Cross-Trait Genetic Analyses Indicate Pleiotropy and Complex Causal Relationships between Headache and Thyroid Function Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview
2.2. Summary Statistics
2.3. Genome-Wide Genetic Correlation
2.4. Local Genetic Correlation between the Traits
2.5. Cross-Trait GWAS Meta-Analysis and Its Characterization
2.6. Causal Analyses between Headache and Thyroid Traits
2.7. Gene-Based Association Study
2.8. Independent Gene-Based Test
2.9. Gene-Level Genetic Overlap
2.10. Pathway Analysis
3. Results
3.1. Univariate SNP Heritability across Headache and Thyroid Traits
3.2. Genetic Correlation between Headache and Thyroid Traits
3.3. Shared Local Variants between Headache and Thyroid Traits
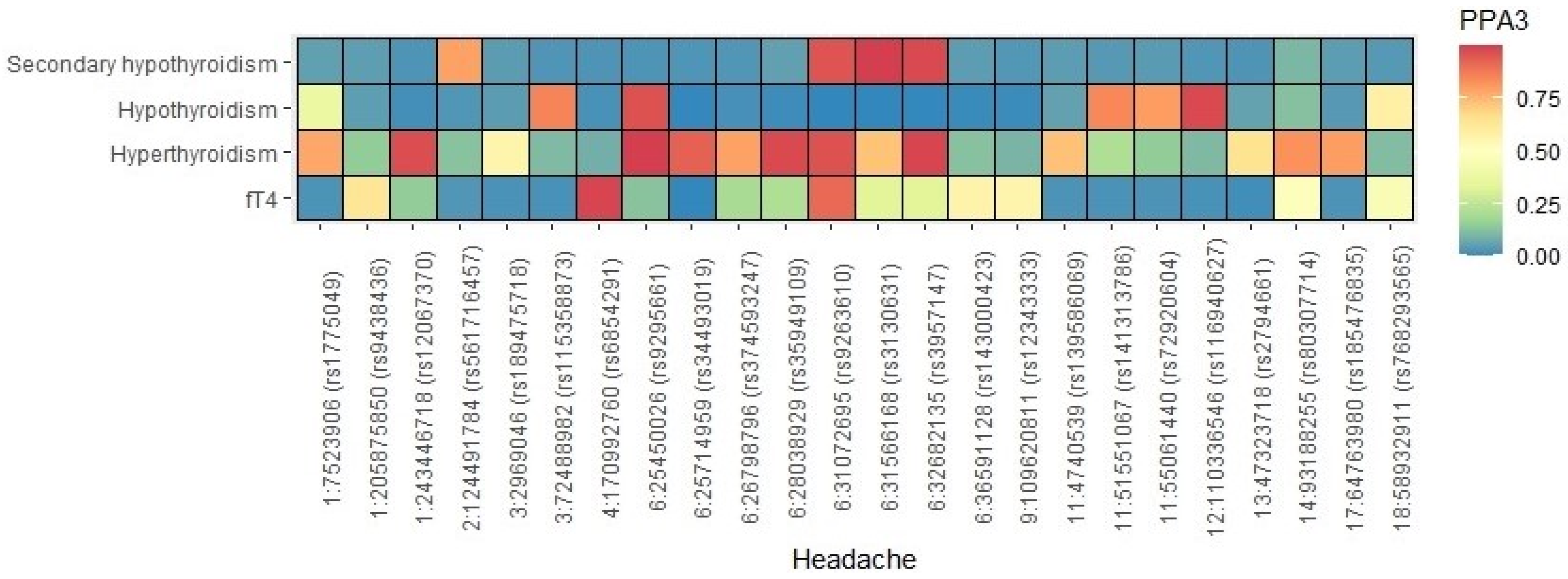
3.4. Shared Loci between Headache and Thyroid Traits
3.5. Causal Relationships
3.6. Gene-Level Genetic Overlap
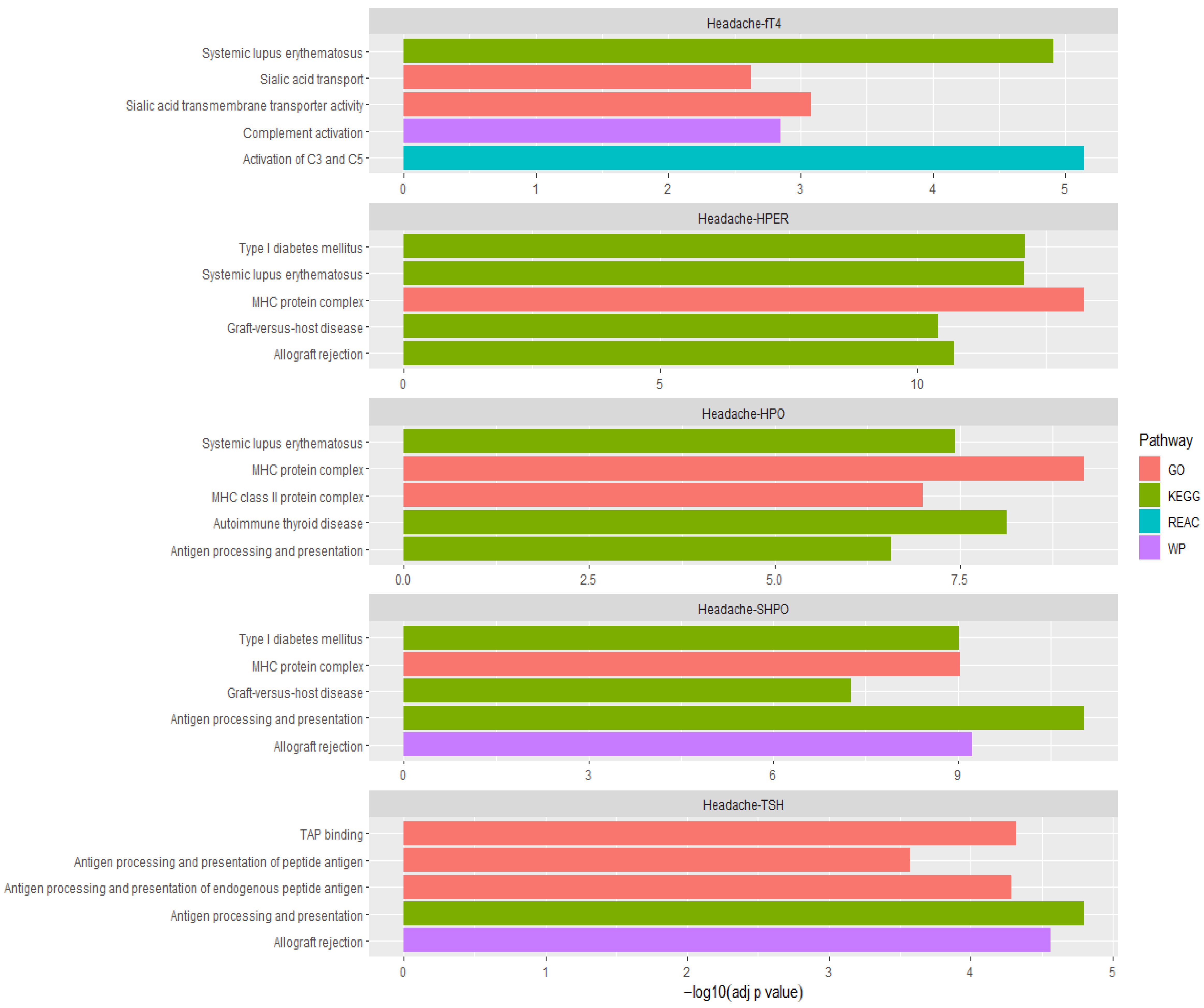
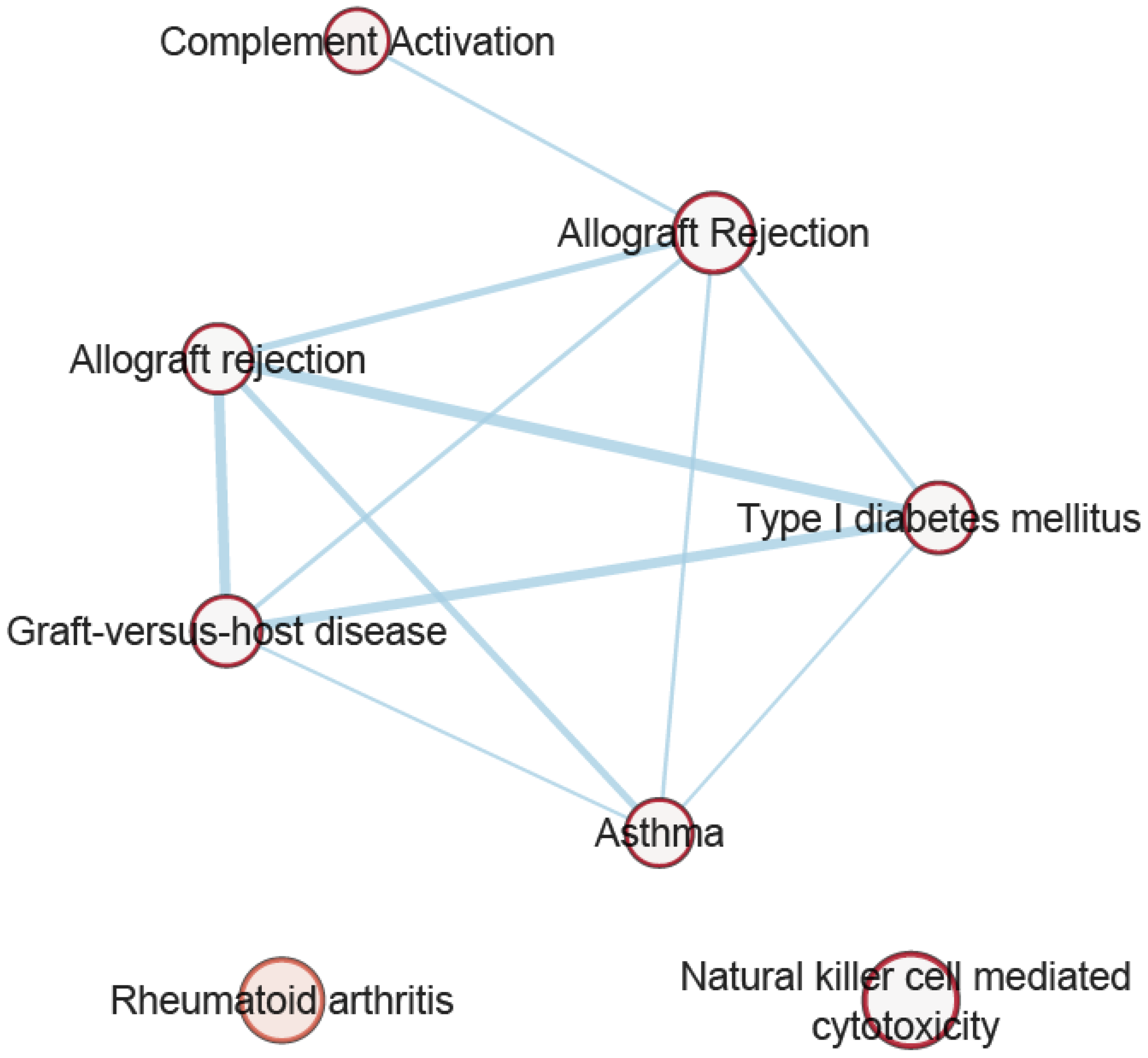
3.7. Pathway Analysis of Overlapping Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stovner, L.J.; Hagen, K.; Linde, M.; Steiner, T.J. The global prevalence of headache: An update, with analysis of the influences of methodological factors on prevalence estimates. J. Headache Pain 2022, 23, 34. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F. Headache disorders: Differentiating and managing the common subtypes. Br. J. Pain 2012, 6, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Biscetti, L.; De Vanna, G.; Cresta, E.; Corbelli, I.; Gaetani, L.; Cupini, L.; Calabresi, P.; Sarchielli, P. Headache and immunological/autoimmune disorders: A comprehensive review of available epidemiological evidence with insights on potential underlying mechanisms. J. Neuroinflammation 2021, 18, 259. [Google Scholar] [CrossRef]
