Comparison of Marker Effects and Breeding Values at Two Levels at THI for Milk Yield and Quality Traits in Brazilian Holstein Cows
Abstract
1. Introduction
2. Material and Methods
2.1. Pseudophenotypes and THI from Previous Study
2.2. Genotypes
2.3. Model and GWAS
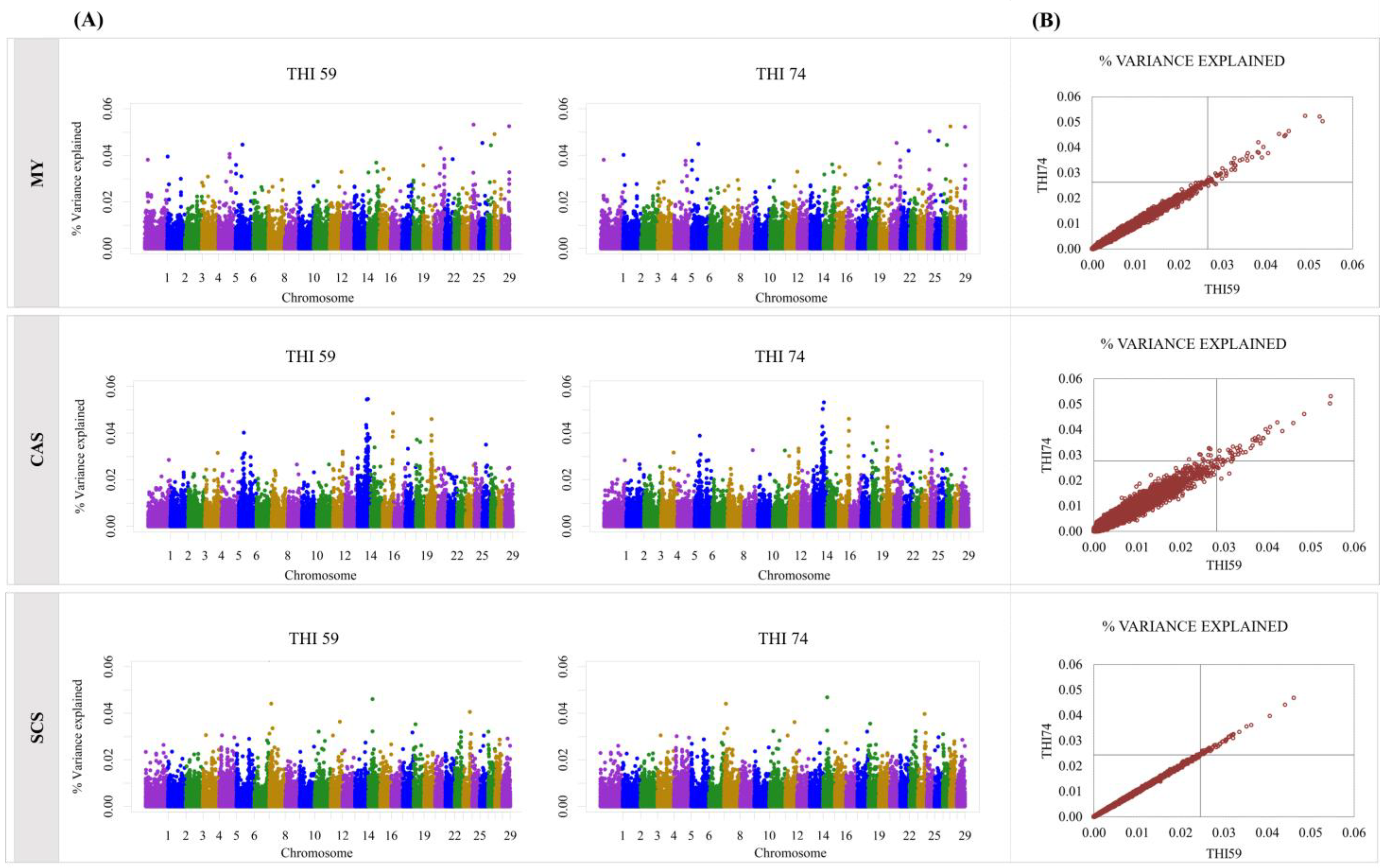
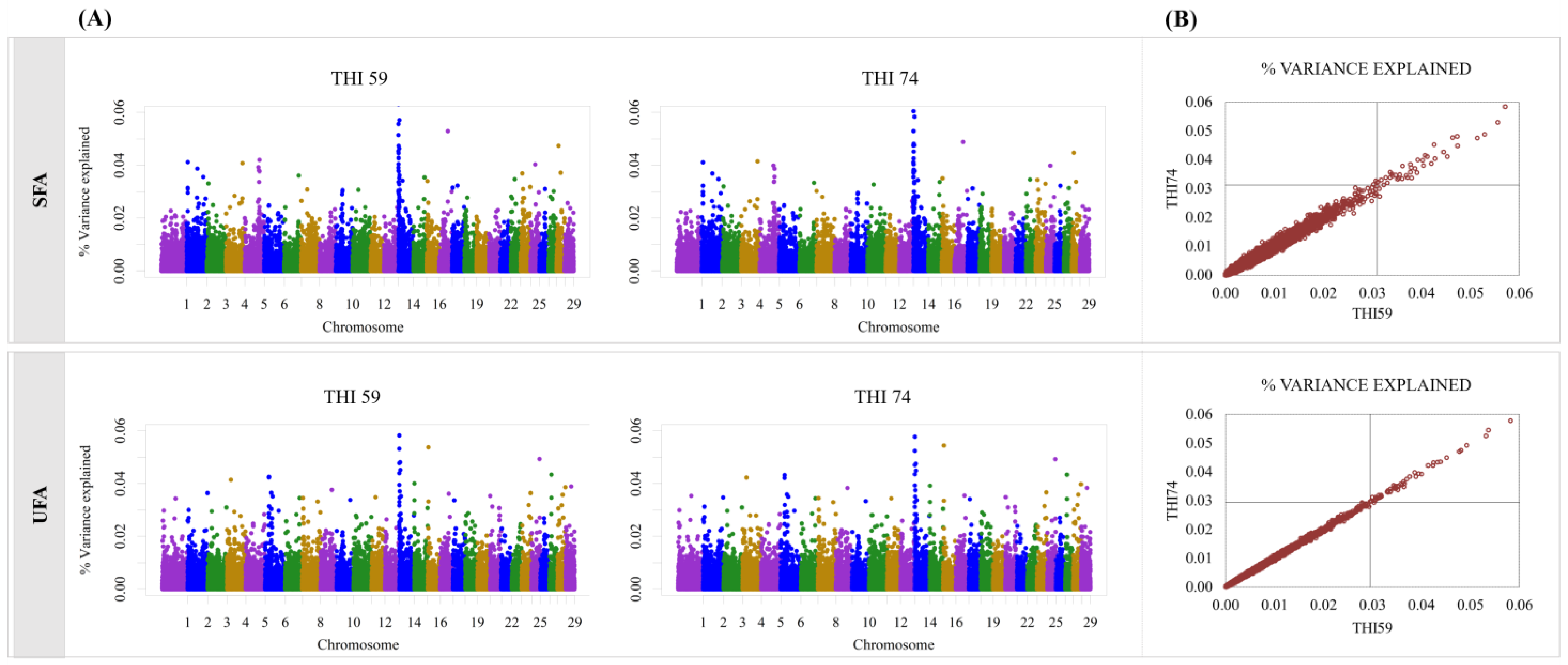
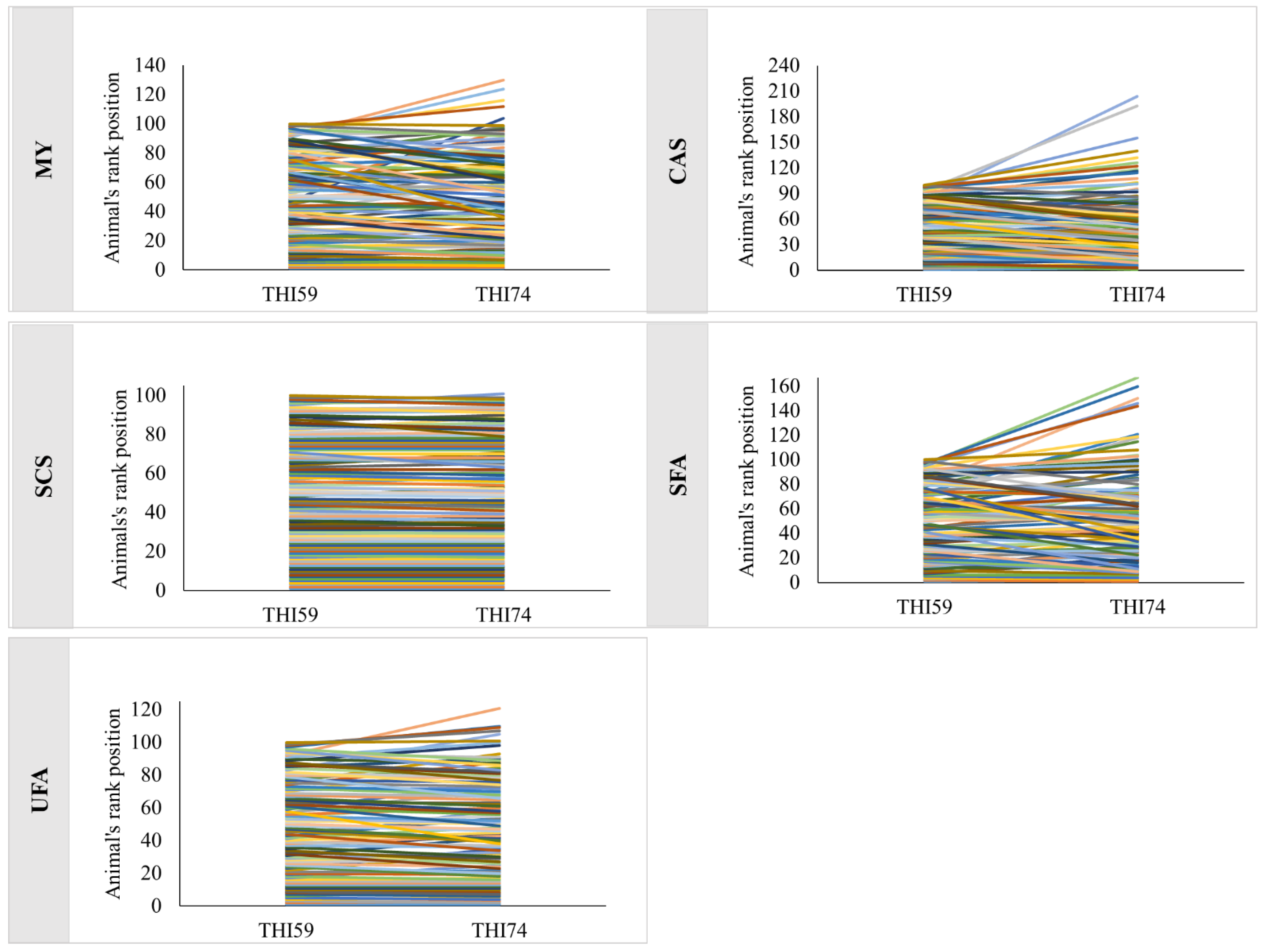
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Renaudeau, D.; Collin, A.; Yahav, S.; de Basilio, V.; Gourdine, J.L.; Collier, R.J. Adaptation to Hot Climate and Strategies to Alleviate Heat Stress in Livestock Production. Animal 2012, 6, 707–728. [Google Scholar] [CrossRef] [PubMed]
- Maia, G.G.; Siqueira, L.G.B.; de Paula Vasconcelos, C.O.; Tomich, T.R.; de Almeida Camargo, L.S.; Rodrigues, J.P.P.; de Menezes, R.A.; Gonçalves, L.C.; Teixeira, B.F.; de Oliveira Grando, R.; et al. Effects of Heat Stress on Rumination Activity in Holstein-Gyr Dry Cows. Livest. Sci. 2020, 239, 104092. [Google Scholar] [CrossRef]
- Santana, M.L.; Bignardi, A.B.; Pereira, R.J.; Menéndez-Buxadera, A.; El Faro, L. Random Regression Models to Account for the Effect of Genotype by Environment Interaction Due to Heat Stress on the Milk Yield of Holstein Cows under Tropical Conditions. J. Appl. Genet. 