. Genetic risk association from SARS patients and controls. | [24,51,52,53] |
| ANPEP, ENPEP, DPP4 (or CD26) | ACE2 studies, HCoV-22944, MERS-CoV45 | Closest co-expression of these three peptidases (R > 0.8) with ACE2 in different human tissues. HCoV-22944 binds to ENPEP while MERS to DPP4. | Single cell in silico ligand-receptor affinity assays. Data sourced from GEO, Human Cell Atlas, Viral Receptor and Membranome databases. | [2,49,54] |
| Cathepsin-B-L | SARS-CoV-1, SARS-CoV-2, MERS-CoV | Facilitates SARS-CoV-2 cell entry by virus–cell membrane fusion mechanisms but its inhibition does not disable virus entry. | In vitro. SARS-CoV-2 S protein pseudovirus system in the human lung cell model. | [13,14] |
| Candidate Genes | Virus | Host Cell Response/Viral Mechanism | Research Assay | Ref. |
|---|---|---|---|---|
| 40s | Nsp1 studies | Encoding for ribosomal protein S3, interacts with viral Nsp1, inhibiting the host’s protein translation by capping the 5′mRNA. | In vitro. Reporter gene assays followed by transcriptomics, RNA immunoprecipitation and proteomics assays. | [55,56] |
| CCL5, CCL3, CXCL10 | SARS-CoV-1, | These genes encode for IP-10 * protein. Increased levels in lung epithelial cells after Nsp1-direct activation of the NF-kB pathway. IP-10 showed specific uP-regulation in the COVID-19 lung model (when compared to SARS patients). | In vitro. Gene cloning, mRNA and protein expression analysis. Ex vivo. Lung tissue transfected with COVID-19 and gene expression analysis. | [10,57] |
| STING1, TRAF3, TBK1, IKKε | SARS-CoV-1, HCoV (NL63) | The SARS-CoV-1 PLP transmembrane protein interacts with STING, TRAF3, TBK1 and IKKε, disrupting the STING/TBK1/IKKε complex formation and suppressing the production of IFN-α and IFN-β, vital for initial innate immune response. PLP protein is highly conserved in both SARS-CoV viruses, highlighting the use of potential agonists for this protein as treatments. | In vitro. SARS-CoV-1 propagation, and plasmids expressing genes of interest’s co-transduction. Co-IP and ubiquitination signalling detection. In silico. Homology alignments of both SARS viruses, approved compounds database screening and homology models predictions. | [58,59] |
| ADP-ribose | ssRNA | After binding to Nsp3, post-translational modification of PARP15, PARP14 and PARP10 is associated with anti-viral response. | In vitro. Cloning, gene expression, mutagenesis, protein purification and crystallization. In silico, sequence alignments, glycosylation sites’ predictions and 3D mapping. | [60,61,62] |
| Candidate genes | Disease | Reported observations | Ref. |
|---|---|---|---|
| IL1β, IFN-γ, CXCL10, and MCP-1 | SARS and COVID-19 | Increased in SARS and COVID-19 patients. Associated with Th1 cell immunity aberrant response and ARDS | [20] |
| IL4 and IL10 | COVID-19 | Increased in COVID-19 patients. Associated with Th2 cell immunity response, facilitating further ARDS | [20] |
| IL2R and IL6 | COVID-19 | High expression levels positively correlated with the severity of the disease | [10,20,63] |
| CXCL10, MCP-1 and TNF-α. | COVID-19 | COVID-19 ICU patients have increased serum levels of these genes when compared to non-ICU COVID-19 patients | [10,64] |
| IL1β, IL6, cRP | COVID-19 | High levels of IL1β are associated with a poor prognosis. IL6 and cRP are potential early risk biomarkers | [63] |