- Martin, A.T.; Pinney, S.M.; Xie, C.; Herrick, R.L.; Bai, Y.; Buckholz, J.; Martin, V.T. Headache Disorders May Be a Risk Factor for the Development of New Onset Hypothyroidism. Headache J. Head Face Pain 2016, 57, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Spanou, I.; Christidi, F.; Liakakis, G.; Rizonaki, K.; Bougea, A.; Anagnostou, E.; Kararizou, E. Primary headache subtypes and thyroid dysfunction: Is there any association? Arq. Neuro-Psiquiatr. 2020, 78, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Moreau, T.; Manceau, E.; Giroud-Baleydier, F.; Dumas, R.; Giroud, M. Headache in hypothyroidism. Prevalence and outcome under thyroid hormone therapy. Cephalalgia 1998, 18, 687–689. [Google Scholar] [CrossRef]
- Carvalho, M.D.F.L.; De Medeiros, J.S.; Valença, M.M. Headache in recent onset hypothyroidism: Prevalence, characteristics and outcome after treatment with levothyroxine. Cephalalgia 2016, 37, 938–946. [Google Scholar] [CrossRef]
- Hagen, K.; Bjoro, T.; Zwart, J.-A.; Vatten, L.; Stovner, L.J.; Bovim, G. Low headache prevalence amongst women with high TSH values. Head-HUNT—A large population-based study. Eur. J. Neurol. 2001, 8, 693–699. [Google Scholar] [CrossRef]
- Uslar, V.; Becker, C.; Weyhe, D.; Tabriz, N. Thyroid disease-specific quality of life questionnaires—A systematic review. Endocrinol. Diabetes Metab. 2022, 5, e357. [Google Scholar] [CrossRef]
- Wilson, K.L.; Casey, B.M.; McIntire, D.D.; Halvorson, L.M.; Cunningham, F.G. Subclinical Thyroid Disease and the Incidence of Hypertension in Pregnancy. Obstet. Gynecol. 2012, 119, 315–320. [Google Scholar] [CrossRef]