2016, 57, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Dauria, B.D.; Sigdel, A.; Petrini, J.; Bóscollo, P.P.; Pilonetto, F.; Salvian, M.; Rezende, F.M.; Pedrosa, V.B.; Bittar, C.M.M.; Machado, P.F.; et al. Genetic Effects of Heat Stress on Milk Fatty Acids in Brazilian Holstein Cattle. J. Dairy Sci. 2022, 105, 3296–3305. [Google Scholar] [CrossRef]
- Carrara, E.R.; Petrini, J.; Salvian, M.; de Oliveira, H.R.; Rovadoscki, G.A.; de Souza Iung, L.H.; Miquilini, M.; Machado, P.F.; Mourão, G.B. Genetic Parameters for Milk Yield and Quality Traits of Brazilian Holstein Cows as a Function of Temperature and Humidity Index. J. Anim. Breed. Genet. 2021, 138, 643–654. [Google Scholar] [CrossRef]
- Sigdel, A.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Whole Genome Mapping Reveals Novel Genes and Pathways Involved in Milk Production Under Heat Stress in US Holstein Cows. Front. Genet. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Genetic Component of Heat Stress in Dairy Cattle, Parameter Estimation. J. Dairy Sci. 2000, 83, 2126–2130. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Smith, T.; Rude, B.J.; Ward, S.H. Short Communication: Comparison of the Effects of Heat Stress on Milk and Component Yields and Somatic Cell Score in Holstein and Jersey Cows. J. Dairy Sci. 2013, 96, 3028–3033. [Google Scholar] [CrossRef]
- Bernabucci, U.; Biffani, S.; Buggiotti, L.; Vitali, A.; Lacetera, N.; Nardone, A. The Effects of Heat Stress in Italian Holstein Dairy Cattle. J. Dairy Sci. 2014, 97, 471–486. [Google Scholar] [CrossRef]