| SERPINE1 * | COVID-19 | High levels of this protein is associated with vascular inflammation and a higher risk of thrombosis. | [63] |
| LDH-hsCRP-lymphocyte | COVID-19 | Very accurate (>90%) predictive mortality biomarker signature | [65] |
| LD Gene | SNP | Chromosome Position | p-Value | Odds Ratio | CHISQ® | Regulome DB Score |
|---|---|---|---|---|---|---|
| (GRCh37/hg19 Built) | ||||||
| IL1B | rs79750333 | chr2:114515437–114515438 | 2.68 × 10−10 | 2.96 | 37.73 | 0.67 |
| TNF | rs2853982 | chr6:31378750–31378751 | 4.59 × 10−10 | 3.15 | 36.98 | 0.6 |
| IL6 | rs10237482 | chr7:22475177–22475178 | 2.57 × 10−8 | 0.46 | 31.87 | 0.8 |
| LDHA, LDHC | rs56357050 | chr11:18785334–18785335 | 5.68 × 10−8 | 0.46 | 30.56 | 0.6 |
| CXCL10 | rs114493545 | chr4:123766808–123766809 | 3.11 × 10−7 | 6.24 | 22.82 | 0.8 |
| IFNAR1, IFNAR2 | rs60075147 | chr21:33660824–33660825 | 4.31 × 10−7 | 2.74 | 24.48 | 1 |
| LDHB | rs10841699 rs2196017 | chr12:21060248–21060249, chr12:21061314–21061315 | 1.51 × 10−6 | 0.39 | 24.77 | 0.59,0.72 |
| IL10 | rs7530746 | chr1:206712542–206712543 | 6.92 × 10−6 | 1.82 | 20.72 | 0.6 |
| IFNG | rs741347 | chr12:68631372–68631373 | 9.37 × 10−6 | 0.42 | 20.33 | 0.98 |
| VEGFA | rs9381273 | chr6:43976267–43976268 | 9.75 × 10−6 | 2.29 | 19.08 | 0.6 |
| IL4 | rs2243268 | chr5:132013962–132013963 | 1.19 × 10−5 | 0.47 | 19.83 | 0.6 |
| PLAUR | rs2356437 rs7258485 | chr19:44352664–44352665, chr19:44353241–44353242 | 1.46 × 10−5 | 2.03 | 18.65 | 0.6 |
| CRP | rs3806187 | chr1:159750628–159750629 | 2.14 × 10−5 | 0.48 | 18.69 | 0.7 |
| CCL2 | rs1431994 | chr17:32771454–32771455 | 3.01 × 10−5 | 1.9 | 17.14 | 0.6 |
| APOA2 | rs17381453 | chr1:160514986–160514987 | 3.05 × 10−5 | 1.85 | 17.55 | 0.6 |
| IL2 | rs11937337 | chr4:122373527–122373528 | 3.99 × 10−5 | 0.32 | 17.73 | 0.6 |
| LDHD | rs147230411 | chr16:74390997–74390998 | 7.65 × 10−5 | 6.9 | 13.77 | 0.69 |
| SERPINE1 * | rs75339477 | chr7:101012169–101012190 | 2.17 × 10−4 | 0.6 | 13.91 | 0.59 |
| rs79520712 | ||||||
| rs62465617 | ||||||
| rs62465619 | ||||||
| rs62465620 | ||||||
| rs376313468 | ||||||
| CDH1 | rs696587 | chr16:68546471–68546472 | 5.78 × 10−3 | 0.67 | 7.841 | 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moya, L.; Farashi, S.; Suravajhala, P.; Janaththani, P.; Batra, J. Severe COVID-19 May Impact Hepatic Fibrosis /Hepatic Stellate Cells Activation as Indicated by a Pathway and Population Genetic Study. Genes 2023, 14, 22. https://doi.org/10.3390/genes14010022
Moya L, Farashi S, Suravajhala P, Janaththani P, Batra J. Severe COVID-19 May Impact Hepatic Fibrosis /Hepatic Stellate Cells Activation as Indicated by a Pathway and Population Genetic Study. Genes. 2023; 14(1):22. https://doi.org/10.3390/genes14010022
Chicago/Turabian StyleMoya, Leire, Samaneh Farashi, Prashanth Suravajhala, Panchadsaram Janaththani, and Jyotsna Batra. 2023. "Severe COVID-19 May Impact Hepatic Fibrosis /Hepatic Stellate Cells Activation as Indicated by a Pathway and Population Genetic Study" Genes 14, no. 1: 22. https://doi.org/10.3390/genes14010022
APA StyleMoya, L., Farashi, S., Suravajhala, P., Janaththani, P., & Batra, J. (2023). Severe COVID-19 May Impact Hepatic Fibrosis /Hepatic Stellate Cells Activation as Indicated by a Pathway and Population Genetic Study. Genes, 14(1), 22. https://doi.org/10.3390/genes14010022