- Liu, D.; Jiang, F.; Shan, Z.; Wang, B.; Wang, J.; Lai, Y.; Chen, Y.; Li, M.; Liu, H.; Li, C.; et al. A cross-sectional survey of relationship between serum TSH level and blood pressure. J. Hum. Hypertens. 2009, 24, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Siu, C.-W.; Yeung, C.-Y.; Lau, C.-P.; Kung, A.W.C.; Tse, H.-F. Incidence, clinical characteristics and outcome of congestive heart failure as the initial presentation in patients with primary hyperthyroidism. Heart 2007, 93, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Völzke, H.; Ittermann, T.; O Schmidt, C.; Dörr, M.; John, U.; Wallaschofski, H.; Stricker, B.H.C.; Felix, S.B.; Rettig, R. Subclinical hyperthyroidism and blood pressure in a population-based prospective cohort study. Eur. J. Endocrinol. 2009, 161, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.N.; Albrecht, D.; Scholz, A.; Gutierrez-Buey, G.; Lazarus, J.H.; Dayan, C.M.; Okosieme, O.E. Global epidemiology of hyperthyroidism and hypothyroidism. Nat. Rev. Endocrinol. 2018, 14, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Beck-Peccoz, P.; Rodari, G.; Giavoli, C.; Lania, A. Central hypothyroidism—A neglected thyroid disorder. Nat. Rev. Endocrinol. 2017, 13, 588–598. [Google Scholar] [CrossRef]
- Hadlow, N.C.; Rothacker, K.M.; Wardrop, R.; Brown, S.J.; Lim, E.M.; Walsh, J. The Relationship Between TSH and Free T4 in a Large Population Is Complex and Nonlinear and Differs by Age and Sex. J. Clin. Endocrinol. Metab. 2013, 98, 2936–2943. [Google Scholar] [CrossRef]
- Bashir, H.; Shah, P.; Bhat, M.; Mddm, B.; Imran, A. Association of hypothyroidism in patients with migraine and tension-type headache disorders in Kashmir, North India. Neurol. Asia 2015, 20, 257–261. [Google Scholar]
- PAN-UKB Team. Available online: https://pan.ukbb.broadinstitute.org (accessed on 29 August 2020).