- Recce, S.; Huber, E.; Notaro, U.S.; Rodríguez, F.M.; Ortega, H.H.; Rey, F.; Signorini, M.L.; Salvetti, N.R. Association between Heat Stress during Intrauterine Development and the Calving-to-Conception and Calving-to-First-Service Intervals in Holstein Cows. Theriogenology 2021, 162, 95–104. [Google Scholar] [CrossRef]
- VanRaden, P.M. Findhap.F90–Find Haplotypes and Impute Genotypes Using Multiple Chip Sets and Sequence Data. Available online: http://aipl.arsusda.gov/software/findhap (accessed on 22 January 2018).
- Petrini, J.; Iung, L.H.S.; Rodriguez, M.A.P.; Salvian, M.; Pértille, F.; Rovadoscki, G.A.; Cassoli, L.D.; Coutinho, L.L.; Machado, P.F.; Wiggans, G.R.; et al. Genetic Parameters for Milk Fatty Acids, Milk Yield and Quality Traits of a Holstein Cattle Population Reared under Tropical Conditions. J. Anim. Breed. Genet. 2016, 133, 384–395. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs. Available online: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all7.pdf (accessed on 14 February 2022).
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, V.B.; Schenkel, F.S.; Chen, S.Y.; Oliveira, H.R.; Casey, T.M.; Melka, M.G.; Brito, L.F. Genomewide Association Analyses of Lactation Persistency and Milk Production Traits in Holstein Cattle Based on Imputed Whole-Genome Sequence Data. Genes 2021, 12, 1830. [Google Scholar] [CrossRef] [PubMed]
- Ringseis, R.; Gessner, D.K.; Eder, K. Molecular Insights into the Mechanisms of Liver-Associated Diseases in Early-Lactating Dairy Cows: Hypothetical Role of Endoplasmic Reticulum Stress. J. Anim. Physiol. Anim. Nutr. 2015, 99, 626–645. [Google Scholar] [CrossRef]
- Atashi, H.; Salavati, M.; De Koster, J.; Ehrlich, J.; Crowe, M.; Opsomer, G.; Hostens, M.; McLoughlin, N.; Fahey, A.; Matthews, E.; et al. Genome-wide Association for Milk Production and Lactation Curve Parameters in Holstein Dairy Cows. J. Anim. Breed. Genet. 2020, 137, 292–304. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Sardina, M.T.; Tolone, M.; Di Gerlando, R.; Sutera, A.M.; Fontanesi, L.; Portolano, B. Genome-Wide Identification of Runs of Homozygosity Islands and Associated Genes in Local Dairy Cattle Breeds. Animal 2018, 12, 2480–2488. [Google Scholar] [CrossRef]
- Dikmen, S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Genome-Wide Association Mapping for Identification of Quantitative Trait Loci for Rectal Temperature during Heat Stress in Holstein Cattle. PLoS ONE 2013, 8, e69202. [Google Scholar] [CrossRef]
- Fugate, R.T.; Dauten, L.H.; Wiggans, G.R.; White, H.M. Determination of Single Nucleotide Polymorphisms Associated with Subclinical Ketosis in Jersey Cattle. Available online: https://heatherwhite.dysci.wisc.edu/wp-content/uploads/sites/164/2014/07/ADSA-Jersey-SNP-poster-FINAL.pdf (accessed on 25 November 2022).