- Teumer, A.; LifeLines Cohort Study; Chaker, L.; Groeneweg, S.; Li, Y.; Di Munno, C.; Barbieri, C.; Schultheiss, U.T.; Traglia, M.; Ahluwalia, T.S.; et al. Genome-wide analyses identify a role for SLC17A4 and AADAT in thyroid hormone regulation. Nat. Commun. 2018, 9, 4455. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.-R.; Duncan, L.; Perry, J.R.B.; Patterson, N.; Robinson, E.B.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Berisa, T.; Liu, J.Z.; Ségurel, L.; Tung, J.Y.; Hinds, D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016, 48, 709–717. [Google Scholar] [CrossRef]
- Han, B.; Eskin, E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am. J. Hum. Genet. 2011, 88, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. elife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, Z.; Zhang, F.; Wu, Y.; Trzaskowski, M.; Maier, R.; Robinson, M.R.; McGrath, J.J.; Visscher, P.M.; Wray, N.R.; et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018, 9, 224. [Google Scholar] [CrossRef]
- O’Connor, L.J.; Price, A.L. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet. 2018, 50, 1728–1734. [Google Scholar] [CrossRef]
- Li, M.-X.; Gui, H.-S.; Kwan, J.S.; Sham, P.C. GATES: A Rapid and Powerful Gene-Based Association Test Using Extended Simes Procedure. Am. J. Hum. Genet. 2011, 88, 283–293. [Google Scholar] [CrossRef]
- Chanda, P.; Huang, H.; Arking, D.E.; Bader, J.S. Fast Association Tests for Genes with FAST. PLoS ONE 2013, 8, e68585. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Marioni, J.C.; Pai, A.A.; Degner, J.F.; Engelhardt, B.E.; Nkadori, E.; Veyrieras, J.-B.; Stephens, M.; Gilad, Y.; Pritchard, J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 2010, 464, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-X.; Yeung, J.M.Y.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2011, 131, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Adewuyi, E.O.; Sapkota, Y.; International Endogene Consortium (IEC); 23 and Me Research Team; International Headache Genetics Consortium (IHGC); Auta, A.; Yoshihara, K.; Nyegaard, M.; Griffiths, L.R.; Montgomery, G.W.; et al. Shared Molecular Genetic Mechanisms Underlie Endometriosis and Migraine Comorbidity. Genes 2020, 11, 268. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; The International Headache Genetics Consortium; Zhao, H.; I Boomsma, D.; Ligthart, L.; Belin, A.C.; Smith, G.D.; Esko, T.; Freilinger, T.M.; Hansen, T.F.; et al. Molecular genetic overlap between migraine and major depressive disorder. Eur. J. Hum. Genet. 2018, 26, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler—A web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Meng, W.; Adams, M.J.; Hebert, H.L.; Deary, I.J.; McIntosh, A.M.; Smith, B.H. A Genome-Wide Association Study Finds Genetic Associations with Broadly-Defined Headache in UK Biobank (N = 223,773). Ebiomedicine 2018, 28, 180–186. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Noseda, R.; Burstein, R. Migraine pathophysiology: Anatomy of the trigeminovascular pathway and associated neurological symptoms, cortical spreading depression, sensitization, and modulation of pain. Pain 2013, 154, S44–S53. [Google Scholar] [CrossRef]
- Stankewitz, A.; Keidel, L.; Rehm, M.; Irving, S.; Kaczmarz, S.; Preibisch, C.; Witkovsky, V.; Zimmer, C.; Schulz, E.; Toelle, T.R. Migraine attacks as a result of hypothalamic loss of control. NeuroImage Clin. 2021, 32, 102784. [Google Scholar] [CrossRef]