- Blott, S.; Kim, J.-J.; Moisio, S.; Schmidt-Küntzel, A.; Cornet, A.; Berzi, P.; Cambisano, N.; Ford, C.; Grisart, B.; Johnson, D.; et al. Molecular Dissection of a Quantitative Trait Locus: A Phenylalanine-to-Tyrosine Substitution in the Transmembrane Domain of the Bovine Growth Hormone Receptor Is Associated with a Major Effect on Milk Yield and Composition. Genetics 2003, 163, 253–266. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Snell, R. Positional Candidate Cloning of a QTL in Dairy Cattle: Identification of a Missense Mutation in the Bovine DGAT1 Gene with Major Effect on Milk Yield and Composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Bovenhuis, H.; Visker, M.H.P.W.; Poulsen, N.A.; Sehested, J.; van Valenberg, H.J.F.; van Arendonk, J.A.M.; Larsen, L.B.; Buitenhuis, A.J. Effects of the Diacylglycerol O-Acyltransferase 1 (DGAT1) K232A Polymorphism on Fatty Acid, Protein, and Mineral Composition of Dairy Cattle Milk. J. Dairy Sci. 2016, 99, 3113–3123. [Google Scholar] [CrossRef] [PubMed]
- Bayless, K.J.; Davis, G.E.; Meininger, G.A. Isolation and Biological Properties of Osteopontin from Bovine Milk. Protein Expr. Purif. 1997, 9, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Nemir, M.; Bhattacharyya, D.; Li, X.; Singh, K.; Mukherjee, A.B.; Mukherjee, B.B. Targeted Inhibition of Osteopontin Expression in the Mammary Gland Causes Abnormal Morphogenesis and Lactation Deficiency. J. Biol. Chem. 2000, 275, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-Van Der Wind, A.; Lee, J.H.; Drackley, J.K.; Band, M.R.; Hernandez, A.G.; Shani, M.; et al. Identification of a Missense Mutation in the Bovine ABCG2 Gene with a Major Effect on the QTL on Chromosome 6 Affecting Milk Yield and Composition in Holstein Cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.J.; Savin, K.; van Tassell, C.P.; Sonstegard, T.S.; Goddard, M.E. A Validated Genome Wide Association Study to Breed Cattle Adapted to an Environment Altered by Climate Change. PLoS ONE 2009, 4, e6676. [Google Scholar] [CrossRef] [PubMed]
- Dikmen, S.; Wang, X.-Z.; Ortega, M.S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Single Nucleotide Polymorphisms Associated with Thermoregulation in Lactating Dairy Cows Exposed to Heat Stress. J. Anim. Breed. Genet. 2015, 132, 409–419. [Google Scholar] [CrossRef] [PubMed]
| Traits | N | Mean | SD | MIN | MAX | |
|---|---|---|---|---|---|---|
| MY (kg/day) | THI59 | 1013 | 1.13 | 2.13 | −5.63 | 8.51 |
| THI74 | 1.00 | 2.01 | −5.59 | 7.93 | ||
| CAS (%) | THI59 | 1013 | −0.04 | 0.11 | −0.39 | 0.27 |
| THI74 | −0.04 | 0.12 | −0.41 | 0.28 | ||
| SCS | THI59 | 1013 | −0.13 | 0.53 | −1.57 | 1.46 |
| THI74 | −0.11 | 0.45 | −1.33 | 1.22 | ||
| SFA (%) | THI59 | 1013 | −0.03 | 0.19 | −0.67 | 0.57 |
| THI74 | −0.02 | 0.17 | −0.59 | 0.55 | ||
| UFA (%) | THI59 | 1013 | 0.001 | 0.05 | −0.17 | 0.13 |
| THI74 | 0.001 | 0.05 | −0.19 | 0.15 |
| SNP Name | Chr:Pos | THI59 | THI74 | Gene | Gene Position |
|---|---|---|---|---|---|
| MY | |||||