- Hoermann, R.; Midgley, J.E.M.; Larisch, R.; Dietrich, J.W. Recent Advances in Thyroid Hormone Regulation: Toward a New Paradigm for Optimal Diagnosis and Treatment. Front. Endocrinol. 2017, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, S.P.; Bean, N.G.; Falhammar, H.; Tuke, J. Clinical Parameters Are More Likely to Be Associated with Thyroid Hormone Levels than with Thyrotropin Levels: A Systematic Review and Meta-Analysis. Thyroid 2020, 30, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Hautakangas, H.; Winsvold, B.S.; Ruotsalainen, S.E.; Bjornsdottir, G.; Harder, A.V.E.; Kogelman, L.J.A.; Thomas, L.F.; Noordam, R.; Benner, C.; Gormley, P.; et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat. Genet. 2022, 54, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Schenk, T.M.H.; Shears, S.B. ITPK1 (Inositol 1,3,4-Triphosphate 5/6 Kinase). In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2012; pp. 984–990. [Google Scholar]
- Porcu, E.; Medici, M.; Pistis, G.; Volpato, C.B.; Wilson, S.; Cappola, A.R.; Bos, S.D.; Deelen, J.; Heijer, M.D.; Freathy, R.; et al. A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function. PLoS Genet. 2013, 9, e1003266. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Brumpton, B.; Kabil, O.; Gudmundsson, J.; Thorleifsson, G.; Weinstock, J.; Zawistowski, M.; Nielsen, J.B.; Chaker, L.; Medici, M.; et al. GWAS of thyroid stimulating hormone highlights pleiotropic effects and inverse association with thyroid cancer. Nat. Commun. 2020, 11, 3981. [Google Scholar] [CrossRef] [PubMed]
- Vandervore, L.V.; Schot, R.; Milanese, C.; Smits, D.J.; Kasteleijn, E.; Fry, A.E.; Pilz, D.T.; Brock, S.; Börklü-Yücel, E.; Post, M.; et al. TMX2 Is a Crucial Regulator of Cellular Redox State, and Its Dysfunction Causes Severe Brain Developmental Abnormalities. Am. J. Hum. Genet. 2019, 105, 1126–1147. [Google Scholar] [CrossRef]
- Vijayakumar, E.C.; Bhatt, L.; Prabhavalkar, K.S. High Mobility Group Box-1 (HMGB1): A Potential Target in Therapeutics. Curr. Drug Targets 2019, 20, 1474–1485. [Google Scholar] [CrossRef]
- Krashin, E.; Piekiełko-Witkowska, A.; Ellis, M.; Ashur-Fabian, O. Thyroid Hormones and Cancer: A Comprehensive Review of Preclinical and Clinical Studies. Front. Endocrinol. 2019, 10, 59. [Google Scholar] [CrossRef]
- Famularo, G.; Polchi, S.; Tarroni, P. Headache as a presenting symptom of acute myocardial infarction. Headache J. Head Face Pain 2002, 42, 1025–1028. [Google Scholar] [CrossRef]
- Asvestas, D.; Vlachos, K.; Salachas, A.; Letsas, K.P.; Sideris, A. Headache: An unusual presentation of acute myocardial infraction. World J. Cardiol. 2014, 6, 514–516. [Google Scholar] [CrossRef]
- von Hafe, M.; Neves, J.S.; Vale, C.; Borges-Canha, M.; Leite-Moreira, A. The impact of thyroid hormone dysfunction on ischemic heart disease. Endocr. Connect. 2019, 8, R76–R90. [Google Scholar] [CrossRef] [PubMed]
- Lyngberg, A.C.; Rasmussen, B.K.; Jensen, R. Has the prevalence of migraine and tension-type headache changed over a 12-year period? A Danish population survey. Eur. J. Epidemiol. 2005, 20, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Tasnim, S.; Wilson, S.G.; Walsh, J.P.; Nyholt, D.R.; The International Headache Genetics Consortium (IHGC). Shared genetics and causal relationships between migraine and thyroid function traits. Cephalalgia 2022, accepted. [Google Scholar] [CrossRef]
- Dalakas, M.C.; Alexopoulos, H.; Spaeth, P.J. Complement in neurological disorders and emerging complement-targeted therapeutics. Nat. Rev. Neurol. 2020, 16, 601–617. [Google Scholar] [CrossRef]
- Zhao, C.; Yu, Y.; Liu, J.; Lu, G.; Li, T.; Gao, Y.; Zhang, J.; Guo, X. Diversity of complement activation in different thyroid diseases. Int. Immunopharmacol. 2022, 106, 108636. [Google Scholar] [CrossRef]
- Uygun, Ö.; Ertaş, M.; Ekizoğlu, E.; Bolay, H.; Özge, A.; Orhan, E.K.; Çağatay, A.A.; Baykan, B. Headache characteristics in COVID-19 pandemic-a survey study. J. Headache Pain 2020, 21, 121. [Google Scholar] [CrossRef]
- Naguib, R. Potential relationships between COVID-19 and the thyroid gland: An update. J. Int. Med. Res. 2022, 50, 3000605221082898. [Google Scholar] [CrossRef]
- Damara, F.A.; Muchamad, G.R.; Ikhsani, R.; Hendro; Syafiyah, A.H.; Bashari, M.H. Thyroid disease and hypothyroidism are associated with poor COVID-19 outcomes: A systematic review, meta-analysis, and meta-regression. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102312. [Google Scholar] [CrossRef]
- Li, G.H.; Tang, C.M.; Cheung, C.L. COVID-19 and Thyroid Function: A Bi-Directional Two-Sample Mendelian Randomization Study. Thyroid 2022, 32, 1037–1050. [Google Scholar] [CrossRef]
- May, A.; Burstein, R. Hypothalamic regulation of headache and migraine. Cephalalgia 2019, 39, 1710–1719. [Google Scholar] [CrossRef]
- Takizawa, T.; Kurihara, I.; Suzuki, N.; Nakahara, J.; Shibata, M. Painless Thyroiditis Presenting with Headache. Intern. Med. 2021, 60, 2693–2696. [Google Scholar] [CrossRef] [PubMed]
| Trait1 (t1) | Cases/Controls | h2 | Trait2 (t2) | Cases/Controls | h2 | Valid SNPs | rg | SE | Z | p | Intercept t1(SE)/t2(SE) | Intercept (Genecov) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Headache | 84,036/335,552 | 0.10 | Hypothyroidism | 20,563/399,910 | 0.22 | 1181077 | 0.09 | 0.02 | 3.79 | 2.00 × 10−4 | 1.03(0.01)/1.03(0.02) | 0.0160 |
| Headache | 84,036/335,552 | 0.10 | Hyperthyroidism | 3197/417,276 | 0.15 | 1181077 | −0.14 | 0.04 | −3.12 | 1.80 × 10−3 | 1.03(0.01)/1.01(0.01) | −0.0038 |
| Headache | 84,036/335,552 | 0.10 | Secondary Hypothyroidism | 1430/399,034 | 0.04 | 1181077 | 0.20 | 0.10 | 1.94 | 5.24 × 10−2 | 1.03(0.01)/1.01(0.01) | 0.0061 |
| Headache | 84,036/335,552 | 0.10 | TSH | 72,167 | 0.11 | 1164428 | −0.04 | 0.03 | −1.13 | 0.2597 | 1.03(0.01)/1.04(0.01) | Constrained to 0 |
| Headache | 84,036/335,552 | 0.10 | fT4 | 72,167 | 0.15 | 1164126 | 0.08 | 0.03 | 2.78 | 5.50 × 10−3 | 1.03(0.01)/1.01(0.01) | Constrained to 0 |
| Trait 1 | Trait 2 | SNP | EA | NEA | chr | pos | Meta-Analysis | Trait 1 | Trait 2 | Nearest Gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | β | SE | p | β | SE | p | β | SE | ||||||||
| Headache | HPO | rs187359614 | A | G | 4 | 29763484 | 2.12 × 10−8 | 0.36 | 0.064 | 6.63 × 10−7 | 0.36 | 0.073 | 9.78 × 10−3 | 0.35 | 0.134 | AC109351.1 |
| rs3960788 | C | T | 4 | 103915618 | 3.53 × 10−8 | −0.03 | 0.005 | 4.53 × 10−7 | −0.03 | 0.006 | 2.42 × 10−2 | −0.03 | 0.011 | SLC9B1 | ||
| rs215695 | T | C | 7 | 32397908 | 1.29 × 10−9 | −0.03 | 0.006 | 1.16 × 10−7 | −0.03 | 0.006 | 3.10 × 10−3 | −0.04 | 0.012 | PDE1C | ||
| rs7485814 | T | A | 12 | 84326974 | 2.96 × 10−8 | 0.03 | 0.005 | 4.12 × 10−7 | 0.03 | 0.006 | 2.23 × 10−2 | 0.03 | 0.011 | SNORA3 | ||
| rs12912956 | C | T | 15 | 72568275 | 4.94 × 10−8 | 0.03 | 0.006 | 3.76 × 10−6 | 0.03 | 0.007 | 3.55 × 10−3 | 0.04 | 0.012 | CELF6 | ||
| SHPO | rs59382356 | T | C | 2 | 100484479 | 3.50 × 10−8 | −0.03 | 0.006 | 1.33 × 10−7 | −0.03 | 0.006 | 4.70 × 10−2 | −0.08 | 0.039 | Aff3 | |
| rs42854 | C | G | 5 | 74963277 | 4.35 × 10−8 | −0.03 | 0.006 | 2.33 × 10−7 | −0.03 | 0.006 | 1.62 × 10−2 | −0.10 | 0.041 | ANKDD1B | ||
| rs28540738 | G | A | 14 | 93591673 | 2.57 × 10−8 | −0.03 | 0.006 | 1.14 × 10−7 | −0.03 | 0.006 | 3.15 × 10−2 | −0.09 | 0.041 | ITPK1 | ||
| TSH | rs12723104 | C | T | 1 | 33760197 | 1.42 × 10−8 | −0.03 | 0.005 | 4.57 × 10−3 | −0.02 | 0.007 | 1.34 × 10−7 | −0.04 | 0.007 | ZNF362 | |
| rs6672112 | C | T | 1 | 51188594 | 1.73 × 10−9 | 0.03 | 0.005 | 6.44 × 10−4 | 0.02 | 0.006 | 2.58 × 10−7 | 0.03 | 0.007 | FAF1 | ||