| BovineHD0800009461 | 8:31223530 | 0.03 | 0.04 | MPDZ | 31108126:31283422 |
| BovineHD1200021758 | 12:77021076 | 0.03 | 0.04 | DNAJC3 | 76958978:77024110 |
| ARS-BFGL-NGS-103064 | 14:2754909 | 0.03 | 0.04 | LY6K | 2755205:2759123 |
| ARS-BFGL-NGS-17362 | 15:75215172 | 0.03 | 0.02 | EXT2 | 75074940:75265719 |
| CAS | |||||
| BTA-147298 | 6:23183379 | 0.04 | 0.03 | SLC9B1 | 23187624:23254634 |
| BTA-75698 | 6:31570212 | 0.04 | 0.03 | PDLIM5 | 31333660:31568270 |
| ARS-BFGL-NGS-108245 | 6:96841919 | 0.02 | 0.03 | C4orf22 | 96794834:97503371 |
| BovineHD0900019961 | 9:71844127 | 0.04 | 0.03 | VNN1 | 71832156:71851981 |
| BovineHD0900019961 | 9:71844127 | 0.03 | 0.04 | U6 snRNA | 71844589:71844686 |
| ARS-BFGL-NGS-85411 | 14:66339300 | 0.03 | 0.04 | SPAG1 | 66319303:66406220 |
| BovineHD1400018564 | 14:66400092 | 0.04 | 0.03 | SPAG1 | 66319303:66406220 |
| BTA-131179 | 16:75462069 | 0.04 | 0.03 | HSD11B1 | 75463273:75533162 |
| BovineHD2500000707 | 25:3293120 | 0.03 | 0.04 | ADCY9 | 3233104:3344619 |
| BovineHD2500001657 | 25:6561255 | 0.04 | 0.03 | RBFOX1 | 4964122:6689485 |
| SCS | |||||
| BovineHD1800000631 | 18:2324775 | 0.03 | 0.02 | WDR59 | 2290822:2371069 |
| SFA | |||||
| ss46527095 | 2:7321910 | 0.04 | 0.03 | COL3A1 | 7317287:7356904 |
| UFA | |||||
| BTC-033565 | 6:38286952 | 0.03 | 0.04 | MEPE | 38279473:38293661 |
| Traits * | Top 10% | Top 40% | All Animals |
|---|---|---|---|
| MY | 0.90 | 0.95 | 0.99 |
| CAS | 0.88 | 0.93 | 0.98 |
| ECS | 1.00 | 1.00 | 1.00 |
| SFA | 0.88 | 0.94 | 0.98 |
| UFA | 0.97 | 0.99 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrara, E.R.; Dauria, B.D.; Gervásio, I.C.; Silveira, R.M.F.; Rovadoski, G.A.; Petrini, J.; Salvian, M.; Machado, P.F.; Mourão, G.B. Comparison of Marker Effects and Breeding Values at Two Levels at THI for Milk Yield and Quality Traits in Brazilian Holstein Cows. Genes 2023, 14, 17. https://doi.org/10.3390/genes14010017
Carrara ER, Dauria BD, Gervásio IC, Silveira RMF, Rovadoski GA, Petrini J, Salvian M, Machado PF, Mourão GB. Comparison of Marker Effects and Breeding Values at Two Levels at THI for Milk Yield and Quality Traits in Brazilian Holstein Cows. Genes. 2023; 14(1):17. https://doi.org/10.3390/genes14010017
Chicago/Turabian StyleCarrara, Eula Regina, Brayan Dias Dauria, Izally Carvalho Gervásio, Robson Mateus Freitas Silveira, Gregori Alberto Rovadoski, Juliana Petrini, Mayara Salvian, Paulo Fernando Machado, and Gerson Barreto Mourão. 2023. "Comparison of Marker Effects and Breeding Values at Two Levels at THI for Milk Yield and Quality Traits in Brazilian Holstein Cows" Genes 14, no. 1: 17. https://doi.org/10.3390/genes14010017
APA StyleCarrara, E. R., Dauria, B. D., Gervásio, I. C., Silveira, R. M. F., Rovadoski, G. A., Petrini, J., Salvian, M., Machado, P. F., & Mourão, G. B. (2023). Comparison of Marker Effects and Breeding Values at Two Levels at THI for Milk Yield and Quality Traits in Brazilian Holstein Cows. Genes, 14(1), 17. https://doi.org/10.3390/genes14010017