| rs7590268 | G | T | 2 | 43540125 | 3.33 × 10−9 | −0.03 | 0.005 | 1.94 × 10−3 | −0.02 | 0.007 | 8.10 × 10−8 | −0.04 | 0.007 | THADA | ||
| rs12549150 | T | C | 8 | 11422936 | 1.17 × 10−8 | −0.02 | 0.004 | 1.17 × 10−7 | −0.03 | 0.006 | 7.90 × 10−3 | −0.02 | 0.006 | BLK | ||
| rs71460549 | C | G | 11 | 57484130 | 9.68 × 10−9 | 0.03 | 0.005 | 2.47 × 10−7 | 0.03 | 0.006 | 4.33 × 10−3 | 0.02 | 0.007 | TMX2:TMX2-CTNND1 | ||
| rs16958839 | T | A | 16 | 82934879 | 2.94 × 10−8 | −0.06 | 0.011 | 1.56 × 10−5 | −0.06 | 0.014 | 5.11 × 10−4 | −0.06 | 0.018 | CDH13 | ||
| rs4793187 | A | G | 17 | 41121263 | 8.30 × 10−9 | −0.03 | 0.005 | 5.64 × 10−7 | −0.03 | 0.006 | 1.91 × 10−3 | −0.02 | 0.007 | PTGES3L-AARSD1:PTGES3L | ||
| rs4986170 | A | G | 17 | 43207858 | 5.20 × 10−10 | −0.03 | 0.006 | 6.94 × 10−6 | −0.03 | 0.008 | 1.79 × 10−5 | −0.04 | 0.008 | PLCD3 | ||
| rs6504608 | A | C | 17 | 47424681 | 1.31 × 10−8 | 0.02 | 0.004 | 7.19 × 10−3 | 0.02 | 0.006 | 7.16 × 10−8 | 0.03 | 0.006 | ZNF652 | ||
| rs116855522 | T | C | 19 | 5293048 | 1.03 × 10−8 | −0.03 | 0.005 | 2.47 × 10−4 | −0.02 | 0.006 | 5.61 × 10−6 | −0.03 | 0.007 | PTPRS | ||
| rs8108474 | T | C | 19 | 46301479 | 9.65 × 10−9 | −0.03 | 0.004 | 5.62 × 10−6 | −0.03 | 0.006 | 3.90 × 10−4 | −0.02 | 0.006 | RSPH6A | ||
| rs6141766 | G | A | 20 | 31222769 | 1.55 × 10−9 | 0.04 | 0.006 | 9.22 × 10−7 | 0.04 | 0.008 | 3.38 × 10−4 | 0.03 | 0.009 | C20orf203 | ||
| fT4 | rs2356864 | A | G | 1 | 50839740 | 6.93 × 10−9 | −0.03 | 0.004 | 2.15 × 10−7 | −0.03 | 0.006 | 3.82 × 10−3 | −0.02 | 0.007 | HMGB1P45 | |
| rs6668959 | T | C | 1 | 174057626 | 6.04 × 10−10 | −0.03 | 0.004 | 6.09 × 10−6 | −0.03 | 0.006 | 2.37 × 10−5 | −0.03 | 0.007 | RPL30P1 | ||
| rs143000423 | A | C | 6 | 36591128 | 1.30 × 10−8 | 0.09 | 0.016 | 6.15 × 10−6 | 0.09 | 0.020 | 5.64 × 10−4 | 0.09 | 0.027 | MIR3925 | ||
| rs17055186 | G | A | 8 | 26260910 | 4.70 × 10−8 | 0.04 | 0.007 | 1.13 × 10−4 | 0.03 | 0.009 | 1.03 × 10−4 | 0.04 | 0.010 | BNIP3L | ||
| rs12343333 | C | T | 9 | 109620811 | 5.90 × 10−10 | 0.03 | 0.005 | 2.12 × 10−5 | 0.03 | 0.006 | 5.58 × 10−6 | 0.03 | 0.007 | ZNF462 | ||
| rs2926664 | G | A | 11 | 57400495 | 1.32 × 10−8 | −0.03 | 0.004 | 3.49 × 10−3 | −0.02 | 0.006 | 1.20 × 10−7 | −0.04 | 0.007 | AP000662.4 | ||
| rs580223 | A | T | 11 | 57541120 | 2.00 × 10−9 | −0.03 | 0.005 | 8.89 × 10−6 | −0.03 | 0.007 | 5.42 × 10−5 | −0.03 | 0.008 | TMX2-CTNND1:RP11-691N7.6:CTNND1 | ||
| rs12883201 | T | C | 14 | 93570702 | 4.55 × 10−10 | −0.03 | 0.005 | 7.92 × 10−5 | −0.03 | 0.007 | 6.98 × 10−7 | −0.04 | 0.008 | ITPK1 | ||
| rs8029914 | T | C | 15 | 49304651 | 2.99 × 10−8 | 0.02 | 0.004 | 5.90 × 10−5 | 0.02 | 0.006 | 1.33 × 10−4 | 0.03 | 0.007 | SECISBP2L | ||
| (a) 2SMR | ||||||||||||||
| Exposure | Outcome | IVW | Weighted median | MR-Egger | MR-Egger Intercept | MR-PRESSO | ||||||||
| β | p | β | p | β | p | Intercept | p | Global test p | Raw β | Raw p | Corr-β | Corr- p | ||
| HPO | Headache | −0.01 | 0.12 | −0.02 | 1.99 × 10−2 | −0.01 | 0.53 | −0.001 | 0.93 | < 1 × 10−4 | −0.01 | 0.22 | −0.01 | 0.21 |
| Headache | HPO | 0.04 | 0.69 | −0.03 | 0.65 | −0.30 | 0.24 | 0.019 | 0.16 | < 1 × 10−4 | 0.04 | 0.66 | 0.04 | 0.49 |
| HPER | Headache | −0.04 | 4.76 × 10−2 | −0.03 | 8.71 × 10−3 | −0.09 | 4.13 × 10−2 | 0.015 | 0.17 | < 1 × 10−4 | −0.04 | 0.06 | −0.01 | 0.35 |
| Headache | HPER | −0.17 | 0.5 | 0.11 | 0.43 | −0.37 | 0.6 | 0.011 | 0.76 | < 1 × 10−4 | −0.15 | 0.53 | 0.17 | 0.09 |
| SHPO | Headache | 0.02 | 0.14 | 0.03 | 0.13 | 0.001 | 0.97 | 0.007 | 0.53 | 0.3581 | −0.0001 | 0.97 | - | - |
| Headache | SHPO | 0.37 | 4.19 × 10−3 | 0.47 | 9.81 × 10−3 | 0.46 | 0.21 | −0.005 | 0.8 | 0.2104 | 0.4 | 1.60 × 10−3 | - | - |
| TSH | Headache | 0.004 | 0.83 | 0.02 | 0.38 | 0.03 | 0.46 | −0.002 | 0.47 | 0.0499 | 0.02 | 0.18 | 0.03 | 3.88 × 10−2 |
| Headache | TSH | 0.008 | 0.83 | −0.04 | 0.24 | −0.11 | 0.29 | 0.007 | 0.22 | < 1 × 10−4 | 0.02 | 0.56 | 0.04 | 0.09 |
| fT4 | Headache | 0.012 | 0.49 | 0.07 | 0.22 | 0.08 | 0.66 | −0.001 | 0.8 | 0.0027 | 0.01 | 0.69 | −0.01 | 0.63 |
| Headache | fT4 | 0.04 | 0.28 | 0.04 | 0.37 | −0.02 | 0.81 | 0.004 | 0.51 | 0.0014 | −0.01 | 0.76 | −0.02 | 0.48 |
| (b) GSMR | ||||||||||||||
| Exposure | Outcome | β | SE | p | ||||||||||
| HPO | Headache | 0.01 | 0.004 | 2.45 × 10−3 | ||||||||||
| Headache | HPO | −0.05 | 0.044 | 0.28 | ||||||||||
| HPER | Headache | −0.04 | 0.005 | 1.16 × 10−13 | ||||||||||
| Headache | HPER | 0.19 | 0.1 | 4.89 × 10−2 | ||||||||||
| SHPO | Headache | −0.03 | 0.097 | 0.76 | ||||||||||
| Headache | SHPO | 0.5 | 0.14 | 3.64 × 10−4 | ||||||||||
| TSH | Headache | 0.001 | 0.014 | 0.94 | ||||||||||
| Headache | TSH | −0.01 | 0.0235 | 0.64 | ||||||||||
| fT4 | Headache | −0.004 | 0.0199 | 0.82 | ||||||||||
| Headache | fT4 | 0.04 | 0.0255 | 0.16 | ||||||||||
| (c) LCV | ||||||||||||||
| Trait 1 | Trait 2 | gcp | SE | p | ||||||||||
| Headache | HPO | 0.01 | 0.58 | 0.99 | ||||||||||
| HPER | 0.17 | 0.48 | 0.89 | |||||||||||
| SHPO | −0.50 | 0.08 | 0.56 | |||||||||||
| TSH | −0.02 | 0.57 | 0.89 | |||||||||||
| fT4 | −0.14 | 0.25 | 0.64 | |||||||||||
| (a) Effective number of independent genes in headache and hypothyroidism | |||||||||||
| Disorder | Total genes | p < 0.1 | p < 0.05 | p < 0.01 | |||||||
| Raw 3 | Effective 4 | Raw 3 | Effective 4 | Proportion 5 | Raw 3 | Effective 4 | Proportion 5 | Raw 3 | Effective 4 | Proportion 5 | |
| Headache 1 | 33,264 | 22,365 | 8107 | 5381 | 0.241 | 5378 | 3555 | 0.159 | 2394 | 1536 | 0.069 |
| Hypothyroidism 2 | 33,264 | 22,567 | 8677 | 5832 | 0.258 | 6148 | 4133 | 0.183 | 3218 | 2123 | 0.094 |
| (b) Number of overlapping genes and binomial test p-value for gene-level genetic overlap | |||||||||||
| Discovery set | Target set | Overlapping genes | Proportion of overlap | Binomial test p-value | |||||||
| Raw | Effective | Expected | Observed | ||||||||
| p < 0.1 | |||||||||||
| Headache | Hypothyroidism | 2790 | 1819 | 5832/22,567= 0.258 | 1819/5381 = 0.338 | 4.72 × 10−39 | |||||
| p < 0.05 | |||||||||||
| Headache | Hypothyroidism | 1638 | 1034 | 4133/22,567= 0.183 | 1034/3555 = 0.291 | 2.83 × 10−55 | |||||
| p < 0.01 | |||||||||||
| Headache | Hypothyroidism | 709 | 392 | 2123/22,567= 0.094 | 392/1536 = 0.255 | 6.83 × 10−75 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasnim, S.; Wilson, S.G.; Walsh, J.P.; Nyholt, D.R. Cross-Trait Genetic Analyses Indicate Pleiotropy and Complex Causal Relationships between Headache and Thyroid Function Traits. Genes 2023, 14, 16. https://doi.org/10.3390/genes14010016
Tasnim S, Wilson SG, Walsh JP, Nyholt DR. Cross-Trait Genetic Analyses Indicate Pleiotropy and Complex Causal Relationships between Headache and Thyroid Function Traits. Genes. 2023; 14(1):16. https://doi.org/10.3390/genes14010016
Chicago/Turabian StyleTasnim, Sana, Scott G. Wilson, John P. Walsh, and Dale R. Nyholt. 2023. "Cross-Trait Genetic Analyses Indicate Pleiotropy and Complex Causal Relationships between Headache and Thyroid Function Traits" Genes 14, no. 1: 16. https://doi.org/10.3390/genes14010016
APA StyleTasnim, S., Wilson, S. G., Walsh, J. P., & Nyholt, D. R. (2023). Cross-Trait Genetic Analyses Indicate Pleiotropy and Complex Causal Relationships between Headache and Thyroid Function Traits. Genes, 14(1), 16. https://doi.org/10.3390/genes14010016








